Abstract
A vacuolar acid phosphatase (APase) that accumulates during phosphate (Pi) starvation of Arabidopsis (Arabidopsis thaliana) suspension cells was purified to homogeneity. The final preparation is a purple APase (PAP), as it exhibited a pink color in solution (Amax = 520 nm). It exists as a 100-kD homodimer composed of 55-kD glycosylated subunits that cross-reacted with an anti-(tomato intracellular PAP)-IgG. BLAST analysis of its 23-amino acid N-terminal sequence revealed that this PAP is encoded by At5g34850 (AtPAP26; one of 29 PAP genes in Arabidopsis) and that a 30-amino acid signal peptide is cleaved from the AtPAP26 preprotein during its translocation into the vacuole. AtPAP26 displays much stronger sequence similarity to orthologs from other plants than to other Arabidopsis PAPs. AtPAP26 exhibited optimal activity at pH 5.6 and broad substrate selectivity. The 5-fold increase in APase activity that occurred in Pi-deprived cells was paralleled by a similar increase in the amount of a 55-kD anti-(tomato PAP or AtPAP26)-IgG immunoreactive polypeptide and a >30-fold reduction in intracellular free Pi concentration. Semiquantitative reverse transcription-PCR indicated that Pi-sufficient, Pi-starved, and Pi-resupplied cells contain similar amounts of AtPAP26 transcripts. Thus, transcriptional controls appear to exert little influence on AtPAP26 levels, relative to translational and/or proteolytic controls. APase activity and AtPAP26 protein levels were also up-regulated in shoots and roots of Pi-deprived Arabidopsis seedlings. We hypothesize that AtPAP26 recycles Pi from intracellular P metabolites in Pi-starved Arabidopsis. As AtPAP26 also exhibited alkaline peroxidase activity, a potential additional role in the metabolism of reactive oxygen species is discussed.
Acid phosphatases (APases; E.C. 3.1.3.2) catalyze the hydrolysis of phosphate (Pi) from phosphate monoesters and anhydrides within the acidic pH range. APases function in the production, transport, and recycling of Pi, a crucial macronutrient for cellular metabolism and bioenergetics. Pi starvation-inducible (PSI) intracellular and secreted APases are a widespread plant response to nutritional Pi deficiency, a major limitation to plant growth and agricultural productivity (Duff et al., 1994; Vance et al., 2003; Plaxton, 2004; Raghothama and Karthikeyan, 2005). Secreted APases are likely involved in Pi scavenging from extracellular organic P monoesters, whereas PSI intracellular APases are believed to remobilize and scavenge Pi from intracellular P monoesters and anhydrides in Pi-deficient (−Pi) plants. This is accompanied by marked reductions in cytoplasmic P-metabolite pools during extended Pi stress (Lee and Ratcliffe, 1993; Plaxton, 2004).
The induction of APase activity in −Pi plants and yeast (Saccharomyces cerevisiae) has been correlated with de novo APase synthesis in several systems, including Brassica nigra, Brassica napus, and tomato (Lycopersicon esculentum) suspension cells (Duff et al., 1991b; Carswell et al., 1997; Bozzo et al., 2002, 2004a, 2006), proteoid lupin roots (Miller et al., 2001), and yeast (Oshima et al., 1996). Molecular analysis of the yeast Pi starvation response indicated the synchronized control of genes encoding APases, plasma membrane Pi transporters, and Pi-sensor protein kinases, termed the pho regulon (Oshima et al., 1996). A similar Pi-sensing system has been proposed for plants that leads to the induction of Pi-scavenging, recycling, transport, and metabolism-related genes (Hammond et al., 2003, 2004; Vance et al., 2003; Wu et al., 2003; Ticconi and Abel, 2004; Raghothama and Karthikeyan, 2005; Amtmann et al., 2006). By contrast, Pi resupply to −Pi plants results in the rapid repression of PSI APase genes (Miller et al., 2001; Müller et al., 2004), while simultaneously inducing proteases that appear to specifically target PSI intracellular and extracellular APases (Bozzo et al., 2004b). Recent transcript profiling and biochemical/immunological studies have revealed the differential expression of plant PSI APase genes and proteins in both a temporal and tissue-specific fashion (Wu et al., 2003; Zimmermann et al., 2004; Amtmann et al., 2006; Bozzo et al., 2006).
Three PSI APase isozymes that demonstrated distinctive physical and kinetic properties were recently purified and biochemically and immunologically characterized from −Pi tomato suspension cells (Bozzo et al., 2002, 2004a). Two are secreted monomeric APases having subunit molecular masses of 84 and 57 kD, respectively, whereas the third is a 142-kD heterodimeric intracellular APase composed of a 1:1 ratio of 63- and 57-kD subunits. All three PSI tomato APase isozymes are purple APases (PAPs; Bozzo et al., 2002, 2004a), which represent a specific APase class that contains a bimetallic active center endowing them with a characteristic purple or pink color in solution. Although the primary structures of different PAPs vary extensively, domains involved in coordinating the bimetal active site are highly conserved. This allowed Li et al. (2002) to identify 29 putative PAP genes in the Arabidopsis (Arabidopsis thaliana) genome. However, very few Arabidopsis PAPs have been biochemically characterized to date (del Pozo et al., 1999; Zhu et al., 2005). Several mammalian and plant PAPs exist as multifunctional proteins that also display alkaline peroxidase activity, hypothesized to function in the metabolism of reactive oxygen species (ROS) during senescence or pathogen infection (Hayman and Cox, 1994; del Pozo et al., 1999; Bozzo et al., 2002, 2004a). In addition to induction by Pi deprivation, the best-studied Arabidopsis PAP, AtPAP17 (originally classified as AtACP5), is also induced by oxidative stress and is highly expressed in senescing leaves (del Pozo et al., 1999). The APase and peroxidation activities displayed by AtPAP17 support the view that this PAP may function to both mobilize Pi as well as participate in ROS metabolism during leaf senescence.
Suspension cell cultures are a useful model system for assessing plant Pi starvation responses because: (1) there is a homogeneous population of cells that are equally exposed to the conditions prevalent in liquid culture; (2) a large quantity of cells (and their surrounding media containing secreted proteins) at a defined nutritional state can be obtained over a relatively short time period; and (3) they demonstrate many molecular and biochemical adaptations to Pi deprivation that otherwise occur in planta (Theodorou and Plaxton, 1994; Bozzo et al., 2006). However, there are few reports describing the use of cell cultures for the investigation of Arabidopsis Pi starvation responses (Li et al., 2002). Owing to the availability of functional genomic tools and resources, we recently initiated biochemical studies of PSI Arabidopsis APases. The aims of this study were to: (1) investigate the utility of suspension cell cultures as a model system for assessing the biochemical adaptations of −Pi Arabidopsis; and (2) identify and characterize the biochemical, immunological, and molecular features of the predominant PSI intracellular APase of Arabidopsis suspension cells.
RESULTS
Optimization of Phosphate Concentration in the Culture of Pi-Sufficient Arabidopsis Suspension Cells
One week following subculture of the Arabidopsis suspension cells into fresh Murashige and Skoog (MS) media containing 1.25 or 2.5 mm Pi, exogenous cell culture filtrate (CCF) Pi was undetectable. However, the CCF Pi concentration of 7-d-old cells initially subcultured into media containing 5 mm Pi was 0.25 ± 0.11 mm, and their fresh weight accumulation was about 25% and 11% greater than cells subcultured into media containing 1.25 and 2.5 mm Pi, respectively (Fig. 1). All subsequent experiments were conducted using Arabidopsis suspension cells cultured in MS media containing 0 or 5 mm Pi (−Pi and +Pi, respectively).
Figure 1.
The 1.25 mm Pi concentration of conventional MS liquid media is insufficient to maintain Arabidopsis suspension cells fully Pi sufficient over 7 d in batch culture. At 0 d, 10-mL aliquots of cells cultured for 7 d in 5 mm Pi were subcultured into 90 mL of fresh MS media containing the indicated Pi concentration. At 7 d, biomass yields and Pi concentration in the CCF were determined. All values represent means ± se of n = 3 separate flasks.
Influence of Pi Starvation on the Growth, Pi Content, Intracellular APase Activity, and Immunoreactive APase Polypeptides of Arabidopsis Cell Cultures
Arabidopsis suspension cells cultured for 7 d in −Pi media had approximately 45% of the fresh weight of corresponding +Pi cells (approximately 22 g and 50 g of cells were obtained per 500 mL of culture of 7-d −Pi and +Pi cells, respectively). Their reduced growth was correlated with depletion of CCF Pi to undetectable levels within 1 d following subculture of the +Pi cells into −Pi media (Fig. 2A). The Pi concentration of the CCF of +Pi cells steadily decreased from about 5 mm at 0 d to 0.22 mm by 9 d (Fig. 2A). By 7 d, intracellular free Pi content of the −Pi cells decreased by about 35-fold, whereas free Pi levels only decreased by about 15% in the 7-d-old +Pi cells (Fig. 2B).
Figure 2.
Up-regulation of intracellular APase in Arabidopsis suspension cells becoming Pi deficient. At 0 d, 100-mL aliquots of cells cultured for 7 d in 5 mm Pi were subcultured into 400 mL of fresh MS media containing 5 mm +Pi or 0 mm −Pi. Time courses for CCF of extracellular Pi (A), intracellular Pi (B), and APase activity of clarified extracts (C) of the +Pi and −Pi cells were determined. All values represent means ± se of n = 3 separate flasks. Where invisible, the error bars are too small to be seen. D, Immunological detection of APase in clarified extracts from the Arabidopsis suspension cells. Purified intracellular PAP (50 ng) from −Pi tomato cells (Bozzo et al., 2004a) and clarified cell-extract proteins (15 μg) from the Arabidopsis cells were resolved by SDS-PAGE and blot transferred to a PVDF membrane. Native intracellular APase of −Pi tomato cells exists as a heterodimer composed of 63- and 57-kD subunits (Bozzo et al., 2004a). The immunoblot was probed with a 1:50 dilution of affinity-purified anti-(tomato intracellular PAP)-IgG, and immunoreactive peptides were detected using an alkaline-phosphatase linked secondary antibody as in Bozzo et al. (2006). “−Pi→+Pi” denotes extracts from 6-d −Pi cells that were resupplied with 2.5 mm Pi and cultured for an additional 2 d. Relative amounts of the 55-kD antigenic polypeptide were quantified by laser densitometry. O, Origin; TD, tracking dye front.
Figure 2 also shows the APase time-course activity profiles and corresponding anti-(PSI tomato intracellular PAP)-IgG immunoblots of the +Pi versus −Pi cells. Extractable APase activity remained relatively low and constant in the +Pi cells. By contrast, marked up-regulation of APase activity in the −Pi cells from an initial level of 0.13 units mg−1 protein began at approximately 4 d, attaining a maximum of 0.66 units mg−1 protein by 7 d (Fig. 2C), but was subsequently reduced to about 0.51 units mg−1 by 9 d (Fig. 2C). The APase activity decreased to that of +Pi cells within 2 d of resupply of 2.5 mm Pi to the 6-d −Pi cultures. Laser densitometric quantification of the immunoblot (Fig. 2D) revealed a good correlation between extractable APase activity and the relative amount of an anti-(PSI tomato intracellular PAP)-IgG immunoreactive polypeptide of approximately 55 kD.
Purification and Physical and Immunological Properties of the Principal Intracellular APase from Pi-Starved Arabidopsis Suspension Cells
As outlined in Table I, an APase from 7-d-old −Pi Arabidopsis cells was purified 957-fold to a final phosphoenolpyruvate (PEP)-hydrolyzing specific activity of 421 units mg−1 protein and an overall recovery of 20%. A single peak of APase activity was resolved during all chromatographic steps. Binding of the enzyme onto the Concanavlin A (Con A) resin suggested that it is glycosylated. Similar to the intracellular but not secreted PAP isozymes from −Pi tomato cells (Bozzo et al., 2002, 2004a), the purified APase was relatively heat labile, losing 25%, 80%, and 100% of its original activity when incubated for 3 min at 50°C, 60°C, and 70°C, respectively. The final APase preparation exhibited a pink color. The physical basis for this color is apparent from its visible absorption spectrum, which demonstrates that it is a PAP because it absorbs maximally in the middle of the visible region, peaking at about 520 nm (Fig. 3).
Table I.
Purification of an intracellular PAP (AtPAP26) from 400 g of Pi-starved Arabidopsis suspension cells
| Step | Activity | Protein | Specific Activity | Purification | Yield |
|---|---|---|---|---|---|
| units | mg | units mg−1 | -fold | % | |
| Clarified extract | 1,300 | 2,940 | 0.44 | – | 100 |
| PEG fractionation | 800 | 810 | 0.99 | 2.2 | 62 |
 fractogel fractogel |
650 | 60 | 10.8 | 24.5 | 50 |
| Con A-sepharose | 400 | 2.6 | 154 | 350 | 31 |
| Phenyl superose | 265 | 0.63 | 421 | 957 | 20 |
Figure 3.
Visible absorbance spectrum of APase isolated from −Pi Arabidopsis suspension cells. The spectrum was obtained using a solution of 5 mg mL−1 of the purified APase.
The native molecular mass of the purified PAP was estimated to be approximately 100 kD via analytical gel-filtration FPLC on a calibrated Superose 12 HR10/30 column. When the final PAP preparation was denatured and subjected to SDS-PAGE, a single Coomassie Blue-staining polypeptide of 55 kD was resolved that strongly cross-reacted with the anti-(PSI tomato intracellular PAP)-IgG (Fig. 4, A and C). This result was obtained when SDS-PAGE was performed under both reducing (dithiothreitol [DTT] present in the sample buffer; Fig. 4) or nonreducing (no DTT in the sample buffer) conditions (results not shown). The 55-kD polypeptide was detected with periodic acid-Schiff staining, confirming that it is a glycoprotein (Fig. 4B).
Figure 4.
SDS-PAGE and immunoblot analysis of purified PAP from −Pi Arabidopsis suspension cells. A, SDS-PAGE (10% separating gel) of purified Arabidopsis APase. Lane 1 contains 6.5 μg of various protein molecular mass standards, whereas lane 2 contains 2 μg of the purified PAP. Protein staining was performed using Coomassie Blue R-250. B, SDS-PAGE of 5 μg of the purified PAP was followed by glycoprotein staining using a periodic acid-Schiff procedure (Gradilone et al., 1998). C and D, Immunoblotting was performed using a 1:50 dilution of affinity-purified anti-(tomato intracellular PAP)-IgG (C; Bozzo et al., 2006) or a 1:1,000 dilution of anti-(AtPAP26)-immune serum (D), and antigenic polypeptides were visualized using an alkaline-phosphatase-linked secondary antibody. Lanes 1 and 2 of each segment contain 15 ng of purified PAP from −Pi Arabidopsis and tomato suspension cells, respectively. Lane 3 and 4 of segment D contain clarified cell-extract proteins (15 μg) from the +Pi and −Pi 7-d Arabidopsis cells, respectively. O, Origin; TD, tracking dye front.
Polyclonal antibodies were raised in rabbits against the PAP purified from the −Pi Arabidopsis cells. This protein was subsequently identified as AtPAP26 (see below). The anti-(AtPAP26)-immune serum readily cross-reacted with immunoblots of 15 ng of homogeneous PAP isolated from −Pi Arabidopsis or tomato suspension cells (Fig. 4D, lanes 1 and 2). Immunoblots of clarified extracts from 7-d +Pi versus −Pi Arabidopsis suspension cells indicated that the anti-(AtPAP26)-immune serum was monospecific for the 55-kD AtPAP26 polypeptide (Fig. 4D, lanes 3 and 4). Laser densitometric quantification of the immunoblots indicated that the −Pi extracts contained approximately 5.5-fold more of the 55-kD AtPAP26 polypeptide relative to extracts of +Pi cells.
Amino Acid Sequencing and Bioinformatics Analysis of AtPAP26
The 23 N-terminal amino acids of the 55-kD subunit of the purified PAP from −Pi Arabidopsis cells were sequenced by Edman degradation (Fig. 5). Interrogation of The Arabidopsis Information Resource (TAIR) database matched this sequence to the N-terminal portion of the deduced sequence encoded by a putative Arabidopsis PAP gene (At5g34850; gi: 56788343). Although originally annotated as a calcineurin-like phosphoesterase, At5g34850 was subsequently reclassified as AtPAP26, one of the 29 putative PAPs encoded by the Arabidopsis genome (Li et al., 2002). Figure 5 shows a multiple alignment of the deduced amino acid sequence of AtPAP26 with other plant PAPs. The AtPAP26 sequence best aligned with several PAP orthologs from other plant species (Fig. 5). The deduced amino acid sequence of AtPAP26 is maximally (74%–80%) identical to the PAP orthologs (Table II) aligned in Figure 5, whereas the most closely related paralogs from Arabidopsis (AtPAP10 and AtPAP12) only show 58% identity. Using TargetP, it was predicted that the N terminus of the deduced AtPAP26 polypeptide contains a 22-amino acid signal peptide. This is close to the actual signal peptide of 30 amino acids that was demonstrated by comparing the N-terminal sequence of the 55-kD AtPAP26 polypeptide with its deduced full-length amino acid sequence (Fig. 5). TargetP (and several other algorithms) strongly predicted that AtPAP26 is localized to the secretory pathway, which includes both vacuolar and secreted proteins. The subunit molecular mass of the purified AtPAP26 as estimated by SDS-PAGE was 55 kD (Fig. 4), whereas the predicted size of the mature polypeptide was 51 kD. The 4-kD discrepancy can be explained by the addition of glycan groups.
Figure 5.
Deduced amino acid sequence alignment of Arabidopsis AtPAP26 with orthologs from various plants. The PAP sequences included are from Arabidopsis (AAW29950.1), tomato (BT014303), soybean (AAN85417), rice (BAD37373), and onion (BAB60719). Identical and similar amino acids are indicated by shaded black and gray, respectively. Conserved sequence motifs containing potential metal-ligating residues (Li et al., 2002) are marked by asterisks. The arrow indicates the predicted cleavage site of the AtPAP26 signal peptide. The 23-amino acid N-terminal sequence obtained by automated Edman degradation of the 55-kD subunit of the PAP purified from the −Pi Arabidopsis suspension cells appears in bold font above the deduced sequences. The N-terminal sequence and a tryptic peptide sequence obtained for the 57-kD subunit of the PAP purified from −Pi tomato suspension cells (Bozzo et al., 2004a) are also indicated.
Table II.
AtPAP26 ortholog protein accession and their predicted molecular mass and localization
| Plant Source | Protein Accessiona | Percent Identity to AtPAP26 | Percent Similarity to AtPAP26 | Predicted Mr | Predicted Localizationb |
|---|---|---|---|---|---|
| kD | |||||
| Arabidopsis (AtPAP26) | AAW29950.1 | 100 | 100 | 55.01 | Secretory |
| Tomato | BT014303 | 76 | 89 | 54.88 | Secretory |
| Soybean | AAN85417 | 80 | 89 | 54.86 | Secretory |
| Rice | BAD37373 | 75 | 86 | 55.16 | Secretory |
| Onion | BAB60719 | 74 | 85 | 55.97 | Secretory |
Given by NCBI Protein Database.
Predicted by the TargetP program; secretory denotes presence of putative signal peptides for the secretory pathway.
Semiquantitative Reverse Transcription-PCR Analysis of AtPAP17 and AtPAP26 Transcript Levels in +Pi versus −Pi Arabidopsis Suspension Cells
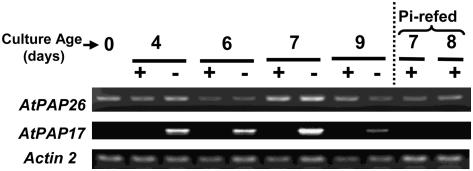
To analyze AtPAP26 expression in more detail, total RNA extracted from +Pi, −Pi, and Pi-resupplied Arabidopsis suspension cells was subjected to semiquantitative reverse transcription (RT)-PCR. AtPAP17 was employed as a positive control because transcripts encoding the 34-kD AtPAP17 polypeptide are markedly increased in roots and shoots of −Pi Arabidopsis seedlings (del Pozo et al., 1999; Li et al., 2002) but rapidly disappear following Pi-resupply (Müller et al., 2004). AtPAP17 transcripts were detected following RT-PCR of total RNA extracted from the −Pi, but not +Pi, Arabidopsis suspension cells (Fig. 6). Resupply of 2.5 mm Pi to the −Pi cells resulted in negligible expression of AtPAP17. By contrast, AtPAP26-derived transcripts were present at similar levels in Arabidopsis cell cultures, irrespective of their nutritional Pi status (Fig. 6).
Figure 6.
Semiquantitative RT-PCR analysis of AtPAP26 gene expression in Pi-sufficient (+), Pi-starved (−), and Pi-resupplied Arabidopsis suspension cells. The cells were cultured as described in the legend for Figure 2. Levels of mRNA were analyzed by RT-PCR using primers specific for the genes AtPAP26, AtPAP17, and Actin 2. AtPAP17 was used as a positive control (del Pozo et al., 1999; Li et al., 2002; Müller et al., 2004), whereas Arabidopsis Actin 2 was used as a reference control to ensure equal template loading. All PCR products were taken at cycle numbers determined to be nonsaturating. Control RT-PCR reactions lacking RT did not show any bands. The AtPAP17 and AtPAP26 primers amplified 469-bp and 610-bp fragments of the 5′ regions of their respective cDNAs, as expected. “Pi-refed” denotes extracts from 6-d −Pi cells that were resupplied with 2.5 mm Pi and cultured for an additional 24 and 48 h.
Influence of Pi Nutrition on APase Activity and AtPAP26 Accumulation in Arabidopsis Seedlings
Two-week-old +Pi Arabidopsis seedlings were cultivated for an additional 10 d on +Pi or −Pi liquid media prior to shoot and root harvesting and extraction. The APase activity of clarified shoot or root extracts from the −Pi seedlings was about 2-fold greater than that of the +Pi controls (Fig. 7). An anti-(AtPAP26)-immune serum immunoreactive 55-kD polypeptide that comigrated with homogeneous AtPAP26 was observed on immunoblots of shoot or root extracts from the +Pi and −Pi plants. The amount of the immunoreactive 55-kD polypeptide was estimated by densitometry to be about twice as abundant in root or shoot extracts from the −Pi seedlings relative to the corresponding tissues of the +Pi seedlings (Fig. 7).
Figure 7.
Immunological PAP detection and corresponding APase activity of clarified extracts from roots and shoots of +Pi versus −Pi Arabidopsis seedlings. The seedlings were germinated and cultivated on MS media containing 1.25 mm Pi for 14 d, transferred to fresh MS media containing 1.25 or 0 mm Pi (+Pi and −Pi, respectively), and cultivated for an additional 10 d prior to extraction and analysis of root and shoot tissues. Purified AtPAP26 (20 ng) from −Pi Arabidopsis suspension cells and clarified extract proteins (15 μg) from shoots and roots of the +Pi and −Pi Arabidopsis seedlings were resolved by SDS-PAGE and blot transferred onto a PVDF membrane. The immunoblot was probed with a 1:1,000 dilution of anti-(AtPAP26)-immune serum and immunoreactive peptides detected using an alkaline-phosphatase linked secondary antibody. Relative amounts of the antigenic 55-kD polypeptide were quantified by laser densitometry. O, Origin; TD, tracking dye front. Corresponding specific APase activities are indicated below each lane. APase activities were determined using assay A, represent the means of n = 4 separate extractions, and are reproducible to within ±20% of the mean value.
AtPAP26 Kinetic Properties
Unless otherwise stated, all kinetic studies were performed using assay A. The purified AtPAP26 exhibited a relatively broad pH-APase activity profile, with a maximum centered at about pH 5.6 (Fig. 8). Half-maximal activity was obtained at pH 4.7 and 6.6. All subsequent APase kinetic studies were performed at pH 5.6. Hyperbolic PEP saturation kinetics were observed [Km(PEP) = 0.8 mm]. AtPAP26 was activated by Mg2+, Co2+, Mn2+, and Ba2+, with Mg2+ being the preferred divalent cation activator (Table III). There was no effect on AtPAP26 APase activity when the reaction mixture (lacking Mg2+) contained 5 mm KCl or 5 mm EDTA. However, AtPAP26 was potently inhibited by 5 mm Fe2+, Cu2+, or Zn2+ (Table III).
Figure 8.
Phosphatase versus peroxidase activity of purified AtPAP26 as a function of assay pH. Assays were performed as described in the “Materials and Methods” except that they were buffered by a mixture of 25 mm sodium acetate, 25 mm MES, and 25 mm Bis-Tris propane. All values represent the means of n = 3 separate determinations and are reproducible to within ±10% of the mean value.
Table III.
Effect of various substances on the activity of purified AtPAP26
Enzyme activity was determined using assay A in the presence of 5 mm of each substance and is expressed relative to the control set at 100%.
| Addition | Relative Activity |
|---|---|
| % | |
| MgCl2 | 190 |
| CoCl2 | 147 |
| MnCl2 | 144 |
| BaCl2 | 121 |
| ZnCl2 | 10 |
| CuSO4 | 2 |
| FeCl2 | 0 |
| NaF | 53 |
| Pi | 31 |
| Arsenate | 8 |
| Molybdate | 7 |
| Vanadate | 0 |
Metabolite and Ion Effects
A variety of compounds were examined for their effects on the APase activity of AtPAP26 under subsaturating concentrations of PEP (0.4 mm). The following compounds exerted no effect (±10% control activity): tartrate, Asp, Gln, glutathione, Asn, ascorbic acid, and phosphite (5 mm each). The most notable inhibitors were NaF, molybdate, arsenate, vanadate, and Pi (Table III). The I50 value for Pi was determined to be 2.1 mm.
Substrate Specificity
APase activity of AtPAP26 was determined using assay B and a wide range of phosphorylated compounds, tested at a concentration of 5 mm. PEP, inorganic pyrophosphate, phenyl-P, and p-nitrophenyl-P (pNPP) were the most effective substrates (Table IV). By contrast, the purified enzyme showed no APase activity with phytic acid, phosphocholine, or bis-pNPP (5 mm each).
Table IV.
Substrate specificity of purified AtPAP26
APase activity was determined with 5 mm of each compound using assay B as described in the “Materials and Methods.” APase activity is expressed relative to the rate of Pi hydrolysis from PEP set at 100%.
| Substrate | Relative Activity |
|---|---|
| % | |
| PEP | 100 |
| Phenyl-P | 100 |
| pNPP | 89 |
| Inorganic pyrophosphate | 86 |
| NADP+ | 55 |
| Phosphotyrosine | 51 |
| Glc-6-P | 46 |
| Ribose-5-P | 40 |
| Phospho-Thr | 39 |
| Fru-6-P | 19 |
| ATP | 17 |
| GTP | 16 |
| dAMP | 16 |
| AMP | 9 |
Peroxidase Activity of AtPAP26
The ability of AtPAP26 to catalyze the peroxidation of luminol was investigated using a chemiluminescence assay. In the presence of luminol and hydrogen peroxide, AtPAP26 induced a striking chemiluminescence. Photon emission due to this peroxidase activity was proportional to assay time and AtPAP26 concentration. A fairly sharp pH/peroxidase activity peak in the alkaline range was observed, with maximal activity occurring at pH 8.8 (Fig. 8). Calibration of the luminometer with known amounts of horseradish peroxidase (116 units mg−1 protein) allowed estimation of the specific peroxidase activity of APase to be approximately 5 units mg−1 protein at pH 8.8. Molybdate, vanadate, or Pi (5 mm each) exerted no influence on the peroxidase activity of the purified AtPAP26.
DISCUSSION
Plant suspension cells are typically subcultured into standard MS media containing 1.25 mm Pi (Murashige and Skoog, 1962). However, MS media containing at least 5 mm Pi was necessary to avoid eventual Pi limitation of the Arabidopsis cells while optimizing their growth over the subsequent 7-d culture period (Fig. 1). The inability of 1.25 mm Pi to maintain suspension cells Pi sufficient for at least 7 d in batch culture has been noted for several other plant species, including B. nigra (Duff et al., 1989; Lefebvre et al., 1990), B. napus (Carswell et al., 1997), and tomato (Bozzo et al., 2002, 2004a). Nevertheless, there are numerous reports describing biochemical, molecular, and/or proteomic studies of Arabidopsis suspension cells that were routinely cultured for at least 7 d in MS media that initially contained ≤1.25 mm Pi (for example, see Li et al., 2002; Ndimba et al., 2003; Shimaoka et al., 2004).
The reduced biomass accumulation of the 7-d −Pi Arabidopsis cell cultures was correlated with a large (>30-fold) reduction in their intracellular free Pi content (Fig. 2B). This magnitude of Pi decline upon Pi deprivation is consistent with observations from shoots of Arabidopsis seedlings (Müller et al., 2004), as well other plant systems, including tomato and B. nigra suspension cells and seedlings (Duff et al., 1989; Theodorou and Plaxton, 1994; Bozzo et al., 2006) and maize roots (Lee and Ratcliffe, 1993). Time-course analyses revealed that 7 d following subculture of the Arabidopsis cells into the −Pi media, extractable APase activity increased by a maximum of about 5-fold (Fig. 2C). This was correlated with a similar increase in the relative amount of an anti-(PSI tomato intracellular PAP)-IgG immunoreactive 55-kD polypeptide (Fig. 2D) that comigrated with the AtPAP26 purified from the −Pi cells (Fig. 4, C and D). The marked lag between the elimination of extracellular Pi and enhanced APase activity and levels of the immunoreactive AtPAP26 polypeptide (Fig. 2) was likely coincident with depletion of the vacuolar Pi pool (Rebeille et al., 1983) and points to an intracellular mechanism of Pi sensing. Pi resupply to 6-d-old −Pi Arabidopsis cells caused parallel decreases in APase activity and amount of the immunoreactive 55-kD AtPAP26 polypeptide to that of +Pi cells 48 h later (Fig. 2, C and D).
The predominant intracellular APase from −Pi Arabidopsis cells was purified about 1,000-fold to apparent homogeneity and a final PEP-hydrolyzing specific activity of 421 units mg−1 protein (Table I; Fig. 4A). This specific activity is comparable to values reported for a range of homogenous plant APases including AtPAP17 (del Pozo et al., 1999), as well as PSI APases from −Pi tomato and B. nigra suspension cells (Duff et al., 1989; Bozzo et al., 2002, 2004a). The 23-amino acid N-terminal sequence of the purified Arabidopsis APase matched an N-terminal portion of the deduced sequence of a putative Arabidopsis PAP gene (gene locus At5g34850: AtPAP26; Fig. 5). Unlike other APases, the activity of PAPs is insensitive to tartrate. Furthermore, several native plant PAPs exist as homodimers composed of 55-kD glycosylated subunits (LeBansky et al., 1992; Olczak and Watorek, 1998; Schenk et al., 2000). AtPAP26 displayed all these characteristics (Fig. 4, A and B), including an Amax at 520 nm (Fig. 3), a pink color in solution, and insensitivity to tartrate. A PSI PAP, AtPAP17, was purified and characterized from −Pi Arabidopsis seedlings by del Pozo et al. (1999). It displayed a subunit size of 34 kD, corresponding to the low-Mr, type-5 PAPs typical of mammalian PAPs (Schenk et al., 2000). Originally designated as AtACP5 (del Pozo et al., 1999), this PAP was reclassified as AtPAP17 (Li et al., 2002). In this study, we have biochemically characterized an intracellular Arabidopsis PAP (AtPAP26) belonging to the large molecular mass (approximately 55 kD) PAP family (Schenk et al., 2000; Li et al., 2002). AtPAP26 shares antigenic determinants with the principal PSI intracellular PAP from −Pi tomato suspension cells (Figs. 4, C and D), which exists as a heterodimer composed of an equivalent ratio of structurally related but nonidentical 63- and 57-kD subunits (Figs. 2D and 4, C and D; Bozzo et al., 2004a).
In silico analysis of the AtPAP26 gene indicated that it is translated as a 475-amino acid polypeptide that exhibits significant sequence identity with several other plant PAPs (including the 57-kD β-subunit of the PSI intracellular PAP from tomato; Bozzo et al., 2004a, 2006), all of which contain conserved domains involved in coordinating the dimetal nuclear center characteristic of the PAP active site (Fig. 5). This high degree of sequence similarity across dicots and monocots (Fig. 5; Table II) implies an important and conserved function for AtPAP26 orthologs in vascular plants. It was predicted that the N terminus of the deduced AtPAP26 polypeptide contains a 22-amino acid signal peptide. The N-terminal sequence of purified AtPAP26 begins at position 31, implying an actual signal peptide length of 30 amino acids (Fig. 5). Comparison of the N-terminal sequence of the 57-kD β-subunit of the PSI intracellular tomato PAP (Bozzo et al., 2004a) with its deduced full-length sequence (Fig. 5) revealed that its signal peptide is cleaved at an identical position as AtPAP26. AtPAP26's acidic pH optima suggested that it may be localized in the cell vacuole because various plant APases, including the 55-kD intracellular PSI APase of B. nigra suspension cells, are vacuolar (Duff et al., 1991a, 1994). The vacuolar localization of AtPAP26 was demonstrated by the proteomics study of Shimaoka et al. (2004), who identified 163 different proteins of the vacuolar membrane (tonoplast) of heterotrophic Arabidopsis suspension cells. AtPAP26 (i.e. protein encoded by gene locus At5g34850) was one of 91 different peripheral (loosely bound) tonoplast-associating proteins identified in this study, consistent with the prediction that the AtPAP26 polypeptide lacks any membrane-spanning domains (Shimaoka et al., 2004). A similar examination of the vegetative vacuole proteome of Arabidopsis rosette leaf tissue has confirmed that AtPAP26 is a vacuolar protein (Carter et al., 2004).
It is notable that semiquantitative RT-PCR revealed that in contrast to AtPAP26 polypeptide levels (Figs. 2 and 4), AtPAP26-derived transcripts were present at similar levels in the Arabidopsis suspension cells irrespective of their nutritional Pi status (Fig. 6). Although unusual, several other proteins are known to display significant environmentally induced variations in Arabidopsis without concomitant changes in the corresponding mRNA transcripts (Bonaventure and Ohlrogge, 2002; Valverde et al., 2004; Wang et al., 2006). By contrast, AtPAP17 transcripts could only be detected following RT-PCR of total RNA extracted from the −Pi, but not +Pi or Pi-resupplied, Arabidopsis suspension cells (Fig. 6), as previously documented for shoots and roots of Arabidopsis seedlings (del Pozo et al., 1999; Li et al., 2002; Müller et al., 2004). Our AtPAP26 RT-PCR results corroborate recent transcript profiling studies that have not detected AtPAP26 as being a PSI gene in −Pi Arabidopsis seedlings (Hammond et al., 2003; Wu et al., 2003; Misson et al., 2005). Specific transcript profiling of 28 members of the Arabidopsis PAP family (Zhu et al., 2005) as well as our examination of two of the most comprehensive microarray sites for Arabidopsis (Botany Array Resource, http://bbc.botany.utoronto.ca/; Genevestigator, https://www.genevestigator.ethz.ch/) confirmed that basal transcript levels for AtPAP26 are generally high and that AtPAP26 is constitutively transcribed in all tissues examined to date. The only significant (over 2-fold) increases in AtPAP26 transcript levels that we are aware of occurred following treatment of Arabidopsis seedlings with the pesticide isoxaben (2.4-fold) or during programmed cell death/senescence (2.9-fold; Gepstein et al., 2003). Similarly, GmPAP3 is a soybean (Glycine max) PAP gene whose transcription is unresponsive to Pi deprivation but is significantly induced during oxidative stress (Liao et al., 2003). AtPAP26 is a Group I PAP (subgroup Ia-2) as described by Li et al. (2002). In contrast to AtPAP26, other members of this subgroup (i.e. AtPAP10, AtPAP12) are moderately induced at the transcript level during Pi stress (Misson et al., 2005). Several other Arabidopsis PAPs display strong gene induction (i.e. AtPAPs 5, 17, 22, and 25) or are repressed (i.e. AtPAPs 3 and 4) during Pi starvation (Misson et al., 2005), giving a complex picture of regulation at both the transcriptional and the translational level. It would be useful to establish whether these dramatic changes in AtPAP transcripts correlate with alterations in the activity and amounts of the corresponding PAP proteins.
Our results also indicated that accumulation of the 55-kD AtPAP26 polypeptide coincided with an approximate 2-fold increase in shoot and root APase activity of Arabidopsis seedlings that had been subjected to 10 d of Pi deprivation (Fig. 7). A weakly cross-reactive anti-(AtPAP26)-immune serum antigenic polypeptide of approximately 37 kD was detected on immunoblots of shoot extracts from the −Pi seedlings (Fig. 7). Further studies are required to determine whether this polypeptide corresponds to a PSI PAP. Although AtPAP17 was transcriptionally up-regulated in the −Pi suspension cells (Fig. 6), no APases other than AtPAP26 were isolated during APase purification from the −Pi cells. The relatively low sequence identity (<20%) between AtPAP17 and AtPAP26 suggests that negligible immunological cross-reactivity would occur between these proteins.
Kinetic Properties of AtPAP26
Most PAPs characterized to date are considered to be nonspecific APases (Schenk et al., 2000). Similarly, purified Arabidopsis AtPAP26 hydrolyzed Pi from a wide range of phosphorylated compounds (Table IV). However, AtPAP26 is not a phytase nor a PAP with phosphodiesterase activity because it displayed no activity with phytic acid or bis-pNPP. AtPAP26 was activated by Mg2+, Co2+, Mn2+, and Ba2+, but potently inhibited by Zn2+, Cu2+, Fe2+, molybdate, vanadate, and fluoride (Table II). Similar findings have been reported for other plant APases (Duff et al., 1989, 1991a, 1994; Turner and Plaxton, 2001; Bozzo et al., 2002, 2004a). AtPAP26 also displayed significant inhibition by Pi (I50 = 2.1 mm). The intracellular Pi concentration of the 0- to 2-d −Pi cells (equals approximately 2.0–5 mm, assuming 1 g fresh weight equals approximately 1 mL; Fig. 2B) should exert significant inhibition of AtPAP26's APase activity in vivo. Conversely, the >20-fold reductions in intracellular Pi levels of the 4- to 11-d −Pi cells should relieve AtPAP26 from Pi inhibition and contribute to its enhanced in vivo APase activity in the −Pi cells.
Plant PSI PAPs displaying both APase and alkaline peroxidase activity have been reported for Arabidopsis (AtPAP17; del Pozo et al., 1999) and tomato (Bozzo et al., 2002, 2004a). In mammals, PAP peroxidase activity appears to play a role in the ROS formation in osteoclasts and macrophages, which are linked with bone resorption and macrophage killing (Hayman and Cox, 1994). AtPAP26 also displayed alkaline peroxidase activity (Fig. 8) that was insensitive to potent APase inhibitors. Plant PAPs displaying peroxidation activity have been hypothesized to function in ROS production (del Pozo et al., 1999; Bozzo et al., 2002, 2004a), a phenomenon that is closely associated with the oxidative burst that occurs during the plant defense response to pathogen infection. AtPAP17 was suggested to play an additional role in ROS metabolism during senescence (del Pozo et al., 1999). Similarly, the peroxidase activity of AtPAP26 might contribute to the metabolism of intracellular ROS following loss of vacuolar integrity that occurs during programmed cell death that accompanies senescence. At the same time, AtPAP26 APase activity may participate in intracellular Pi mobilization from senescing Arabidopsis tissues (in which AtPAP26 transcripts increase by about 3-fold; Gepstein et al., 2003).
Conclusion
Our results indicate that AtPAP26 is likely to be the predominant intracellular PAP up-regulated in −Pi Arabidopsis. Analysis of intact Arabidopsis seedling responses to Pi deficiency indicated enhanced shoot and root synthesis of the same AtPAP26 purified from the −Pi suspension cells. Thus, suspension cell cultures appear to represent a valuable model system for further investigations of the molecular and biochemical adaptations of Arabidopsis to suboptimal Pi nutrition. We are currently examining the development and Pi metabolism of AtPAP26 T-DNA mutant knockouts to test the hypothesis that AtPAP26 functions in vivo to recycle Pi from intracellular P metabolites in −Pi Arabidopsis. Of relevance is the study of Tomscha et al. (2004), who characterized an Arabidopsis APase mutant (pup3) that exhibited about 40% lower APase activity in root and shoot extracts. AtPAP12 was determined to be one of the APase isoforms defective in pup3. However, AtPAP26 was implicated in the pup3 mutant because the pup3 mutation mapped to a region of chromosome 5 within the Arabidopsis genome that includes AtPAP26 (Tomscha et al., 2004).
Recent studies of Arabidopsis Pi starvation responses have focused on identifying genes, including those encoding PAPs, that show altered transcription during Pi deprivation (Li et al., 2002; Hammond et al., 2003, 2004; Wu et al., 2003; Misson et al., 2005; Raghothama and Karthikeyan, 2005; Amtmann et al., 2006). However, our results underscore the importance of parallel biochemical and proteomic studies of the Arabidopsis Pi starvation response and support the recommendation of Li et al. (2002) that any examination of PAP function in plant Pi nutrition should consider both inducible and constitutively expressed members of the PAP family. As transcriptional controls appear to exert little influence on in vivo levels of AtPAP26 polypeptides in −Pi Arabidopsis, it will be of interest to identify translational and proteolytic controls that mediate AtPAP26's accumulation under Pi deficiency and its marked turnover following Pi resupply to the −Pi cells. Although AtPAP26 appears to be transcriptionally induced during senescence (Gepstein et al., 2003), additional studies are also needed to determine whether AtPAP26's peroxidase activity plays an intermediary role between vacuolar autolysis and cellular degenerative processes in response to abiotic or biotic oxidative stresses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Heterotrophic Arabidopsis (Arabidopsis thaliana) Landsberg erecta ecotype suspension cells were maintained at 25°C in the dark on a rotational shaker (125 rpm) in MS media, pH 5.7, containing 3% (w/v) Suc, 0.5 mg L−1 α-naphthalene acetic acid, 0.05 mg L−1 kinetin, and 5 mm K2HPO4. MS media lacking Pi was purchased from Caisson Laboratories. After autoclaving the media, filter-sterilized Pi was added from a 250 mm, pH 6.1, stock solution. Subculturing was routinely performed by transferring 10 mL of a 7-d cell suspension into 90 mL of fresh MS media containing 5 mm K2HPO4 in 500-mL Erlenmeyer flasks. Cells used in time-course studies and for APase purification were obtained by scaling up the culture volume; 100 mL of a 7-d-old culture was used to inoculate 400 mL of fresh MS media containing 0 mm KPi (−Pi) or 5 mm KPi (+Pi) in 3.2-L Fernbach flasks. At various times over a 9-d growth period, the cells were harvested by filtration through Whatman 541 filter paper on a Büchner funnel. Cells and aliquots of the corresponding CCF were frozen in liquid N2 and stored at −80°C.
Arabidopsis seeds (Columbia ecotype) were surface sterilized as described previously (Murley et al., 1998). Approximately 20 seeds were placed in 125-mL Erlenmeyer flasks containing 20 mL of sterile 0.5× MS medium, pH 5.7, containing 1% (w/v) Suc and 1.25 mm Pi. Seeds were stratified for 3 d at 4°C and placed on an orbital shaker at 80 rpm and 24°C under a 16/8-h light/dark regime (light intensity = 80 μmol m−2 s−1). After 7 d, the media were replaced with fresh media. At 14 d, the media were replaced with MS media that contained either 1.25 mm Pi (+Pi) or 0 mm Pi (−Pi). The +Pi seedlings were resupplied with 1.25 mm Pi at 21 d. Seedlings from both groups were harvested at 24 d, rinsed with Milli-Q water, and blotted dry. Shoots and roots were rapidly excised, frozen in liquid N2, and stored at −80°C.
Enzyme Assays
All enzyme assays were linear with respect to time and concentration of enzyme assayed. One unit of activity is defined as the amount of enzyme resulting in the utilization of 1 μmol of substrate min−1 at 25°C.
APase Assay A, Kinetic Studies, and Determination of Protein Concentration
For routine measurements of APase activity, the hydrolysis of PEP to pyruvate was coupled to the lactate dehydrogenase reaction. APase was assayed at 25°C by monitoring the oxidation of NADH at 340 nm using either a Gilford 260 recording spectrophotometer or a Spectromax 250 Microplate spectrophotometer (Molecular Devices). Standard APase assay conditions were 50 mm sodium acetate, pH 5.6, 5 mm PEP, 10 mm MgCl2, 0.2 mm NADH, and 3 units of desalted rabbit muscle lactate dehydrogenase in a final volume of 1 mL (for the Gilford 260) or 0.2 mL (for the Spectramax 250). All assays were initiated by the addition of enzyme preparation and corrected for background NADH oxidation by omitting PEP from the reaction mixture.
Estimation of the purified APase's apparent Km value for PEP and its I50 value for Pi (concentration of Pi producing 50% inhibition of activity) were determined as described by Bozzo et al. (2004a). All kinetic parameters are the means of three separate experiments and are reproducible to within ±10% of the mean value. Protein concentrations were determined according to Bozzo et al. (2002).
APase Assay B and Peroxidase Assay
Substrate selectivity studies were performed by quantifying the Pi released by the APase reaction as previously described (Drueckes et al., 1995; Bozzo et al., 2002). Controls were run for background amounts of Pi present at each substrate concentration tested. To calculate activities, a standard curve over the range of 1 to 133 nmol Pi was constructed for each set of assays.
A chemiluminescence assay was used to determine the capacity of the purified AtPAP26 to catalyze the peroxidation of 5-aminophthalhydrazide (luminol) as described by Bozzo et al. (2002, 2004a). In control experiments, equimolar concentrations of BSA or horseradish peroxidase were substituted for purified APase.
Buffers Used during PAP Purification
Buffer A contained 50 mm potassium acetate, pH 5.6, 1 mm EDTA, 1 mm DTT, 2,2′ dipyridyl disulfide, 1 mm phenylmethylsulfonyl fluoride, 5 mm thiourea, and 1% (w/v) insoluble polyvinylpolypyrrolidone. Buffer B contained 50 mm sodium acetate, pH 5.0, 1.5 mm MgCl2, 1 mm DTT, and 50 mm KCl. Buffer C contained 50 mm sodium acetate, pH 5.0, 1.5 mm MgCl2, 10% (v/v) glycerol, 1 mm DTT, and 50 mm KCl. Buffer D contained 25 mm sodium acetate, pH 5.7, 1.5 mm MgCl2, 50 mm KCl, 0.2 mm CaCl2, 0.2 mm MnCl2, 1 mm DTT, and 10% (v/v) glycerol. Buffer E contained 25 mm MES, pH 6.5, 1 mm DTT, and 30% (saturation) (NH4)2SO4. Buffer F contained 25 mm MES, pH 6.5, 1 mm DTT, and 15% (v/v) ethylene glycol.
PAP Purification
All chromatographic steps were carried out at room temperature using an ÄKTA FPLC system (Amersham). Quick-frozen 7-d −Pi Arabidopsis suspension cells (400 g) were ground to a powder under liquid N2 using a mortar and pestle, homogenized (1:2 w/v) in ice-cold buffer A with a small scoop of acid-washed sand, and centrifuged at 4°C and 14,000g for 20 min. Polyethylene glycol (PEG) 8000 (50% [w/v] dissolved in 20 mm Bis-Tris propane, pH 7.4) was added to the supernatant to a final concentration 2.5% (w/v) and stirred at 4°C for 20 min. Following centrifugation, finely ground PEG 8000 was added to the supernatant fluid to a final concentration of 20% (w/v) and stirred at 4°C for 60 min and centrifuged as above. PEG pellets were resuspended in buffer B to a final protein concentration of about 15 mg mL−1. The solution was stirred for 20 min, centrifuged as above, and absorbed at 1.5 mL min−1 onto a column (1.6 × 5 cm) of  -Fractogel EMD-650 (S) (Merck) preequilibrated with buffer C. The column was washed with buffer C until the A280 decreased to baseline and developed with a linear gradient (250 mL) of 50 to 500 mm KCl in buffer C (fraction size = 8 mL). A single peak of APase activity eluted at approximately 300 mm KCl. Pooled peak fractions were concentrated to about 9 mL using an Amicon Ultra-15 (10,000 Mr cutoff) centrifugal concentrator (Millipore) and applied at 0.5 mL min−1 onto a column (1 × 3.8 cm) of Con A Sepharose (Amersham) preequilibrated with buffer D. APase was eluted using 70 mL of a linear 0 to 500 mm methyl-α d-mannopyranoside gradient in buffer D (fraction size = 1 mL). Pooled peak fractions were concentrated to about 500 μL as described above, brought to 30% (saturation) (NH4)2SO4, and applied at 0.5 mL min−1 onto a Phenyl Superose HR 5/5 column (Amersham) preequilibrated with buffer E. APase activity was eluted using 20 mL of a 0% to 100% linear gradient of buffer F (100%–0% buffer E). Pooled peak fractions were concentrated as above to 110 μL, divided into 7.5-μL aliquots, quick-frozen in liquid N2, and stored at −80°C. APase activity was stable for at least 4 months when stored frozen.
-Fractogel EMD-650 (S) (Merck) preequilibrated with buffer C. The column was washed with buffer C until the A280 decreased to baseline and developed with a linear gradient (250 mL) of 50 to 500 mm KCl in buffer C (fraction size = 8 mL). A single peak of APase activity eluted at approximately 300 mm KCl. Pooled peak fractions were concentrated to about 9 mL using an Amicon Ultra-15 (10,000 Mr cutoff) centrifugal concentrator (Millipore) and applied at 0.5 mL min−1 onto a column (1 × 3.8 cm) of Con A Sepharose (Amersham) preequilibrated with buffer D. APase was eluted using 70 mL of a linear 0 to 500 mm methyl-α d-mannopyranoside gradient in buffer D (fraction size = 1 mL). Pooled peak fractions were concentrated to about 500 μL as described above, brought to 30% (saturation) (NH4)2SO4, and applied at 0.5 mL min−1 onto a Phenyl Superose HR 5/5 column (Amersham) preequilibrated with buffer E. APase activity was eluted using 20 mL of a 0% to 100% linear gradient of buffer F (100%–0% buffer E). Pooled peak fractions were concentrated as above to 110 μL, divided into 7.5-μL aliquots, quick-frozen in liquid N2, and stored at −80°C. APase activity was stable for at least 4 months when stored frozen.
AtPAP26 Antibody Production
Purified AtPAP26 (200 μg) from the −Pi Arabidopsis suspension cells was dialyzed overnight against Pi-buffered saline, filtered through a 0.2-μm membrane, and emulsified (1 mL total volume) in RIBI adjuvant (RIBI ImmunoChem Research). After collection of preimmune serum, the APase was injected into a 2-kg New Zealand rabbit. A secondary injection (100 μg) was administered after 28 d. At 10 d after the final injection, blood was collected by cardiac puncture. After incubation overnight at 4°C, the clotted cells were removed by centrifugation at 1,000g for 10 min. The antiserum was frozen in liquid N2 and stored at −80°C in 0.04% (w/v) NaN3. Affinity-purified rabbit anti-(tomato intracellular PAP)-IgG was obtained as described previously (Bozzo et al., 2006).
Protein Electrophoresis, Immunoblotting, and Estimation of Native Molecular Mass
SDS-PAGE, subunit and native Mr estimates, glycoprotein staining of SDS gels, immunoblotting onto polyvinylidene difluoride (PVDF) membranes (Immobilon transfer; 0.45-μm pore size; Millipore Canada), and visualization of antigenic polypeptides using an alkaline-phosphatase-tagged secondary antibody was conducted as previously described (Bozzo et al., 2002, 2004a). Densitometric analysis of immunoblots was performed using an LKB Ultroscan XL laser densitometer and GELSCAN software (Version 2.1; Pharmacia LKB Biotech). Derived A663 values were linear with respect to the amount of the immunoblotted extract. Immunological specificities were confirmed by performing immunoblots in which rabbit preimmune serum was substituted for the anti-(tomato intracellular PAP)-IgG or anti-(AtPAP26)-immune sera. All immunoblot results were replicated a minimum of three times. Representative results are shown in the various figures.
Amino Acid Sequencing and Bioinformatics Analysis of AtPAP26
N-terminal sequencing was done by automated Edman degradation at the Protein and Peptide Sequencing Facility of the Biotechnology Research Institute (Montreal). Similarity searches were performed using the BLAST program available on the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/Entrez/). Amino acid sequence alignment was performed with ClustalW (http://www.ebi.ac.uk/clustalw/), whereas TargetP (http://www.cbs.dtu.dk/services/TargetP/) was used to predict AtPAP26's subcellular location and signal peptide cleavage site.
RNA Extraction and Semiquantitative RT-PCR
Total RNA was extracted from Arabidopsis suspension cells using the TRIzol method (Invitrogen). Concentration and purity of the RNA were determined by spectrophotometry and corroborated by gel electrophoresis. RNA was treated with DNase (Amersham-Biosciences), and cDNA was reverse transcribed using SuperScript II RNase H− reverse transcriptase (Invitrogen) in the presence of RNase inhibitor (MBI Fermentas) according to the manufacturer's instructions. Negative reactions lacking reverse transcriptase were run in parallel for each sample to control for genomic DNA contamination. PCR amplification using intron-flanking primers specific to Arabidopsis Actin 2 (At3g18780; forward primer, 5′-TCGGTGGTTCCATTCTTGCT-3′; reverse primer, 5′-GCTTTTTAAGCCTTTGATCTTGAGAG-3′) were used to further verify the absence of genomic contamination and equal amounts of cDNA template in each reaction. Gene-specific primers, also designed to flank introns, were used to amplify AtPAP26 (At5g34850; upstream primer, 5′-GGTGATAATCTCTGTGTTCTTGAGC-3′; reverse primer, 5′-GCTATCCCATCTCACACCAACG-3′) and AtPAP17 (At3g17790; forward primer, 5′-GGTCGTCGATCGTTAATATCC-3′; reverse primer, 5′-TCTACCAACTCTGCATCAACG-3′). The PCR for all samples was performed using Taq Polymerase (MBI Fermentas) at an annealing temperature of 58°C. Aliquots were taken at sequential cycle numbers to empirically determine nonsaturating conditions for individual primer pairs. PCR products were electrophoresed on 1.2% (w/v) agarose gels.
Phosphate Extraction and Assays
Arabidopsis cells were powdered under liquid N2, ground in 10% (w/v) perchloric acid, and centrifuged at 14,000g for 10 min. Supernatants were neutralized with 5 m KOH/1 m triethanolamine. Neutralized extracts were centrifuged as above and supernatants used for intracellular Pi determinations using the spectrophotometric Pi assay described above.
Acknowledgments
We thank Dr. Rob Mullen (University of Guelph) for provision of the Arabidopsis suspension cell cultures, and Drs. Gale Bozzo and Kevin Folta (University of Florida) for useful discussions. We are also grateful to Prof. Daowen Wang (Chinese Academy of Sciences) for his helpful comments on the manuscript.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (research and equipment grants to W.A.S. and W.C.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: William C. Plaxton (plaxton@biology.queensu.ca).
References
- Amtmann A, Hammond JP, Armengaud P, White J (2006) Nutrient sensing in plants: potassium and phosphorus. Adv Bot Res 43: 209–257 [Google Scholar]
- Bonaventure G, Ohlrogge JB (2002) Differential regulation of mRNA levels of acyl carrier protein isoforms in Arabidopsis. Plant Physiol 128: 223–235 [PMC free article] [PubMed] [Google Scholar]
- Bozzo GG, Dunn EL, Plaxton WC (2006) Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant Cell Environ 29: 303–313 [DOI] [PubMed] [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC (2002) Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur J Biochem 269: 6278–6286 [DOI] [PubMed] [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC (2004. a) Structural and kinetic properties of a novel purple acid phosphatase from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Biochem J 377: 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzo GG, Singh VK, Plaxton WC (2004. b) Phosphate or phosphite addition promotes the proteolytic turnover of phosphate-starvation inducible tomato purple acid phosphatase isozymes. FEBS Lett 573: 51–54 [DOI] [PubMed] [Google Scholar]
- Carswell MC, Grant BR, Plaxton WC (1997) Disruption of the phosphate-starvation response of oilseed rape suspension cells by the fungicide phosphonate. Planta 203: 67–74 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Pena A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilizing/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Drueckes P, Schinzel R, Palm D (1995) Photometric microtiter assay of inorganic phosphate in the presence of acid-labile organic phosphates. Anal Biochem 230: 173–177 [DOI] [PubMed] [Google Scholar]
- Duff SMG, Lefebvre DD, Plaxton WC (1989) Purification and characterization of a phosphoenolpyruvate phosphatase from Brassica nigra suspension cells. Plant Physiol 90: 724–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Lefebvre DD, Plaxton WC (1991. a) Purification, characterization, and subcellular localization of an acid phosphatase from Brassica nigra suspension cells. Comparison with phosphoenolpyruvate phosphatase. Arch Biochem Biophys 286: 226–232 [DOI] [PubMed] [Google Scholar]
- Duff SMG, Plaxton WC, Lefebvre DD (1991. b) Phosphate-starvation response in plant cells: de novo synthesis and degradation of acid phosphatases. Proc Natl Acad Sci USA 88: 9538–9542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC (1994) The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant 90: 791–800 [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Gradilone SA, Arranz SE, Cabada MO (1998) Detection of highly glycosylated proteins in polyacrylamide gels. Anal Biochem 261: 224–227 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, White PJ (2004) Genetic responses to phosphorus deficiency. Ann Bot (Lond) 94: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman AR, Cox TM (1994) Purple acid phosphatases of the human macrophage and osteoclast. Comparison, molecular properties, and crystallization of the recombinant di-iron-oxo protein secreted by baculovirus-infected insect cells. J Biol Chem 269: 1294–1300 [PubMed] [Google Scholar]
- LeBansky BR, McKnight TD, Griffing LR (1992) Purification and characterization of a secreted purple acid phosphatase from soybean suspension cells. Plant Physiol 99: 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RB, Ratcliffe RG (1993) Subcellular distribution of inorganic phosphate, and levels of nucleoside triphosphate, in mature maize roots at low external phosphate concentrations: measurements with 31P-NMR. J Exp Bot 44: 587–598 [Google Scholar]
- Lefebvre DD, Duff SMG, Fife CA, Julien-Inalsingh C, Plaxton WC (1990) Response to phosphate deprivation in Brassica nigra suspension cells. Enhancement of intracellular, cell surface, and secreted acid phosphatase activities compared to increases in Pi-absorption rate. Plant Physiol 93: 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D (2002) Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277: 27772–27781 [DOI] [PubMed] [Google Scholar]
- Liao H, Wong F-L, Phang T-H, Cheung M-Y, Li W-YF, Shao G, Yan X, Lam H-M (2003) GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 318: 103–111 [DOI] [PubMed] [Google Scholar]
- Miller SS, Liu J, Allan DL, Menzhuber CJ, Fedorova M, Vance CP (2001) Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiol 127: 594–606 [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Doumas P, et al (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Nilsson L, Krintel C, Nielsen TH (2004) Gene expression during recovery from phosphate starvation in roots and shoots of Arabidopsis thaliana. Physiol Plant 122: 233–243 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murley VR, Theodorou ME, Plaxton WC (1998) Phosphate-starvation-inducible pyrophosphate dependent phosphofructokinase occurs in plants whose roots do not form symbiotic associations with mycorrhizal fungi. Physiol Plant 103: 405–414 [Google Scholar]
- Ndimba BK, Chivasa S, Hamilton JM, Simon WJ, Slabas AR (2003) Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 3: 1047–1059 [DOI] [PubMed] [Google Scholar]
- Olczak M, Watorek W (1998) Oligosaccharide and polypeptide homology of lupin (Lupinus luteus L.) acid phosphatase subunits. Arch Biochem Biophys 360: 645–655 [DOI] [PubMed] [Google Scholar]
- Oshima Y, Ogawa N, Harashima S (1996) Regulation of phosphatase synthesis in Saccharomyces cerevisiae: a review. Gene 179: 171–177 [DOI] [PubMed] [Google Scholar]
- Plaxton WC (2004) Plant response to stress: biochemical adaptations to phosphate deficiency. In R Goodman, ed, Encyclopedia of Plant and Crop Science. Marcel Dekker, New York, pp 976–980
- Raghothama KG, Karthikeyan AS (2005) Phosphate acquisition. Plant Soil 274: 37–49 [Google Scholar]
- Rebeille F, Bligney R, Martin J-B, Douce R (1983) Relationships between the cytoplasm and the vacuole pool in Acer pseudoplatanus cells. Arch Biochem Biophys 225: 143–148 [DOI] [PubMed] [Google Scholar]
- Schenk G, Guddat LW, Carrington LE, Hume DA, Hamilton S, de Jersey J (2000) Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 250: 117–125 [DOI] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki KI, Maeshima M, Yokota A, Tomizawa KI, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45: 672–683 [DOI] [PubMed] [Google Scholar]
- Theodorou ME, Plaxton WC (1994) Induction of PPi-dependent phosphofructokinase by phosphate-starvation in seedlings of Brassica nigra. Plant Cell Environ 17: 287–294 [Google Scholar]
- Ticconi CA, Abel S (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Tomscha JL, Trull MC, Deikman J, Lynch JP, Guiltinan MJ (2004) Phosphatase under-producer mutants have altered phosphorus relations. Plant Physiol 135: 334–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner WL, Plaxton WC (2001) Purification and characterization of banana fruit acid phosphatases. Planta 214: 243–249 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Vance CP, Udhe-Stone C, Allan DL (2003) Phosphorus acquisition and its use: critical adaptations by plants for securing a non-renewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Wang B-C, Wang H-X, Meng D-Z, Qu L-J, Zhu Y-X (2006) Post-translational modifications, but not transcriptional regulation, of major chloroplast RNA-binding proteins are related to Arabidopsis seedling development. Proteomics 6: 2555–2563 [DOI] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations in genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Qian W, Xuzhong L, Li D, Liu X, Liu K, Wang D (2005) Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol Biol 59: 581–594 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Regrierer B, Kossmann J, Frossard E, Amrhein N, Bucher M (2004) Differential expression of three purple acid phosphatases from potato. Plant Biol 6: 519–528 [DOI] [PubMed] [Google Scholar]