Abstract
The identification of a family of NAR2-type genes in higher plants showed that there was a homolog in Arabidopsis (Arabidopsis thaliana), AtNAR2.1. These genes encode part of a two-component nitrate high-affinity transport system (HATS). As the Arabidopsis NRT2 gene family of nitrate transporters has been characterized, we tested the idea that AtNAR2.1 and AtNRT2.1 are partners in a two-component HATS. Results using the yeast split-ubiquitin system and Xenopus oocyte expression showed that the two proteins interacted to give a functional HATS. The growth and nitrogen (N) physiology of two Arabidopsis gene knockout mutants, atnrt2.1-1 and atnar2.1-1, one for each partner protein, were compared. Both types of plants had lost HATS activity at 0.2 mm nitrate, but the effect was more severe in atnar2.1-1 plants. The relationship between plant N status and nitrate transporter expression revealed a pattern that was characteristic of N deficiency that was again stronger in atnar2.1-1. Plants resulting from a cross between both mutants (atnrt2.1-1 × atnar2.1-1) showed a phenotype like that of the atnar2.1-1 mutant when grown in 0.5 mm nitrate. Lateral root assays also revealed growth differences between the two mutants, confirming that atnar2.1-1 had a stronger phenotype. To show that the impaired HATS did not result from the decreased expression of AtNRT2.1, we tested if constitutive root expression of a tobacco (Nicotiana plumbaginifolia) gene, NpNRT2.1, previously been shown to complement atnrt2.1-1, can restore HATS to the atnar2.1-1 mutant. These plants did not recover wild-type nitrate HATS. Taken together, these results show that AtNAR2.1 is essential for HATS of nitrate in Arabidopsis.
Nitrogen (N) is the most important inorganic nutrient in plants, and its availability is a limiting factor for plant growth in most agricultural systems. Limiting N supply influences shoot-root allocation of resources, favoring root system development for exploration of a larger soil volume. Severe N deficiency leads to a general inhibition of plant growth. Nitrate is the main N source available in arable soil, and in this form it can act directly as a signal that regulates shoot-root allocation and modification of the root system architecture (Scheible et al., 1997; Zhang and Forde, 2000).
Many physiological investigations on nitrate uptake by roots led to the conclusion that plants have developed three types of transport system to cope with the variations in nitrate concentrations in cultivated soils (Glass and Siddiqi, 1995). Two saturable high-affinity transport systems (HATS) are able to take up nitrate at low external concentration (1 μm–1 mm). The constitutive system (cHATS) is available even if the plants have never been supplied with nitrate. The inducible system (iHATS) is strongly stimulated by nitrate in the external medium. The low-affinity transport system (LATS) displays linear kinetics and its contribution to global nitrate uptake becomes significant at external nitrate concentrations above 1 mm.
Many studies on the molecular basis of root nitrate uptake systems revealed two genes families, NRT1 and NRT2, potentially coding for nitrate transporters involved in the LATS and HATS systems (Crawford and Glass, 1998; Forde, 2000; Orsel et al., 2002a). The chl1 line, affected in the Arabidopsis (Arabidopsis thaliana) AtNRT1.1 gene, was the first mutant characterized for a deficiency in the nitrate LATS system (Tsay et al., 1993). Extensive studies revealed a complex role for AtNRT1.1, but no phenotype related to plant growth (Guo et al., 2001, 2003). The first members of the NRT2 family were identified in lower eukaryotes. The crnA (nrtA) gene from Aspergillus nidulans was identified from a mutant resistant to chlorate, a toxic nitrate analog (Unkles et al., 1991, 2001). Two other genes, CrNRT2.1 and CrNRT2.2, were identified in the green algae Chlamydomonas reinhardtii using deletion strains deficient in nitrate uptake (Quesada et al., 1994). Most members of the NRT2 family in higher plants were subsequently identified based on their sequence homology to nrtA and CrNRT2.1 (Daniel-Vedele et al., 1998). So far, in each species studied, the NRT2 genes are a small multigenic family. The complete genome analysis of Arabidopsis revealed seven NRT2 members that are differentially expressed in plant tissues with a pattern that can depend on the external N supply (Orsel et al., 2002b; Okamoto et al., 2003).
AtNRT2.1, the first Arabidopsis NRT2 gene to be identified, was cloned on the basis of differential induction under nitrate versus Gln supply (Filleur and Daniel-Vedele, 1999). Expression analysis showed that AtNRT2.1 expression is perfectly coordinated with nitrate HATS regulation: induction by low external nitrate concentration and sudden N deprivation (Filleur and Daniel-Vedele, 1999; Lejay et al., 1999). Root uptake analysis of a knockout mutant, atnrt2.1-1 (atnrt2a), for both genes AtNRT2.1 and AtNRT2.2 provided the first functional evidence supporting the role of NRT2 genes in nitrate-inducible HATS (Filleur et al., 2001). Further investigation of the atnrt2.1-1 mutant has shown that HATS deficiency had a strong impact on plant growth under low nitrate (0.2 mm) but not higher concentrations (6 mm; Orsel et al., 2004b). The shoot biomass and nitrate content of the mutants were strongly decreased compared to the wild type, but the root growth was maintained, leading to a decreased shoot to root ratio, a characteristic feature of N-limited plants. Recent studies have highlighted the role of AtNRT2.1 in the root architecture response to low nitrate availability, especially in lateral root (LR) initiation (Little et al., 2005; Remans et al., 2006).
In C. reinhardtii, two types of genes involved in nitrate transport (CrNRT2 and CrNAR2) are located within a nitrate-regulated gene cluster. Mutants deleted in this genomic region recovered high-affinity nitrate uptake activity only after transformation with plasmids carrying CrNAR2 and either CrNRT2.1 or CrNRT2.2, but not with any of these genes individually (Quesada et al., 1994). The co-injection of two different types of mRNA in Xenopus oocytes revealed that both gene family products were required for functional nitrate uptake (Zhou et al., 2000). Now numerous genes belonging to the NAR2 family have been identified in plants among many different plants species, including AtNAR2.1 in Arabidopsis (Tong et al., 2005). More recently, two AtNAR2 genes have been identified in Arabidopsis: AtNAR2.1 (AtNRT3.1) and AtNAR2.2 (AtNRT3.2) with accession numbers At5g50200 and At4g24720, respectively (Okamoto et al., 2006). We will use the former gene naming system throughout this article. No evidence for the expression of the second gene, AtNAR2.2, was found in the databases (expressed sequence tags, cDNA, or microarray data), but, using several sets of specific primers in a sensitive assay, it was found to be just detectable (Okamoto et al., 2006). In contrast, AtNAR2.1 was strongly expressed in the roots in the AtGenExpress Affymetrix experiments (Weigel et al., 2004).
The presence of both NRT2 and NAR2 families in plant genomes suggested a more general relevance for the NRT2/NAR2 transport model and functional two-component nitrate transport was reconstituted in Xenopus oocytes using the barley (Hordeum vulgare) genes HvNRT2.1/HvNAR2.3 (Tong et al., 2005). As deletion mutants are easier to obtain in Arabidopsis than barley, we have investigated the functional identity of a two-component high-affinity nitrate transport system in planta using this model. Our starting hypothesis was that AtNRT2.1/AtNAR2.1 constitutes a two-component system. We began by testing this hypothesis in two different heterologous expression systems and then moved to a more detailed investigation of the AtNAR2.1/AtNRT2 system by studying a new isolated knockout mutant for AtNAR2.1. The atnar2.1-1 T-DNA insertion mutant isolated and used for this analysis has recently been reported in a parallel but independent study (Atnrt3.1-2). Comparing the atnar2.1-1 mutant and wild-type plants, this work concluded that the nitrate HATS system in higher plants requires a functional NAR2 gene (Okamoto et al., 2006). We are reporting how we have extended this previous work by comparing the growth and N physiology of atnar2.1-1 and atnrt2.1-1 mutants with wild-type plants. The expression of nitrate transporters was first characterized as being induced by the substrate (Crawford and Glass, 1998; Forde, 2000), but, in common with other nutrients such as phosphate and sulfate, it is becoming apparent that N starvation can increase the expression of some family members (Orsel et al., 2004a). We therefore checked the relationship between plant N status and the expression and activity of the AtNAR2.1/AtNRT2 system. Finally, as root morphology changes provide a convenient and sensitive assay for identifying Arabidopsis phenotypes, we have used this method to investigate the function of the two-component system.
RESULTS
Protein-Protein Interaction between AtNRT2.1 and AtNAR2.1
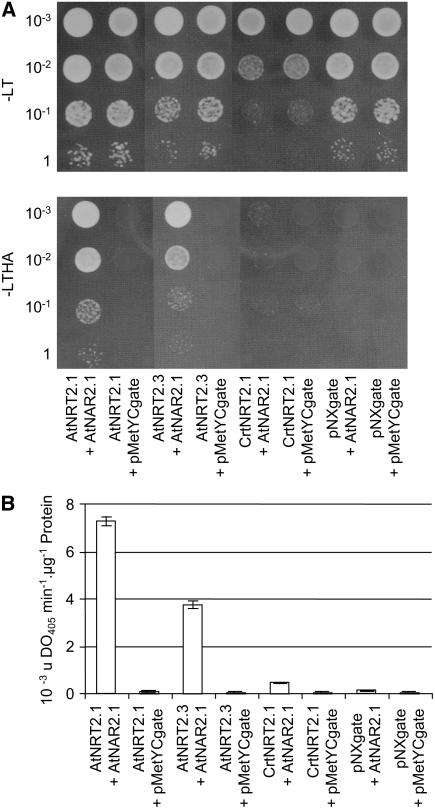
The interaction of AtNRT2.1 and AtNAR2.1 proteins was tested using a mating-based split-ubiquitin system (mbSUS) that allows detection of interacting membrane proteins (Obrdlik et al., 2004). Both CrNRT2.1 and AtNRT2.3 proteins were used to assay the specificity of the interaction. The C. reinhardtii protein CrNRT2.1 is involved in a two-component system with CrNAR2.1 (Quesada et al., 1994) and has 55% sequence similarity with AtNRT2.1 (EMBOSS-Align, program Needle, EBlossum 62). The Arabidopsis AtNRT2.3 protein has more than 76% similarity with AtNRT2.1, but expression of the gene is almost undetectable in all conditions (Orsel et al., 2002b), making it an unlikely partner for AtNAR2.1. Each potential partner was fused to the N (Nub)- or C (Cub)-terminal domain of the ubiquitin protein. In mbSUS, the interaction between the two membrane-bound fusion proteins leads to the reconstruction of the ubiquitin protease activity and release of the protein A-LexA-VP16 (PLV) transcription factor fused to the Cub domain (Obrdlik et al., 2004). Two types of reporter genes are under the control of the PLV transcription factor, allowing growth tests on selective media (His and Ade auxotrophy) and β-galactosidase activity assays (LacZ).
The NRT2 cDNAs were cloned in the pNXgate and pXNgate plasmids and tested in combination with AtNAR2.1 cDNA clones in the pMetYCgate. Growth of diploid cells under selective conditions revealed interaction of AtNAR2.1-CubPLV with NubG-AtNRT2.1 and NubG-AtNRT2.3, but not with NubG-CrNRT2.1 (Fig. 1A). The NRT2-NubG constructs never showed interaction in combination with AtNAR2.1-CubPLV (data not shown). Testing the β-galactosidase activity provides more quantitative results (Fig. 1B). The activity detected when AtNAR2.1-CubPLV was in combination with NubG-AtRNT2.1 was 2 and 16 times higher than in combination with NubG-AtNRT2.3 and NubG-CrNRT2.1, respectively. These results indicate that the strongest interaction occurs between AtNRT2.1 and AtNAR2.1, much more than between any other NRT2 protein and AtNAR2.1. In the yeast system, the AtNRT2.1 and AtNAR2.1 proteins can interact in a specific manner and form a membrane complex.
Figure 1.
Testing the interaction of AtNRT2.1 and AtNAR2.1 using the mbSUS split-ubiquitin system using HIS3, ADE2, and lacZ as reporter genes. Diploid cells carrying pMetAtNAR2.1-Cgate and different pN-NRT2gate plasmid (pNXgate and pMetYCgate are the control vectors with no cloned cDNA) were grown on liquid SD −LT (Trp, Leu) medium containing 50 μm Met. A, Cells growth on control SD-LT or selective SD −LTHA (Trp, Leu, His, Ade) media. B, β-Galactosidase activity with o-nitrophenylglucoside, shown as 10−3 uDO min−1 μg−1 protein. The kinetics of change in absorbance was measured at 405 nm. Values are means ± sd of three independent measurements.
Expressing an AtNAR2.1/AtNRT2 Nitrate Transport System in Xenopus Oocytes
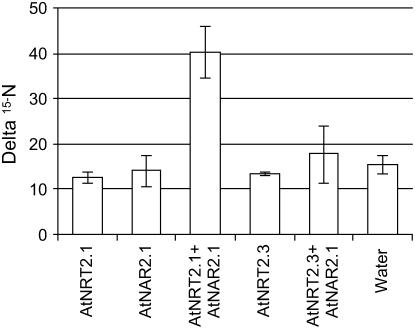
To test if AtNAR2.1 and AtNRT2.1 constituted an Arabidopsis two-component nitrate transport system, both mRNAs were tested in the Xenopus oocyte heterologous system (Tong et al., 2005). Oocytes were injected with various combinations of NRT2/NAR2 mRNA, including single injection with one or the other mRNA. Oocytes were assayed for nitrate transport activity using enriched 15N nitrate (Tong et al., 2005). Measurement of 15N enrichment in single oocytes showed that only the combination of AtNRT2.1 and AtNAR2.1 provided a significant uptake when compared with the water and single injected oocytes (Fig. 2). A very important result is the negative result obtained with AtNRT2.3. Despite some sequence similarity between AtNRT2.1 and AtNRT2.3 (see above), no functional nitrate uptake system could be reconstituted with the AtNRT2.3/AtNAR2.1 combination in oocytes (Fig. 2). These results show that only the co-injection of AtNAR2.1 together with AtNRT2.1 mRNA results in high-affinity nitrate transport in Xenopus oocytes.
Figure 2.
Uptake of 15N nitrate into oocytes injected with water or mRNA mixtures as indicated. Oocytes were incubated for 16 h in ND96 solution (pH 6) enriched with 0.5 mm 15NaNO3. The delta 15N values are means ± sd for five oocytes.
Comparing the Growth and N Pools between Wild Type and the atnrt2.1-1 and atnar2.1-1 Mutants
The atnrt2.1-1 mutant has already been shown to have altered HATS activity and growth at low nitrate concentrations (Orsel et al., 2004b). Plants were grown in hydroponics with only nitrate as the N source at two different concentrations, high (6 mm) and low (0.2 mm). At these two levels of concentration, the nitrate supply was not limiting as the nutrient solutions were frequently replaced and wild-type growth was not significantly different (see Supplemental Fig. S1, a–d). By contrast, growth of the atnar2.1-1 mutant was severely decreased at the low nitrate concentration and this effect was stronger than that measured for atnrt2.1-1 plants (see Supplemental Fig. S1a). The shoot and root biomasses of the atnar2.1-1 mutant were only 10% and 48%, respectively, of the wild-type biomass (Supplemental Fig. S1, c and d), and the shoot to root biomass ratio was strongly reduced to 0.9 (Supplemental Fig. S1b). There was no growth difference between the wild type and both types of mutants when the plants were supplied at the higher nitrate concentration (Supplemental Fig. S1, a–d).
Analysis of shoot and root nitrate content revealed no differences between wild-type and mutant plants when grown at high nitrate concentration (180 and 50 μmol g−1 fresh weight [FW], respectively, for shoot and root; see Supplemental Fig. S1, e and f). When grown at low nitrate concentration, the wild-type shoot and root nitrate content were decreased by half compared to high nitrate, but in contrast nitrate was almost undetectable in either shoot or root of the atnar2.1-1 mutant. For comparison and confirming earlier work (Orsel et al., 2004b), when grown at low nitrate concentration the shoot nitrate content of the atnrt2.1-1 mutant was more decreased than the root nitrate content (43% and 77%, respectively, of the wild type).
The total amino acids of wild-type tissues were increased when plants were grown at low nitrate concentration. Root amino acids increased by 162% of the value for wild-type plants grown at the higher nitrate concentration (Supplemental Fig. S1, g and h). In contrast, when grown at a lower nitrate concentration, the shoot and root amino acids concentrations both decreased to 37% for atnar2.1-1 and 80% for atnrt2.1-1 mutants when compared with the values at higher nitrate. The mutant atnar2.1-1 showed only 40% and 29%, respectively, of the shoot and root wild-type amino acids concentrations when grown at the lower nitrate concentration.
Analysis of total N content revealed no significant difference between the genotypes under high nitrate supply with around 6% and 5% DW, respectively, for shoots and roots (see Supplemental Fig. S1, i and j). But on the low nitrate concentration, while the wild-type total N content was only slightly reduced, the dramatic decrease of nitrate and amino acid content in the root and shoot of the atnar2.1-1 mutant led to decreased total N content, to only 2.2% and 2.7% DW in shoot and root, respectively. The total N content of the atnar2.1-1 plants grown on low nitrate supply is similar to values measured in wild type after 10 d of N starvation (M. Orsel, unpublished data).
Comparing HATS Activity between Wild Type and the atnrt2.1-1 and atnar2.1-1 Mutants
At the end of the experiment used to obtain biomass data (Supplemental Fig. S1), the same plants were used to measure nitrate influxes at 0.2 and 6 mm 15NO3− external concentrations to discriminate between the activities of nitrate HATS and LATS. Growing on 6 mm nitrate, wild type and mutants showed the same HATS activity (see Supplemental Fig. S2a). When grown at the lower nitrate concentration (0.2 mm NO3−), wild-type plants showed increased HATS activity, which reached 120 μmol 15NO3− h−1 g−1 root DW. This classical response of HATS to low nitrate supply was lost in the atnrt2.1-1 mutant, which showed only 25 μmol 15NO3− h−1 g−1 root DW (21% of the wild-type 15NO3− influx). For the atnar2.1-1 mutant, the nitrate HATS activity was only 4 μmol 15NO3− h−1 g−1 root DW, even lower than that measured for the atnrt2.1-1 mutant. This rate was only 3% of the wild-type 15NO3− influx and was less than the atnar2.1-1 HATS activity for plants growing on 6 mm nitrate (20 μmol 15NO3− h−1 g−1 root DW). To evaluate root 15NO3− influx resulting from the activity of the LATS, nitrate influx at 0.2 mm 15NO3− was subtracted from that measured at 6 mm 15NO3− (Supplemental Fig. S2, a and b). The calculated values showed no significant differences between the mutants and the wild type (Supplemental Fig. S2c), indicating that LATS activity at high and low nitrate growth concentrations was not affected in the atnar2.1-1 mutant and as was shown previously for the atnrt2.1-1 mutant (Cerezo et al., 2001).
Comparing the Expression Pattern of Genes Possibly Involved in Nitrate Transport between Wild Type and the atnrt2.1-1 and atnar2.1-1 Mutants
Root expression levels of genes involved or potentially involved in HATS activity were studied by relative quantitative reverse transcription-PCR (see Table I). AtNAR2.1 was highly expressed in wild type and the atnrt2.1-1 mutant, both at the same level as the constitutive reference EF1α. Only residual expression could be detected in the atnar2.1-1 mutant, as the specific primer set used corresponded to the cDNA sequence upstream of the T-DNA insertion. This result appears to contrast with data in the paper by Okamoto et al. (2006), but this difference can be explained by the differing primer sets used for the PCR and/or the slightly different method. AtNRT2.1 transcripts were never detected in the atnrt2.1-1 mutant as AtNRT2.1 gene was deleted by the T-DNA insertion. On low nitrate concentration, root AtNRT2.1 expression was induced in wild-type roots but repressed in the atnar2.1-1 mutant. The expression level of AtNRT2.1 was decreased by 50% compared to the plants grown at the high nitrate concentration. There was no significant difference in AtNRT1.1 expression between wild type and mutants grown on high nitrate (close to 40% of EF1α), and this level was maintained when wild-type plants were grown on 0.2 mm nitrate. But in both atnrt2.1-1 and atnar2.1-1 mutants, AtNRT1.1 expression levels were decreased to 12% and 7% of EF1α, respectively, under low nitrate conditions (Table I). As shown previously, expression of both AtNRT2.4 and AtNRT2.5 on 0.2 mm nitrate in wild type and the atnrt2.1-1 mutant was enhanced (Orsel et al., 2004b). The increase was even greater for the atnar2.1-1 mutant, with AtNRT2.4 and AtNRT2.5 10 and 40 times more strongly expressed when compared with the wild type.
Table I.
Relative gene expression level in roots of wild-type (Ws), atnrt2.1-1, and atnar2.1-1 plants grown under different nitrate regimes
Relative expression level was determined on the same plants as described for Figure 3; values are means ± sd of three replicates (pooling three to five plants). Results are given as a percentage of the EF1α gene used as a constitutively expressed reference. Nd., Not detectable (below 0.01% EF1α).
| Genotype | Nutrition | AtNAR2.1 | AtNRT2.1 | AtNRT2.4 | AtNRT2.5 | AtNRT1.1 |
|---|---|---|---|---|---|---|
| Ws | 6 mm | 98 ± 8 | 189 ± 9 | 0.7 ± 0.1 | Nd. | 37 ± 12 |
| 0.2 mm | 103 ± 18 | 249 ± 27 | 5.3 ± 1.4 | 0.5 ± 0.2 | 33 ± 4 | |
| atnrt2.1-1 | 6 mm | 89 ± 17 | Nd. | 1.1 ± 0.1 | 0.1 ± 0.1 | 39 ± 5 |
| 0.2 mm | 116 ± 19 | Nd. | 8.2 ± 1.7 | 2.7 ± 1.1 | 12 ± 1 | |
| atnar2.1-1 | 6 mm | 11 ± 2 | 278 ± 63 | 1.8 ± 0.1 | 0.1 ± 0.2 | 40 ± 11 |
| 0.2 mm | 4 ± 1 | 132 ± 6 | 57.6 ± 11.0 | 21.0 ± 2.4 | 7 ± 0 |
Response to Short-Term Changes in Plant N Status: Effects on HATS Activity and AtNRT2.1 and AtNAR2.1 Expression between the atnrt2.1-1 and atnar2.1-1 Mutants
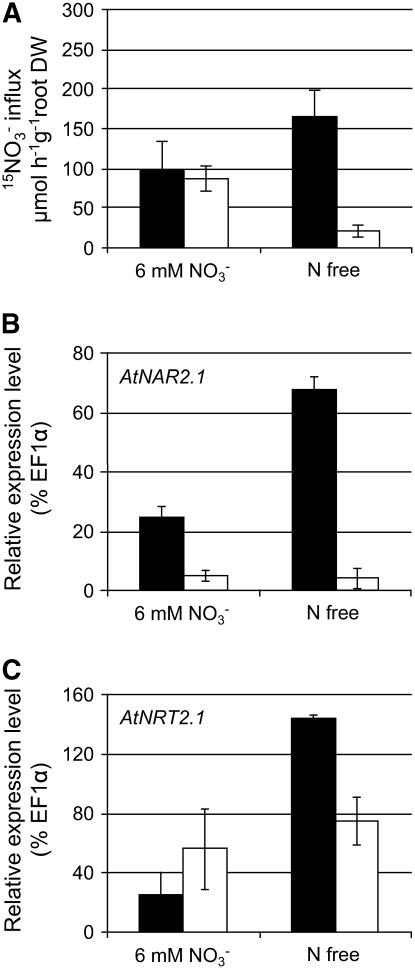
The AtNRT2.1 gene has been shown to be specifically involved in the component of HATS that is inducible by both N starvation and nitrate (Cerezo et al., 2001; Filleur et al., 2001). Results from the first set of hydroponic experiments showed that AtNAR2.1 was a component of HATS (see above). To test if the atnar2.1-1 mutant was impaired in the N starvation-inducible component of HATS, wild-type plants and atnar2.1-1 mutants were grown for 6 weeks in hydroponics at 6 mm nitrate and then transferred for 24 h to an N-free medium (Fig. 3). In wild-type plants, both AtNRT2.1 and AtNAR2.1 expression levels were increased by the transfer to N-free supply (5.8 and 2.7 times, respectively; see Fig. 3). This induction coincided with increased HATS activity; the root 15NO3− nitrate influx rate at 0.2 mm increased from 100 to 166 μmol 15NO3− h−1 g−1 root DW when wild-type plants were transferred to an N-free supply (Fig. 3A). On high nitrate supply, the HATS activity for the atnar2.1-1 mutant was similar to wild-type plants, but there was no induction of this transport system by the transfer to N-free medium. Actually, HATS activity was decreased by 75% when compared with the influx measured on high nitrate supply (Fig. 3A). The expression of AtNRT2.1 was maintained at the same level as that measured on high nitrate supply (Fig. 3C). Therefore, in contrast to wild type, there was repression of HATS in the atnar2.1-1 mutant transferred to N-free medium that cannot be explained by decreased expression of AtNRT2.1.
Figure 3.
Root 15NO3− influx and relative expression levels of AtNAR2.1 and AtNRT2.1 in wild type and atnar2.1-1 after 24-h N starvation. Wild type (black bars) and mutant atnar2.1-1 (white bars) were grown for 41 d in hydroponics in medium containing 6 mm NO3− and then transferred to N-free or 6 mm NO3− medium for 24 h (irradiation 150 μmol photons m−2 s−1). HATS activity was measured as root 15NO3− influx after 5-min labeling with complete nutrient solution containing 0.2 mm 15NO3− (A). B and C, Relative expression level of AtNAR2.1 (B) and AtNRT2.1 (C) in roots was determined on the same plants. Results are given as a percentage of the EF1α gene used as a constitutive reference (detection limit is 0.01% EF1α). The values are means ± sd of five replicates (pooling one to three plants).
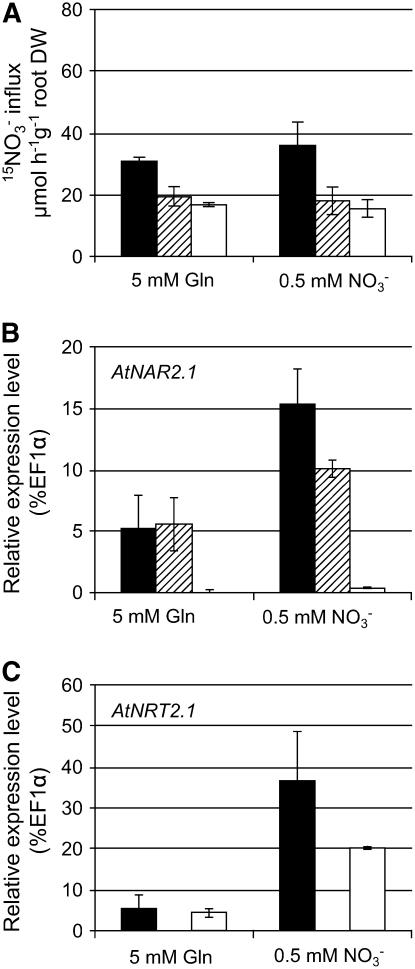
To determine if AtNAR2.1 was involved in both constitutive and nitrate-inducible HATS, wild-type plants and atnrt2.1-1 and atnar2.1-1 mutants were grown in vitro on Gln, a reduced N source, for 10 d and then transferred to a low nitrate concentration supply for 24 h (Fig. 4). When plants were grown on Gln medium, the nitrate HATS activity for both atnrt2.1-1 and atnar2.1-1 mutants was decreased to 63% and 54%, respectively, of the wild-type activity (Fig. 4A), indicating that both AtNRT2.1 and AtNAR2.1 are components of HATS that are not dependent on previous nitrate exposure. After transfer to a low nitrate concentration for 24 h, the root 15NO3− nitrate influx at 0.2 mm was only slightly increased in the wild type and unchanged for both mutants. But both AtNAR2.1 and AtNRT2.1 relative expression levels were increased after transfer to a low nitrate concentration (Fig. 4, B and C). The relative expression level of AtNRT2.1 increased by 6.5- and 4.5-fold in wild-type and atnar2.1-1 plants, respectively. AtNAR2.1 gene expression was increased by 3-fold over that detected in the wild type and doubled in the atnrt2.1-1 mutant background. The high N status of wild-type plants grown on Gln might explain the absence of an increase in HATS by transfer to low nitrate medium even if both AtNRT2.1 and AtNAR2.1 are induced.
Figure 4.
15NO3− influx and relative expression levels of AtNAR2.1 and AtNRT2.1 in wild type, atnrt2.1-1, and atnar2.1-1 after 24 h NO3− induction. Wild-type (black bars) and mutant (atnrt2.1-1, hatched bars; atnar2.1-1, white bars) plants were grown on vertical agar plates containing 5 mm Gln for 10 d and then transferred to either 0.5 mm NO3− or 5 mm Gln for 24 h. A, HATS activity was measured as root 15NO3− influx after 5-min labeling with complete nutrient solution containing 0.2 mm 15NO3−. B and C, Root relative expression level of AtNAR2.1 (B) and AtNRT2.1 (C) was determined on plants from the same batch. Results are given as a percentage of the EF1α gene used as a constitutive reference (detection limit is 0.01% EF1α). The values are means ± sd of four replicates (pooling three to six plants).
In summary, as expected for two partners involved in one system, AtNRT2.1 and AtNAR2.1 are both regulated by nitrate availability: They are induced by sudden N starvation and by low nitrate concentration versus Gln. Both genes are essential for iHATS but also seem to be involved in cHATS.
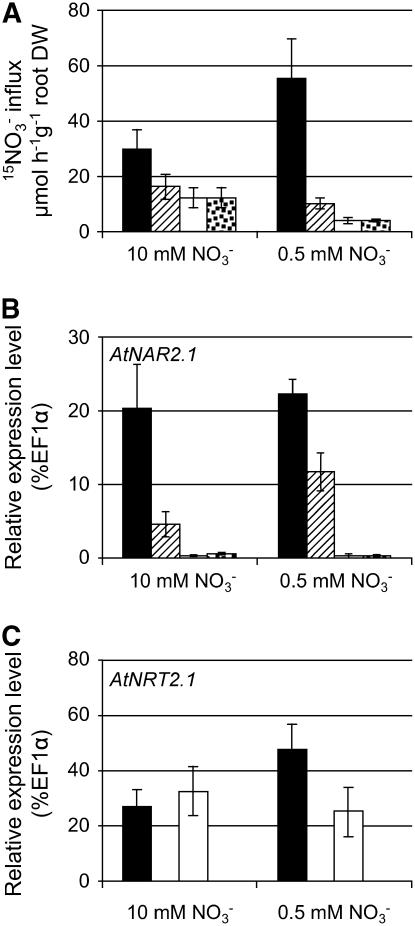
A Double atnrt2.1-1 atnar2.1-1 Mutant
To provide proof that the atnar2.1-1 plant has a stronger phenotype than the atnrt2.1-1, a cross between the two mutants was generated and grown alongside the parent plants. Plants were grown in vitro for 6 d on agar plates containing 10 mm nitrate and then transferred to either 0.5 or 10 mm nitrate (Fig. 5). The double atnrt2.1-1 atnar2.1-1 mutant showed the same phenotype as the atnar2.1-1 mutant (Fig. 5). The root 15NO3− nitrate influx on 0.2 mm was only 4 μmol 15NO3− h−1 g−1 root DW for both genotypes, representing, respectively, 50% and 7% of HATS activity in atnrt2.1-1 and wild type (Fig. 5A). The expression level of AtNAR2.1 was unchanged in the wild type under the two growth conditions, but induced by 2.5-fold in the atnrt2.1-1 mutant background on 0.5 mm nitrate. Only residual expression of AtNAR2.1 could be detected in both the atnar2.1-1 mutant and the double mutant due to the T-DNA insertion (Fig. 5B). Six days after transfer to 0.5 mm, AtNRT2.1 gene expression was increased by 1.7-fold in the wild type, but not detectable under any condition in the atnrt2.1-1 mutant or the double mutant due to the gene deletion (Fig. 5C). There was no significant induction of AtNRT2.1 expression in the atnar2.1-1 mutant background on 0.5 mm nitrate. At low nitrate concentration, both mutants, atnar2.1-1 and the double mutant, had decreased shoot biomass with very similar levels around 1.8 mg FW (Supplemental Fig. S5). The decrease was less severe in the atnrt2.1-1 mutant, with the biomass only changing to 2.6 mg FW (Supplemental Fig. S5). Furthermore, the residual HATS activity remaining in both the double and atnar2.1-1 mutants on low nitrate concentration supply was significantly lower than that measured in the wild type and atnrt2.1-1 mutant. There is evidence for an epistatic interaction, with the atnar2.1-1 phenotype appearing to be stronger than that of the atnrt2.1-1 mutation.
Figure 5.
Root 15NO3− influx and relative expression levels of AtNAR2.1 and AtNRT2.1 in atnrt2.1-1, atnar2.1-1, and the double mutants. Wild-type (black bars) and mutant (atnrt2.1-1, hatched bars; atnar2.1-1, white bars; atnrt2.1-1 atnar2.1-1, dotted bars) plants were grown on vertical agar plates on 10 mm NO3− for 7 d and transferred to 10 mm NO3− or 0.5 mm NO3− for a further 6 d (total of 13 d). A, HATS activity was measured as root 15NO3− influx after 5-min labeling with complete nutrient solution containing 0.2 mm 15NO3−. B and C, Root relative expression level of AtNAR2.1 (B) and AtNRT2.1 (C) was determined on plants from the same batch. Results are given as a percentage of the EF1α gene used as a constitutive reference (detection limit is 0.01% EF1α). The values are means ± sd of four replicates (pooling three to six plants).
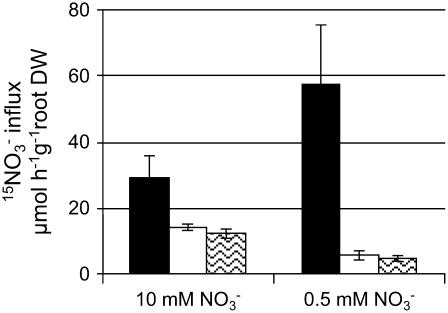
Testing if Constitutive Expression of NpNRT2.1 Can Restore the atnar2.1-1 Phenotype
To determine if NpNRT2.1 from Nicotiana plumbaginifolia complements the atnrt2.1-1 phenotype restoring functional HATS (see Filleur et al., 2001), without interacting with a NAR2.1-type partner, RolDNpNRT2.1X atnar2.1-1 plants were generated by crossing and assayed for root 15NO3− influx (HATS activity) at 0.2 mm nitrate (Fig. 6). Plants were grown in vitro on high nitrate (10 mm) and then transferred to low nitrate (0.5 mm) concentration. The RolDNpNRT2.1X atnar2.1-1 plants displayed the same phenotype as the atnar2.1-1 plants when grown on a low nitrate concentration: the shoot biomass was reduced to 30% of the wild type (Supplemental Fig. S6). Under the same conditions, the nitrate HATS activity was decreased to 5 μmol 15NO3− h−1 g−1 root DW for both RolDNpNRT2.1X atnar2.1-1 and atnar2.1-1 plants, representing only 9% of the wild-type activity (Fig. 6). In the atnar2.1-1 genotype background, while NpNRT2.1 was strongly expressed (data not shown), the HATS activity was not restored. NpNRT2.1 cannot restore functional HATS by itself, and the AtNAR2.1 gene product is necessary for the system. Therefore, functional complementation of HATS in atnrt2.1-1 plants (Filleur et al., 2001) must be explained by the interaction between NpNRT2.1 and AtNAR2.1. The results using RolDNpNRT2.1X atnar2.1-1 plants also show that the impaired HATS of the atnar2.1-1 mutant does not result from the decreased expression of AtNRT2.1.
Figure 6.
Effect of the complementation with the RolDNpNRT2.1 construct on root 15NO3− influx of atnar2.1-1 mutant Arabidopsis seedlings. Wild-type (black bars) and mutant (atnar2.1-1, white bars; atnar2.1-1 RolDNpNRT2.1, waves) plants were grown on vertical agar plates on 10 mm NO3− for 7 d and transferred to 10 mm NO3− or 0.5 mm NO3− for a further 6 d (total of 13 d). HATS activity was measured as root 15NO3− influx after 5-min labeling with complete nutrient solution containing 0.2 mm 15NO3−. The values are means ± sd of four replicates (pooling three plants).
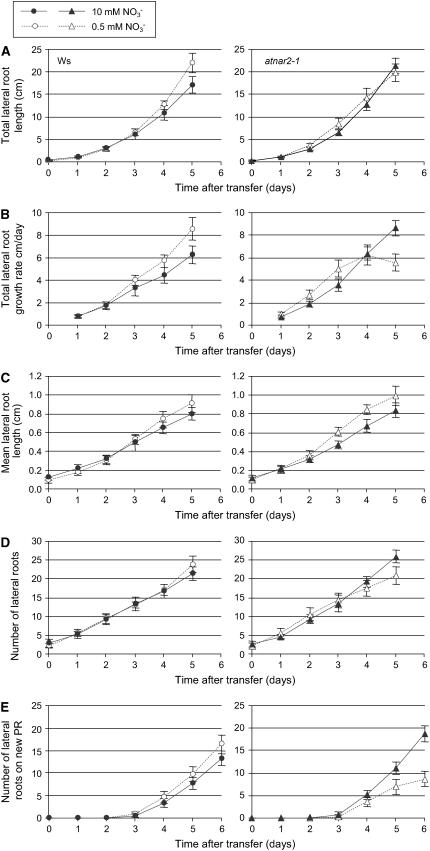
LR Development Reveals a Different Response on Transfer to Low Nitrate Concentration between atnar2.1-1 and the Wild Type
Changes in LR growth are a sensitive and easily measurable response of plants to N limitation (Zhang and Forde, 2000; Remans et al., 2006). To analyze the impact of AtNAR2.1 disruption on the root architecture, wild type and mutants were grown on vertical agar plates on a nitrate-rich medium (10 mm) for 6 d and then transferred to low nitrate (0.5 mm) medium until day 13 (Fig. 7). As previously described (Remans et al., 2006), the primary root growth of the wild type was not affected by the transfer to low nitrate medium (data not shown), but the total LR length was significantly increased 4 d after transfer when compared with plants remaining on 10 mm nitrate supply (Fig. 7A). The increase in the total LR length was due to both increases in the mean length of individual LRs from 3 d after transfer (Fig. 7C) and enhanced LR appearance 5 d after transfer (Fig. 7D). The stimulation of LR appearance was particularly significant on the portion of primary root that was newly developed after the transfer (Fig. 7E).
Figure 7.
Effect of 0.5 mm nitrate supply on LR growth in wild-type and atnar2.1-1 Arabidopsis seedlings. Wild-type (circles) and atnar2.1-1 mutant (triangles) plants were grown on vertical agar plates on 10 mm NO3− for 7 d and then transferred to either 0.5 mm (white symbols) or 10 mm (black symbols) NO3− at t = 0 for a further 6 d (13 d of total growth). Total LR length (A), total LR elongation rate (B), mean length of LRs (C), number of LRs (D), and number of LRs on newly developed primary roots (PR) after transfer (E) were determined by image analysis. The values are means ± sd of 12 seedlings.
When the atnar2.1-1 mutant was transferred to 0.5 mm, the total LR length was slightly increased after 3 d, earlier than for the wild type (Fig. 7A; Supplemental Fig. S7a), but this difference was not statistically significant and was not sustained. After 5 d on 0.5 mm, the total LR growth rate of the atnar2.1-1 mutant was significantly lower than that measured on 10 mm (Fig. 7B) and for wild-type plants on 0.5 mm (see Supplemental Fig. S7b). There was a significant increase in the mean LR length occurring 3 d after transfer of the roots, earlier than for wild type (Fig. 7C). Nonetheless, the mean length of LRs was not statistically different between atnar2.1-1 and wild type during the 5 d after transfer to 0.5 mm (Supplemental Fig. S7c). There was a significant decrease in the number of LRs initiated by the atnar2.1-1 mutant 5 d after transfer to 0.5 mm when compared with 10 mm, but not wild-type plants on 0.5 mm (Fig. 7D; Supplemental Fig. 7d). The decrease was particularly noticeable and statistically significant for the number of LRs initiated on the portion of the primary root newly developed after the transfer when compared to the mutant on 10 mm and wild type on 0.5 mm (Fig. 7E; Supplemental Fig. S7e).
In summary, the atnar2.1-1 mutant has a different phenotype from wild type in the LR response to transfer to low nitrate supply. Although the total LR length increase was not significantly different from the wild type, there were differences in the pattern of LR development. Initially, LR growth was faster than wild type, but by 5 d it was significantly slower. In addition, after 5 d on 0.5 mm nitrate supply, fewer new LR were initiated by the atnar2.1-1 mutant when compared with the wild type.
DISCUSSION
After the identification of the NAR2 homologs in Arabidopsis, one possibility was that AtNAR2.1 alone encoded a nitrate membrane transporter that was itself directly responsible for HATS activity. The gene has recently been renamed AtNRT3.1 (Okamoto et al., 2006). The data obtained from the heterologous expression systems using yeast (Fig. 1) and oocytes (Fig. 2) have shown that AtNRT2.1/AtNAR2.1 is a two-component high-affinity nitrate transport system and that the components interact at the protein level. We have also demonstrated two-component transport activity in planta, by comparing the phenotypes of T-DNA insertion mutants for both components, AtNRT2.1 (Filleur et al., 2001) and AtNAR2.1 (Okamoto et al., 2006). The detailed physiological characteristics of these two mutants (atnar2.1-1 and atnrt2.1-1) provide in planta evidence that when either of the two components are absent, similar but not identical phenotypes result.
Comparing the Physiology of the atnar2.1-1 and atnrt2.1-1 Mutants
In comparison with wild-type plants, there is an easily identifiable dwarf phenotype for both mutants supplied with a low nitrate concentration (0.2 or 0.5 mm) in both hydroponic and in vitro culture. In contrast with the growth of mutant plants supplied with 1 mm NH4NO3 for 4 weeks (see table I in Okamoto et al., 2006), when the atnar2.1-1 mutant is grown on a high nitrate concentration (>6 mm), the growth phenotype can be rescued (Supplemental Fig. S1). The fact that the atnar2.1-1 mutant has a more severely stunted growth phenotype than the atnrt2.1-1 mutant suggests that AtNAR2.1 has a greater role in HATS when compared with AtNRT2.1 (see Supplemental Fig. S1).
Wild-type plants maintain the same growth on both 6 mm and 0.2 mm nitrate concentrations, but tissue concentrations of N compounds are decreased at the lower nitrate supply (Supplemental Fig. S1). Although both mutants show decreased growth on 0.2 mm nitrate, the tissue concentrations of N compounds are not maintained at the levels measured in the wild type (Supplemental Fig. S1). The mutants show N deficiency symptoms when grown on low nitrate concentration. These symptoms include decreased growth and lower tissue N compounds (Supplemental Fig. S1) and are stronger for atnar2.1-1 than for atnrt2.1-1. The atnar2.1-1 mutant growing on low concentrations of nitrate displays a phenotype similar to long-term N starvation in wild-type plants. The mutant plants show very low shoot to root ratio and total N content (Supplemental Fig. S1) that are similar to wild-type plants starved for 10 d (Orsel et al., 2004a).
A direct comparison of HATS activity in the atnar2.1-1 and atnrt2.1-1 mutants has shown that the influx system is more deficient in the former type of plants (Supplemental Fig. S2). This result establishes that AtNAR2.1 is very important for HATS function and, as the atnrt2.1-1 mutant is deficient in both NRT2.1 and NRT2.2, it suggests that there may be other members of the NRT2 family that contribute to HATS and are interacting with AtNAR2.1 to give this activity. The data in Table I show that expression of AtNRT2.4 and AtNRT2.5 increased but AtNRT1.1 decreased in atnar2.1-1. One interpretation of these results is that the former two genes attempt to compensate for the loss of the two-component AtNAR2.1/AtNRT2.1 activity by increasing their expression. Alternatively, the increased expression of these two genes may be part of a general response of the plant to N deficiency (Orsel et al., 2004a, 2004b), and there is evidence that this is the situation. As N deficiency symptoms increase in severity from atnrt2.1-1 to atnar2.1-1 (Supplemental Fig. S2), the expression of AtNRT2.4 and AtNRT2.5 becomes stronger, too, providing indirect evidence supporting this idea. There is also down-regulation of AtNRT2.1 and AtNRT1.1; again, this effect is characteristic of longer-term N starvation. One surprising feature is the lack of response by AtNRT1.1, as the protein has both high- and low-affinity uptake modes (Liu and Tsay, 2003) and should be a good candidate to compensate for the missing HATS activity. When nitrate was resupplied to N-starved plants, in both wild type and AtNAR2.1 knockout mutants, expression of AtNRT1.1 was strongly inducible, but HATS activity was still severely decreased (Okamoto et al., 2006).
As both proteins are required for functional HATS activity, there should be closely coordinated coexpression of both AtNRT2.1/AtNAR2.1 components. One difference between the two mutants concerns the regulation of AtNAR2.1 expression in the atnrt2.1-1 background in hydroponics and in vitro culture. In hydroponics, there is no difference in the expression level of AtNAR2.1 between wild-type and anrt2.1-1 plants at both high and low nitrate concentrations (see Table I). But for in vitro experiments, the expression level of AtNAR2.1 in the atnrt2.1-1 background is lower than for the wild type and is increased by transfer to a low nitrate concentration (Fig. 5B). This result demonstrates the importance of hydroponic experiments, where the nutrient concentration is maintained at a low concentration but actually not limiting for growth (Supplemental Fig. S1, a–f). These data suggest that AtNAR2.1 is not essential for AtNRT2.1 expression, but the level of expression is modulated by the presence of the gene, probably as a consequence of the N status of the plants.
Similarly, AtNRT2.1 expression in the atnar2.1-1 background is repressed compared to the wild type when plants are grown in vitro on low nitrate supply (Fig. 5C). In hydroponic experiments, the expression level of AtNRT2.1 is also lower in the atnar2.1-1 background than for the wild type on low nitrate concentration (Table I). Again, this result may be a consequence of the N-limited status of the plants. The AtNRT2.1 gene is still inducible by nitrate (Fig. 4C) and N starvation (Fig. 3C), but the induction is lower than that measured for the wild type in both cases. During N starvation, the wild type can access and utilize any trace nitrate remaining in the solution using the highly efficient HATS system (Cerezo et al., 2001) and thereby keep AtNRT2.1 induced and maintain expression. This system will not function in the atnar2.1-1 mutant and 24 h is enough to begin the de-induction process giving a decrease in expression due to the lack of nitrate taken up by the roots.
The data from Figure 3 and Table I show that AtNAR2.1 expression is similar in wild-type and atnrt2.1-1 plants growing under differing supplies of nitrate. However, the expression of AtNRT2.1 was significantly increased in wild-type plants treated with 0.2 mm nitrate but decreased in atnar2.1-1 plants at the same nitrate concentration (Table I). These results could be explained by AtNAR2.1 having a direct role in the regulation of transcription.
Feedback by reduced N compounds is often invoked to explain the down-regulation of HATS activity (Vidmar et al., 2000). In vitro experiments on Gln show the same amount of decrease in HATS activity in both mutants (Fig. 4A). If cHATS is defined as the component of HATS that is functional when the plants have not been exposed to nitrate, then both genes are involved in cHATS. These results agree with those of Okamoto et al. (2006), who used nitrate starvation and resupply to show a role for AtNAR2.1 in both iHATS and cHATS. Taken together, these results argue against the idea that there are distinct nitrate transporter genes each responsible for cHATS and iHATS (Okamoto et al., 2003). One part of the cHATS activity seems to be due to the residual expression level of both AtNRT2.1 and AtNAR2.1 in the absence of nitrate (Fig. 3, B and C). In support of this explanation, when wild-type plants were grown with ammonium succinate as the only N source, another noninduced condition for both genes, AtNRT2.1 and AtNAR2.1 expression levels were not zero but were around 7% of EF1α (Wang et al., 2004). In the wild-type plants, both genes are induced by nitrate, but HATS is not increased. This result may be due to the high N status of the plants as Gln has been shown to be a repressor of HATS. This repression of HATS appears to occur by posttranscriptional regulation and has been tested by the addition of Gln to a nitrate medium or using inhibitors (Vidmar et al., 2000; Glass et al., 2002; Fan et al., 2006).
Starving plants of N and subsequent resupply of nitrate is a standard treatment defining nitrate-inducible enes (Crawford and Glass, 1998; Forde, 2000), but the conditions that are used for these experiments are often quite different and this may have important consequences for gene expression and nitrate uptake. As N status of plants is a key factor for determining HATS activity, we have tested the response of the two mutants to differing N treatments. The two mutants differ in the response of HATS to low nitrate concentration or sudden N deprivation (Supplemental Figs. S2 and S3). The residual HATS activity (cHATS) detected 24 h after transfer to N-free supply is lower than the activity detected for the atnar2.1-1 plants remaining on 6 mm (Fig. 3A). Again, this result suggests a greater role for AtNAR2.1 than AtNRT2.1 in HATS. The HATS activity was lower in atnar2.1-1 than in atnrt2.1-1 plants growing on 0.2 mm when compared with that value obtained on 6 mm (see Supplemental Fig. S2). The down-regulation of HATS in the atnar2.1-1 mutant was also observed in vitro when plants were transferred from high to low nitrate supply (see Figs. 5 and 6). This result suggests there are some nitrate uptake proteins whose activity is independent of the presence of AtNAR2 on high nitrate but dependant on it on low nitrate concentrations. AtNAR2 could be an essential element of regulation to maintain the function of these independent systems under low nitrate conditions. This result demonstrates an interesting difference between the two types of mutants. To extend the phenotype characterization, we have compared the LR architecture response to nitrate of wild type and the atnar2.1-1 and atnrt2.1-1 mutants.
A Role for AtNAR2.1 in LR Responses to N Limitation
A role for AtNRT2.1 in the root response to nitrate has recently been demonstrated, and, for both the wild type and the atnrt2.1-1 mutant, the total LR length was increased by transfer to a low nitrate concentration (see figure 7 in Remans et al., 2006). The response of the atnar2.1-1 mutant when compared with the wild type is different, as the increase in total LR length is not so strong (Fig. 7A). However, when the components of total LR length are examined in more detail, the mean length and number of LRs on the newly developed primary root are significantly different. This result may fit with the conclusion that the atnar2.1-1 plant is more rapidly put under an N deficiency stress than the wild type due to a deficient HATS activity. The increased growth of the LR system in response to transfer onto a low nitrate medium is not enough and cannot compensate for the lack of HATS induction in the atnar2.1-1 mutant. Like the atnrt2.1-1 mutant, this response of the root system can be attributed to the lower nitrate uptake rate in atnar2.1-1 plants than in the wild type, creating a stronger N deprivation status. After 4 d at a low nitrate concentration, the atnar2.1-1 mutant cannot sustain an increased growth rate (Fig. 7B; Supplemental Fig. S7b), while no limitation has been reported for the atnrt2.1-1 mutant. This is probably due to a shortage of N occurring earlier than for atnrt2.1-1 due to the greater limitation of the HATS system. The atnar2.1-1 mutant displays the same inhibition of LR initiation on the newly developed primary root. This inhibition has been attributed to the AtNRT2.1 protein itself rather than to decreased nitrate uptake (Remans et al., 2006). The results for LR development in the atnar2.1-1 mutant could fit with this hypothesis because expression of AtNRT2.1 is repressed under these conditions (see Fig. 7C).
One surprising result from the LR growth assays is the finding that the atnar2.1-1 mutant has a phenotype on 10 mm nitrate. The atnar2.1-1 mutant has significantly enhanced LR growth rate when compared with wild type growing on 10 mm nitrate (Fig. 7B). This phenotype is seen 4 to 5 d after transfer to 10 mm nitrate media, and we cannot explain this result except to suggest that AtNAR2.1 might have other functions that have yet to be identified.
Specificity of Two-Component Partners
In this article, we show that root-specific expression of NpNRT2.1 could not complement the atnar2.1-1 mutant (Fig. 6), showing that, specifically, AtNAR2.1 is essential for HATS. Previously published work reported that NpNRT2.1 can complement the atnrt2.1-1 mutant (Filleur et al., 2001), a result that therefore now suggests NpNRT2.1 can interact and form a functional complex with AtNAR2.1. This result in planta is important because all previous work using heterologous expression in the oocyte system had suggested that the protein partners are highly specific (Zhou et al., 2000; Tong et al., 2005). This NpNRT2.1 protein has more similarity to AtNRT2.1 (87%) than with AtNRT2.3 (81%) and CrNRT2.1 (57%; see the NRT2 family tree, figure 3 in Orsel et al., 2002b; EMBOSS-Align, program Needle, EBlossum 62). Taken together, these data show that interspecies complementation between the NAR2/NRT2 protein components can occur to give functional HATS and that NpNRT2.1 is an ortholog of AtNRT2.1. Despite the weak interaction detected between AtNRT2.3 and AtNAR2.1 by mbSUS, we could not detect any nitrate uptake activity in the oocyte system. This finding suggests that either a functional complex cannot be formed in oocytes or AtNRT2.3 is a paralog of AtNRT2.1 with a different function and perhaps may not even be a nitrate transporter.
The mbSUS results show that the two-component interaction between NRT2 and NAR2 occurs between proteins when the N terminus of AtNAR2.1 protein is outside the plasma membrane, while the C-terminal-fused transcription factor must be inside the cell to activate the reporter genes (Obrdlik et al., 2004). The AtNAR2.1 protein is classified in the databases as an endomembrane system (GO:0012505) and an N-terminal “secretory pathway signal” is predicted from the sequence (Aramemnon database). Taken together, this information suggests that NAR2s may be involved in targeting the NRT2s to the plasma membrane like the recent report for a phosphate transporter traffic facilitator (Gonzalez et al., 2005). However, NAR2s share no sequence similarities to SEC12 proteins (Gonzalez et al., 2005).
In contrast to the large multigenic family of nitrate transporters (seven AtNRT2s, 52 AtNRT1s), there are only two AtNAR2 genes, and only one of these seems to be functional in nitrate transport. In many other plant species, except in barley where at least three genes have been found, only one NAR2 gene has been identified (Tong et al., 2005). The finding that the interaction between NRT2/NAR2 components may not be as specific as suggested by oocyte experiments is important.
In conclusion, we have demonstrated that the AtNAR2.1 gene is essential for high-affinity nitrate uptake by Arabidopsis roots, confirming the result of another simultaneous study (Okamoto et al., 2006). In addition, we have shown that AtNRT2.1 and AtNAR2.1 proteins are two essential partners of a two-component HATS system. But in many conditions, the atnar2.1-1 mutant displays a more severe phenotype than the atnrt2.1-1 mutant, suggesting that AtNAR2.1 is interacting with other unidentified proteins. Double-hybrid screening systems adapted to membrane proteins might allow their identification (Obrdlik et al., 2004). The effect of the disruption of AtNAR2.1 is drastic for the plants, even more than a disruption of AtNRT2.1 and AtNRT2.2 genes. Moreover, in a complex soil environment with presumably mixed N sources, the mutant displays a strong growth phenotype (data not shown). Therefore, this gene may be an interesting target for approaches to modify N-use efficiency of plants by genetic manipulation or to take advantage of natural variation within cultivars.
MATERIALS AND METHODS
Split-Ubiquitin Analysis
The interaction of the NRT2 transporters and the AtNAR2 proteins was tested using mbSUS (Obrdlik et al., 2004). Full-length cDNAs of NRT2 transporters were cloned in frame with the Nub subdomain of ubiquitin in pNXgate and pXNgate plasmids (TRP1, AmpR), and introduced in the yeast strain THY.AP4 (MATa ura3 leu2 lexA∷lacZ∷trp1 lexA∷HIS3 lexA∷ADE2). The AtNAR2.1 cDNA was cloned in frame with the Cub subdomain of ubiquitin in pMetYCgate (LEU2, AmpR) and introduced in the yeast strain THY.AP5 (MATα URA3 leu2 trp1 his3 loxP∷ade2). Diploid cells were created by mating, and interaction between the Nub and Cub fusions was tested by analysis of reporters (His and Ade auxotrophy, and β-galactosidase activity).
For growth assays, diploid cells were grown in liquid synthetic dextrose (SD) −LT (Leu, Trp) minimal media containing 50 μm Met (Minimal SD Base and DO Supplement; BD Bioscience) at 28°C overnight. Culture concentrations were adjusted at OD600 = 1 and diluted 10, 100, and 1,000 times. Five microliters of each dilution was plated on solid agar plate SD −LT or SD −LTHA (Leu, Trp, His, Ade) minimal media containing 50 μm Met and incubated at 28°C for 3 d.
Quantitative β-galactosidase assays were performed by harvesting 4-mL cultures at OD600 = 0.6 and resuspending cells in 400 μL of Z buffer. Cells were lysed by three cycles of incubation at 37°C and subsequent freezing in liquid nitrogen, and then agitated with glass beads using the TissueLyser (Qiagen). Cell debris was pelleted at 14,000 rpm for 2 min and the supernatant was used for assays. In microtiter plates, different volumes of protein aliquots were incubated with 40 μL o-nitrophenylglucoside (4 mg mL−1). The kinetics of change in absorbance was measured at 405 nm (accumulation of o-nitrophenyl β-d-galactopyranoside). Total protein content of the aliquots was determined according to Bradford using the Bio-Rad reagent as described previously (Orsel et al., 2004b). The values are means ± sd of three replicates; results from a representative experiment are shown.
Xenopus Oocyte Expression System
Full-length cDNA were cloned by PCR in pGEM-T Easy vector (Promega), fully sequenced, and digested with NotI. cDNA fragments were blunted using the Klenow fragment, and subcloned in the EcoRV site of the pT7TS expression vector containing the 5′-untranslated region (UTR) and 3′-UTR of the Xenopus β-globin gene (Cleaver et al., 1996). For in vitro synthesis of mRNA, pT7TS clones were linearized by digestion with BamHI. Capped full-length mRNAs were synthesized using a T7 RNA transcription kit (mMESSAGE mMACHINE; Ambion).
Xenopus oocytes were prepared as described previously (Zhou et al., 1998) and stored in ND96 solution (96 mm NaCl, 2 mm KCl, 1.80 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, adjusted at pH 7.4 with NaOH). Healthy oocytes at stage V or VI were injected with 50 nL of water (nuclease free) or different mRNAs at 1 μg μL−1 each. After 3-d incubation at 18°C, five to 10 oocytes were incubated in 3 mL of ND96 solution enriched with 0.5 mm Na15NO3 (atom% 15N: 98%) during 16 h at 18°C. For experiments done at acidic external pH, HEPES buffer (Sigma) was replaced by MES buffer (Sigma) at the same concentration. The oocytes were then thoroughly washed four times with ice-cooled 0.5 mm NaNO3 ND96 solution and dried at 60°C. The 15N to 14N ratio of single dried oocyte was measured using an isotope ratio mass spectrometer (model Integra CN; PDZ Europa). The delta 15N was calculated as described previously (Tong et al., 2005). The values are means ± sd of five replicates; results from a representative experiment are shown.
Plant Material
Seed stocks of Arabidopsis (Arabidopsis thaliana L. Heynh) from the Wasselewskija (Ws) ecotype were used for all experiments. The mutant atnrt2.1-1 (formerly atnrt2a, renamed according to the nomenclature proposed by Little et al. [2005]) was isolated in the T-DNA insertion collection of INRA Versailles (Filleur et al., 2001). This mutant provides a functional knockout of the genes AtNRT2.1 (At1g080900) and AtNRT2.2 (At1g08100) due to a T-DNA insertion. The mutant atnar2.1-1 was isolated in the same T-DNA insertion collection using the FLAGdb search tool for the At5g50200 gene. The isolated line was backcrossed with wild-type (Ws) plants and crossed with the mutant atnrt2.1-1 to obtain the double mutant atnrt2.1-1 atnar2.1-1. The atnar2.1-1RolDNpNRT2.1 plants have been obtained by crossing the atnar2.1-1 mutant with wild-type (Ws) plants transformed with the RolDNpNRT2.1 construct described by Filleur et al. (2001). Primer sets for the T-DNA (Tail A: AAA TTG CCT TTT CTT ATC GA) and At5g50200 (Forward: CCC ACA CAA GAT CAT AGC C; Reverse: CAA AAG GAA TTG GTA AAC AAG) have been used for genotyping the plants and sequencing the T-DNA flanking region.
Using the AtNAR2.1 gene sequence, two FST (568D06 and 598H04) were identified in the FLAGdb database. The sequences correspond, respectively, to the left-border and right-border flanking sequences of the T-DNA insertion. The T-DNA insertion led to a 73-bp deletion including the last 68 bp of the coding sequence and 5 bp of the 3′-UTR. We note that a 71-bp deletion size is described for the previously described Atnrt3.1-2 mutant (Okamoto et al., 2006), but we believe it is the same mutation. The deletion was in the sequence encoding the last 22 amino acids (over 210), containing half of the predicted transmembrane domain, Aramemnon database (Schwacke et al., 2003). The mutant, named atnar2.1-1, was backcrossed to the wild type (Ws ecotype) and the physiological impact of AtNAR2.1 disruption was studied in comparison to the disruption of AtNRT2.1 and AtNRT2.2 genes in the atnrt2.1-1 mutant (Filleur et al., 2001; Orsel et al., 2004b).
Plant Growth Conditions
For the atnar2.1-1 growth phenotype analysis (Supplemental Figs. S1 and S2; Table I), plants were grown under hydroponic culture conditions on 6 mm NO3− or 0.2 mm NO3− medium in Sanyo growth chamber with 8-h-light/16-h-dark cycle at 21°C/17°C, respectively, 70% relative humidity, and 150 μmol m−2 s−1 irradiation as described (Orsel et al., 2004b). Note that the light conditions are slightly different from those used previously, so the tissue amino acid concentrations of wild type (see Supplemental Fig. S1) are therefore slightly higher than reported previously (Orsel et al., 2004b), presumably as more carbon is available. For each genotype and nutritional condition, three to five plants were pooled and 15NO3− influx was assayed as described below. Immediately after, roots were separated from the shoots and frozen in liquid nitrogen. Samples were homogenized to a powder to allow 15N and metabolite analysis as well as total RNA extractions. Three independent experiments were performed and results from a representative one are shown.
For the 24-h N-starvation experiments (Fig. 3), plants were grown on 6 mm NO3− for 41 d in the same conditions as described above and transferred into N-free culture medium or new 6 mm NO3− medium for 24 h. The N-free solution contains 2.5 mm K2SO4, 2.2 mm CaCl2 instead of 1 mm K2SO4, 0.7 mm CaCl2. Pools of one to three plants were harvested 24 h after transfer, and 15NO3− influx was assayed as described below. Samples were homogenized to a powder to allow 15N analysis and total RNA extractions. Two independent experiments were performed and results from a representative one are shown.
For the in vitro experiments (Figs. 4–7), the basic medium contained 0.5 mm CaSO4, 0.5 mm MgCl2, 1 mm H2PO4, 2.5 mm MES (Sigma), 72 μm NaFeEDTA, 10 μm MnSO4, 24 μm H3BO3, 3 μm ZnSO4, 0.9 μm CuSO4, 0.04 μm (NH4)6Mo7O24, adjusted to pH 5.7 with KOH. This basic medium was supplemented with 10 mm KNO3, 0.5 mm KNO3, or 5 mm l-Gln (Sigma) for each type of experiment. The K+ concentration was adjusted to 10 mm by addition of K2SO4 in media with 0.5 mm KNO3 or 5 mm l-Gln. These nitrate concentrations differ from those used in the hydroponics and were chosen because they enable a direct comparison of Arabidopsis phenotypes in previously published work (e.g. Remans et al., 2006). The Arabidopsis seeds were sterilized, sown on 10- × 10-cm plate on 50 mL of solid medium (1% Difco BACTO AGAR; BD Biosciences), and stored for 3 d at 4°C in the dark. Plates were incubated vertically at 22°C, with 16-h/8-h light/dark cycle and a light intensity of 140 μmol m−2 s−1. A 1-cm band of solid media was removed at the top of each plate to facilitate shoot growth, and six to eight plantlets were transferred onto fresh growth media as indicated. Plants were harvested at the indicated stage; 15NO3− influx was assayed as described below on pool of three plants, and total RNA was extracted from a pool of six plants from the same experiment. Two independent experiments were performed and results from a representative one are shown.
Root 15NO3− Influx and Metabolite Analysis
Influx of 15NO3− was assayed as described previously (Orsel et al., 2004b). The plants were transferred first to 0.1 mm CaSO4 for 1 min, then to complete nutrient solution containing either 0.2 mm or 6 mm 15NO3− (atom% 15N: 99%) for 5 min, and finally to 0.1 mm CaSO4 for 1 min (300 mL for plants grown in hydroponics, and 20 mL for plants grown in vitro). After homogenization, an aliquot of the frozen powder was dried overnight at 80°C and analyzed using an isotope ratio mass spectrometer (model Integra CN; PDZ Europa). Influx of 15NO3− was calculated from the total N and 15N content of the roots (1 mg DW). An aliquot of the corresponding shoot powder was also analyzed to determine total N content. The values are means ± sd of four to five replicates.
An aliquot of the powder was weighed (50 mg FW) and extracted in a four-step ethanol water procedure for determination of the nitrate content (μmol g−1 FW) and a Rosen evaluation of free-amino acid concentration (μmol g−1 FW) as already described (Orsel et al., 2004b). The values are means ± se of three replicates.
Total RNA Extraction and Gene Expression Analysis
Total root RNA was extracted with the Gen Elute Mammalian Total RNA kit from Sigma-Aldrich, modified by adding a DNase step, which was performed with the Qiagen RNase-free DNase kit. Gene expression was determined by quantitative real-time PCR as described (Orsel et al., 2004b): first strands were synthesized using M-MLV reverse transcriptase (Gibco-BRL) and oligo(dT)15 primers (Promega). The PCR was performed on a LightCycler instrument (Roche) with the LightCycler-FastStart DNA Master SYBR Green I kit for PCR (Roche) according to the manufacturer's protocol. For the 24-h N starvation and the in vitro experiments, total root RNA was extracted with the same Gen Elute Mammalian Total RNA kit but followed by a treatment with the Deoxyribonuclease I kit (amplification grade) from Sigma. Quantitative reverse transcription-PCR analysis was performed on ABI PRISM 7700 using the SuperScript III Platinum Two-Step qRT-PCR kit with SYBR Green from Invitrogen for the first-strand synthesis and the quantitative PCR according to manufacturer's protocol. Specific primer sets were used for each tested gene: AtNRT2.1 (F: AGT CGC TTG CAC GTT ACC TG; R: ACC CTC TGA CTT GGC GTT CTC), AtNRT2.4 (F: CAG TTC CTT CCG ACT CAT CA; R: GCA ACA CCA GCA TTT CCG AC), AtNRT2.5 (F: CTC TGC TTT CGC CGT TCT CTT GTT C; R: CGC TGC TAT AAT CCC TGC TGT CTG G), AtNRT1.1 (F: AGA CCG AAC CAA AAG AAC GA; R: CCA CGA TAA CCG CAG CAA CC), and AtNAR2.1 (F: CCA GAA GAT CCT CTT TGC TTC ACT; R: CCC AAT CGA GCT TAG CGT CCA). Expression levels of tested genes were expressed as a percentage of the constitutive AtEF1A4α (At5g60390) gene expression level (F: CTG GAG GTT TTG AGG CTG GTA T; R: CCA AGG GTG AAA GCA AGA AGA).
Root Growth Analysis
Arabidopsis seedlings were cultured in vitro (see above method) and root growth was analyzed as described previously (Remans et al., 2006). The root systems in vertical agar plates were scanned daily at 300 dpi (ScanJet 6300C; Hewlett-Packard). Root growth parameters were determined after analysis of scanned images using the ImageJ analysis software (http://rsb.info.nih.gov/ij/). Statistical comparisons of means between treatments or genotypes were performed using the pooled Student's t test using Sigmaplot software (Systat Software UK).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Physiological comparison of growth and N status of atnrt2.1-1, atnar2.1-1, and wild-type Arabidopsis grown at two different nitrate concentrations.
Supplemental Figure S2. Root 15NO3− influx in wild-type, atnrt2.1-1, and atnar2.1-1 Arabidopsis grown at two different nitrate concentrations.
Supplemental Figure S3 (Supplement to Fig. 3). Root 15NO3− influx in wild type and atnar2.1-1 after 24-h N starvation.
Supplemental Figure S4 (Supplement to Fig. 4). Shoot FW in wild type, atnrt2.1-1, and atnar2.1-1 after 24-h NO3− resupply (induction).
Supplemental Figure S5 (Supplement to Fig. 5). Comparison of shoot FW of atnrt2.1-1, atnar2.1-1, and the double mutants.
Supplemental Figure S6 (Supplement to Fig. 6). The effect of the complementation with the RolDNpNRT2.1 construct on shoot growth of atnar2.1-1 mutant Arabidopsis seedlings.
Supplemental Figure S7 (Supplement to Fig. 7). The effect of 0.5 mm nitrate supply on LR growth in wild-type and atnar2.1-1 Arabidopsis plants.
Supplementary Material
Acknowledgments
We thank W. Schulze for supplying the mbSUS system; L. Sivilotti for the Xenopus oocytes; and P. Nacry, T. Remans, and D. Wells for their advice on culture methods.
This work was supported by the European Union (grants no. BIO4CT972231, Research Training Network “Plant use of nitrate” HPRN–CT–2002–00247, and Gabi-Génoplante joint project AF2001/092). Rothamsted Research is grant aided by the Biotechnology and Biological Sciences Research Council of the United Kingdom.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Anthony J. Miller (tony.miller@bbsrc.ac.uk).
The online version of this article contains Web-only data.
References
- Cerezo M, Tillard P, Filleur S, Munos S, Daniel-Vedele F, Gojon A (2001) Major alterations of the regulation of root NO3− uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver OB, Patterson KD, Krieg PA (1996) Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development 122: 3549–3556 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3: 389–395 [Google Scholar]
- Daniel-Vedele F, Filleur S, Caboche M (1998) Nitrate transport: a key step in nitrate assimilation. Curr Opin Plant Biol 1: 235–239 [DOI] [PubMed] [Google Scholar]
- Fan X, Gordon-Weeks R, Shen Q, Miller AJ (2006) Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J Exp Bot 57: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Filleur S, Daniel-Vedele F (1999) Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207: 461–469 [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F (2001) An arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett 489: 220–224 [DOI] [PubMed] [Google Scholar]
- Forde BG (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Glass ADM, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, Okamoto M, Rawat S, Siddiqi MY, Unkles SE, et al (2002) The regulation of nitrate and ammonium transport systems in plants. J Exp Bot 53: 855–864 [DOI] [PubMed] [Google Scholar]
- Glass ADM, Siddiqi MY (1995) Nitrogen absorption by plants roots. In HS Srivastava, RP Singh, eds, Nitrogen Nutrition in Higher Plants. Associated Publishing, New Delhi, India, pp 21–56
- Gonzalez E, Solano R, Rubio V, Leyva A, Paz-Ares J (2005) PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Wang RC, Chen MS, Crawford NM (2001) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13: 1761–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Young J, Crawford NM (2003) The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 15: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A (1999) Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509–519 [DOI] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE (2005) The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA 102: 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K-H, Tsay Y-F (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kumar A, Li W, Wang Y, Siddiqi MY, Crawford NM, Glass ADM (2006) High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol 140: 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Vidmar JJ, Glass AD (2003) Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol 44: 304–317 [DOI] [PubMed] [Google Scholar]
- Orsel M, Boivin K, Roussel H, Thibault C, Krapp A, Daniel-Vedele F, Meyer C (2004. a) Functional genomics of plant nitrogen metabolism. In D Leister, ed, Plant Functional Genomics. Haworth Press, Binghamton, NY, pp 431–450
- Orsel M, Eulenburg K, Krapp A, Daniel-Vedele F (2004. b) Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219: 714–721 [DOI] [PubMed] [Google Scholar]
- Orsel M, Filleur S, Fraisier V, Daniel-Vedele F (2002. a) Nitrate transport in plants: which gene and which control? J Exp Bot 53: 825–833 [DOI] [PubMed] [Google Scholar]
- Orsel M, Krapp A, Daniel-Vedele F (2002. b) Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol 129: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada A, Galvan A, Fernandez E (1994) Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J 5: 407–419 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A (2006) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Zhou J-J, Li Z, Miller AJ (2005) A two-component high-affinity nitrate uptake system in barley. Plant J 41: 442–450 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Unkles SE, Hawker KL, Grieve C, Campbell EI, Montague P, Kinghorn JR (1991) crnA encodes a nitrate transporter in Aspergillus nidulans. Proc Natl Acad Sci USA 88: 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkles SE, Zhou D, Siddiqi MY, Kinghorn JR, Glass ADM (2001) Apparent genetic redundancy facilitates ecological plasticity for nitrate transport. EMBO J 20: 6246–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidmar JJ, Zhuo D, Siddiqi MY, Schjoerring JK, Touraine B, Glass AD (2000) Regulation of high-affinity nitrate transporter genes and high-affinity nitrate influx by nitrogen pools in roots of barley. Plant Physiol 123: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutierrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Lohmann J, Schmid M (2004) AtGenExpress: expression atlas of Arabidopsis development. The Arabidopsis Information Resource. http://www.arabidopsis.org (August 18, 2004)
- Zhang H, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51: 51–59 [PubMed] [Google Scholar]
- Zhou JJ, Fernandez E, Galvan A, Miller AJ (2000) A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett 466: 225–227 [DOI] [PubMed] [Google Scholar]
- Zhou JJ, Theodoulou FL, Muldin I, Ingemarsson B, Miller AJ (1998) Cloning and functional characterization of a Brassica napus transporter that is able to transport nitrate and histidine. J Biol Chem 273: 12017–12023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.