Abstract
The sucrose nonfermenting-1 protein kinase (SNF1)/AMP-activated protein kinase subfamily plays a central role in metabolic responses to nutritional and environmental stresses. In yeast (Saccharomyces cerevisiae) and mammals, the β- and γ-noncatalytic subunits are implicated in substrate specificity and subcellular localization, respectively, and regulation of the kinase activity. The atypical βγ-subunit has been previously described in maize (Zea mays), presenting at its N-terminal end a sequence related to the KIS (kinase interacting sequence) domain specific to the β-subunits (Lumbreras et al., 2001). The existence of two components, SNF1-related protein kinase (SnRK1) complexes containing the βγ-subunit and one SnRK1 kinase, had been proposed. In this work, we show that, despite its unusual features, the Arabidopsis (Arabidopsis thaliana) homolog AKINβγ clearly interacts with AKINβ-subunits in vitro and in vivo, suggesting its involvement in heterotrimeric complexes located in both cytoplasm and nucleus. Unexpectedly, a transcriptional analysis of AKINβγ gene expression highlighted the implication of alternative splicing mechanisms in the regulation of AKINβγ expression. A two-hybrid screen performed with AKINβγ as bait, together with in planta bimolecular fluorescence complementation experiments, suggests the existence of interactions in the cytosol between AKINβγ and two leucine-rich repeats related to pathogen resistance proteins. Interestingly, this interaction occurs through the truncated KIS domain that corresponds exactly to a GBD (glycogen-binding domain) recently described in mammals and yeast. A phylogenetic study suggests that AKINβγ-related proteins are restricted to the plant kingdom. Altogether, these data suggest the existence of plant-specific SnRK1 trimeric complexes putatively involved in a plant-specific function such as plant-pathogen interactions.
The Suc nonfermenting-1 protein kinase (SNF1) from yeast (Saccharomyces cerevisiae) and its mammalian homolog, the AMP-activated protein kinase (AMPK), are Ser/Thr kinases implicated in global metabolic regulation in response to cellular and environmental stresses (Hardie, 2004). Yeast SNF1 is implicated in cell adaptation to Glc deprivation (Carlson, 1999) by derepression of Glc-repressed genes in the absence of Glc. It is also involved in developmental processes (Honigberg and Lee, 1998) or cellular functions (Carlson, 1999). In mammals, AMPK is activated by an increase in the cellular AMP to ATP ratio. In response to nutrient starvation, exercise, or hypoxia, AMPK negatively regulates ATP-consuming pathways, including fatty acid and cholesterol synthesis, by phosphorylating metabolic key enzymes and promotes fatty acid β-oxidation to produce ATP (Hardie, 2004). Furthermore, AMPK is implicated in the modulation of Glc-regulated gene expression (Leclerc et al., 1998; Woods et al., 2000).
Some convergent results suggest the implication of SNF1-related protein kinase (SnRK1) in sugar signaling in plants (Bhalerao et al., 1999; Bradford et al., 2003; Halford et al., 2003; Lovas et al., 2003) and in the control of carbohydrate and starch metabolism (Purcell et al., 1998; Geigenberger, 2003; Laurie et al., 2003; Thelander et al., 2004). In vitro kinase assays suggest that SnRK1 might play a role in controlling metabolic pathways by phosphorylating key enzymes such as HMGCoA reductase, Suc phosphate synthase, and nitrate reductase (Sugden et al., 1999). More recently, the SNF1 kinase was also shown to participate in innate antiviral defenses (Hao et al., 2003) and in resource reallocation so that plants better tolerate herbivory (Schwachtje et al., 2006).
It is now well established that these kinases are associated with two types of noncatalytic proteins belonging to heterotrimeric kinase complexes composed of one catalytic subunit, one β-type subunit (SIP1/SIP2/GAL83, AMPKβ, and SnRKβ), and one γ-type protein (SNF4, AMPKγ, and SnRKγ; Halford et al., 2000).
The members of the SIP1/SIP2/GAL83 family might play an essential role in the specificity of recognition between the kinase complex and its targets (Vincent and Carlson, 1999) and also in the subcellular localization of the complex in yeast (Vincent et al., 2001; Warden et al., 2001; Hedbacker et al., 2004). The noncatalytic β-subunits from yeast (Celenza et al., 1989), mammals (Stapleton et al., 1994; Gao et al., 1996), and plants (Bouly et al., 1999; Lakatos et al., 1999; Bradford et al., 2003; Buitink et al., 2004) share a common structure composed of a variable N-terminal domain associated with the two highly conserved KIS (kinase interacting sequence) and ASC (association with SNF1 complex) domains, which mediate the interaction with α- and γ-subunits, respectively (Yang et al., 1992, 1994). Recently, a new domain implicated in glycogen binding and overlapping part of the KIS domain was characterized in AMPKβ1 (Hudson et al., 2003; Polekhina et al., 2003) and GAL83 (Wiatrowski et al., 2004), and thus named GBD (glycogen-binding domain). Interestingly, such a domain has also been identified in the protein phosphatase PTP-KIS (Fordham-Skelton et al., 2002). In Arabidopsis, AKINβ1 and β2 share the classical N-terminal/KIS/ASC structure (Bouly et al., 1999), while AKINβ3 presents only a truncated KIS domain, lacking the region related to the GBD, fused to the ASC domain (Gissot et al., 2004).
In yeast, SNF4 plays an essential role in the Glc regulation of SNF1 activity (Jiang and Carlson, 1996). Results obtained from yeast, mammals, Drosophila melanogaster, and plants (Gao et al., 1996; Stapleton et al., 1996; Bouly et al., 1999; Yoshida et al., 1999; Cheung et al., 2000) have shown that the SNF4/AMPKγ/SnRKγ family of proteins contains four in-tandem CBS (cystathionine β-synthase) motifs (Bateman, 1997). Bateman domains formed by tandems of CBS represent the AMP- and ATP-binding sites of the AMPK complex (Kemp, 2004; Scott et al., 2004). Interestingly, in mammals, several mutations localized within the CBS domains of the three AMPKγ-subunits result in the modification of the kinase activity and of its regulation by AMP (Cheung et al., 2000; Milan et al., 2000; Hamilton et al., 2001; Daniel and Carling, 2002; Scott et al., 2004; Burwinkel et al., 2005). Two unusual maize (Zea mays) SnRK1 γ-related proteins (ZmAKINβγ-1 and -2) have been characterized by Lumbreras et al. (2001). These proteins present in their N-terminal end a sequence related to the KIS domain of the β-subunits (Lumbreras et al., 2001). ZmAKINβγ-1 and -2 have been shown to complement the yeast snf4 mutant and to interact with Arabidopsis AKIN11 (AKINα2) kinase, but at present nothing is known about their partners or their function (Kleinow et al., 2000; Lumbreras et al., 2001). Due to the presence of a KIS domain in AKINβγ, the existence of two SnRK1 complexes containing only the AKINβγ-type subunit and one SnRK1 kinase has been proposed by Lumbreras et al. (2001). In Arabidopsis, AKINβγ is the ortholog of ZmAKINβγ-1 and -2 and corresponds to the initial annotation AtSNF4 (Kleinow et al., 2000).
In this article, we show that AKINβγ interacts with other members of the AKIN complex in the two-hybrid system, suggesting their involvement in heterotrimeric alternative complexes. Bimolecular fluorescence complementation (BiFC) experiments, used to confirm these interactions in planta, have also allowed us to get the first data, to our knowledge, concerning the subcellular localization of plant SnRK1 proteins. Unexpectedly, a transcriptional study of AKINβγ highlights the implication of alternative splicing mechanisms in the regulation of AKINβγ expression. Finally, a two-hybrid screen performed with AKINβγ as bait suggests the interaction of AKINβγ with Leu-rich repeat (LRR)-rich proteins related to pathogen resistance proteins through their truncated KIS domain. Altogether, these data suggest the existence of plant-specific SnRK1 trimeric complexes putatively involved in plant-specific functions such as plant-pathogen interactions.
RESULTS
AKINβγ Gene Is Differentially Spliced
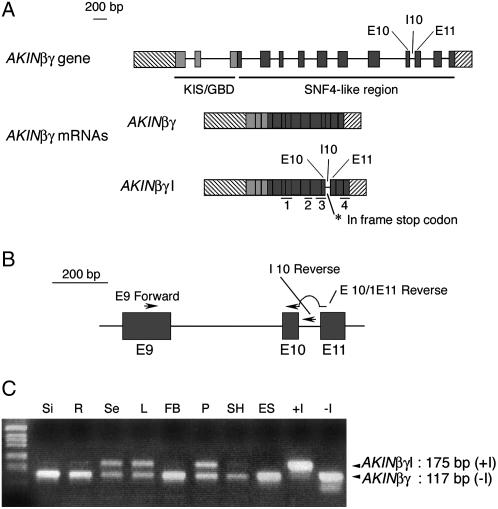
Mammalian AMPKγ1, γ2, γ3, and yeast SNF4 were used as probes to screen the National Center for Biotechnology Information (NCBI) databases (nr and dbest restricted to Arabidopsis). Some of the retrieved sequences correspond partially to the AtSNF4 cDNA initially published by Kleinow et al. (2000), but their 5′ end differs from the annotations of the corresponding bacterial artificial chromosome (BAC) and of AtSNF4 cDNA. Actually, 5′ RACE experiments revealed the existence of two other exons and of a long 5′ untranslated region (UTR) located upstream of the first predicted ATG of AtSNF4 related to a KIS domain. Such a structure has been previously described by Lumbreras et al. (2001) in maize (ZmAKINβγ-1 and ZmAKINβγ-2) and the Arabidopsis ortholog was named AKINβγ. Interestingly, the KIS domain of these subunits is truncated compared to the KIS domains of the β-subunits and corresponds exactly to the GBD domain recently described in AMPKβ1 (Hudson et al., 2003; Polekhina et al., 2003) and GAL83 (Wiatrowski et al., 2004). For this reason, we propose to name this domain KIS/GBD. The AKINβγ Arabidopsis gene contains 13 exons and 12 introns encoding a 2.3-kb long cDNA corresponding to a protein of 487 amino acids (53 kD; Fig. 1A). The length of the AKINβγ cDNA is in accordance with the 2.2-kb length found for ZmAKINβγ-1 and βγ-2 in maize (Lumbreras et al., 2001). Two kinds of PCR products were obtained and cloned after a 3′ RACE PCR, the largest including the unspliced intron 10. The two full-length cDNAs were named AKINβγ and AKINβγI (with and without intron 10, respectively). The protein sequence deduced from the AKINβγI cDNA is reduced to 394 amino acids (43.3 kD) due to the presence of a stop codon located 6 bp downstream of the beginning of intron 10. Therefore, one of the most conserved regions of the protein, mainly corresponding to the fourth CBS domain, is deleted in AKINβγI.
Figure 1.
Structure of the AKINβγ gene and analysis of its expression. A, Intron/exon structure of the AKINβγ gene and the corresponding mRNAs. The 5′ and 3′ UTRs are represented by hatched boxes, exons (E) by colored boxes, and introns (I) by horizontal bars. Exons corresponding to the KIS/GBD domain (light gray boxes) and SNF4 domain (dark gray boxes) are represented by different gray levels. The four CBS domains are positioned on AKINβγ and AKINβγI, the two mRNAs derived from alternate splicing. B, Position of the oligonucleotides on exons 9, 10, and 11 and intron 10. The oligonucleotide E9Forward, positioned on exon 9, is used either with the oligonucleotide I10Reverse on intron 10 to amplify 175 bp of the AKINβγI cDNA or with the oligonucleotide E10/E11Reverse on the exon 10/intron 10 junction to amplify 117 bp of the AKINβγ cDNA. C, Analysis of the presence of AKINβγ and AKINβγI mRNAs. PCR experiments were performed using DNA of several cDNA libraries as template. Si, Siliques; R, roots; Se, seeds; L, leaves; FB, floral buds; P, pollen; SH, hypocotyls; ES, etiolated seedlings; +I, pGEMT vector (Promega) containing the AKINβγI cDNA; −I, pGEMT vector (Promega) containing the AKINβγ cDNA.
AKINβγI and AKINβγ Are Expressed in the Same Organs
To determine if the presence of the two cDNAs corresponds to an artifact product obtained during the building of the shoot library or to an alternative splicing, an analysis of the expression of these two cDNAs has been performed. The small difference in size observed between the two cDNAs (100 bp) did not allow us to separate AKINβγ and AKINβγI transcripts by northern-blot experiments (data not shown). Therefore, PCR experiments were performed using two pairs of oligonucleotides designed to selectively amplify each of the two putative cDNAs. One oligonucleotide (E9Forward), common to both cDNAs, is located on the exon 9, while a second is located either on the exon10/intron10 junction (E10/E11Reverse) to amplify a fragment of 117 bp corresponding to AKINβγ cDNA (without intron 10), or in intron 10 (I10Reverse) to amplify a 175-bp fragment corresponding to AKINβγI cDNA (including intron 10; Fig. 1B). DNA from eight cDNA libraries was used as templates. The sizes of the two PCR products of 117 and 175 bp correspond to the fragments amplified using the two control cDNAs [pGEMT vectors containing either AKINβγ (−I) or AKINβγI (+I) cDNA; Fig. 1C]. AKINβγ mRNA is ubiquitously expressed. On the other hand, a high level of AKINβγI mRNA is observed when using the libraries prepared from seeds, leaves, and pollen, while it is almost undetectable in siliques, roots, hypocotyls, and etiolated seedlings. Nevertheless, whatever the organs or conditions tested, AKINβγI mRNA is always present. These data, also obtained by reverse transcription-PCR experiments (data not shown), confirm the existence of an alternative splicing event.
CBS4 Deletion in AKINβγI Prevents the Interaction with the Kinases and the Complementation of the Yeast snf4 Mutant
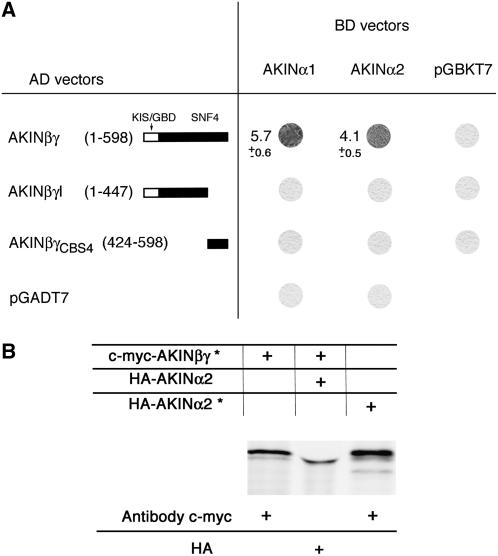
Two-hybrid experiments and in vitro coimmunoprecipitation assays were performed to study protein-protein interactions between AKINβγ/βγI and the AKINα1/α2 kinases (Fig. 2). The whole coding sequences of AKINβγ or AKINβγI were fused in-frame with the HA epitope and the GAL4 activation domain (AD) by cloning into the pGADT7 vector. The AKINα1 and α2 kinases were fused in-frame with the GAL4-binding domain (BD) and c-Myc epitope by cloning into the pGBKT7 vector. The two-hybrid experiments and in vitro binding assays show as expected that AKINβγ interacts with both kinases AKINα1 and α2, while no interaction was detected between AKINβγI and the kinases (Fig. 2). Therefore, the interaction between AKINβγ and the kinases appears to be lost when a region containing the fourth CBS domain (CBS4) is absent. Nevertheless, no interaction with the kinases could be detected using the CBS4 domain alone either in two-hybrid experiments or in in vitro binding assays. Thus, the domain absent in AKINβγI appears necessary but not sufficient for the interaction with the kinases.
Figure 2.
Interactions between AKINβγ/βγI and the AKINα1 and α2 kinases by two-hybrid experiments and in vitro binding assays. A, Qualitative β-galactosidase enzyme assays were performed between full-length AKINβγ/βγI or AKINβγ CBS4 domain and AKINα1 and α2 kinases. B, In vitro binding assays. Due to their similar molecular masses, AKINβγ and AKINα2 proteins were synthesized by in vitro transcription/translation system (Promega) without or with (*) 35S-Met labeling and incubated alone or by pairs, respectively. After coimmunoprecipitation with HA or c-Myc antibodies, proteins were separated by 10% SDS-PAGE and exposed to an x-ray film.
To test whether, despite the absence of interaction with the AKINα kinases, AKINβγI protein was still able to act as a γ-subunit, we performed yeast snf4 complementation experiments. While the full-length AKINβγ protein complements the yeast snf4 mutant on medium lacking Glc as carbon source, AKINβγI protein does not (data not shown).
The SNF4 Domain of AKINβγ Interacts with the AKINβ-Subunits to Form SnRK1 Heterotrimeric Complexes Located Both in the Cytoplasm and in the Nucleus
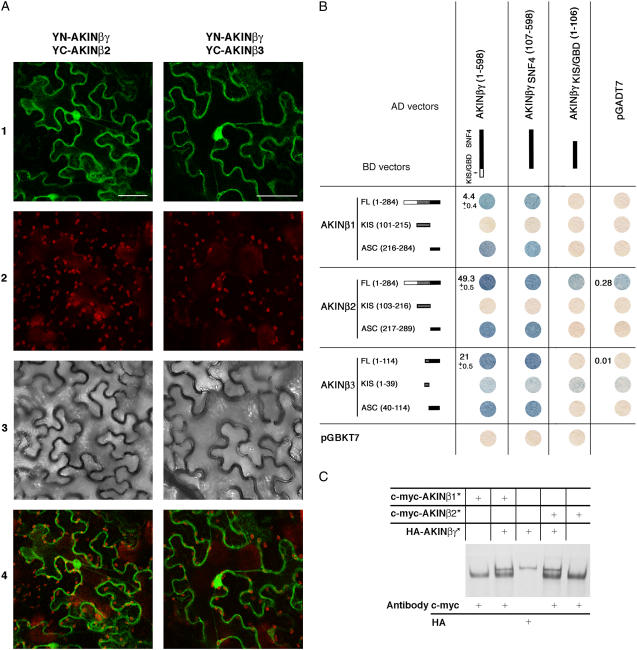
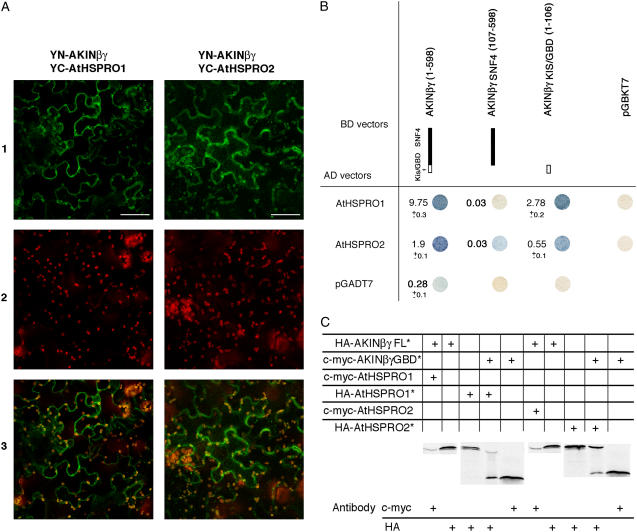
The KIS/GBD of AKINβγ has been previously shown to interact with the kinase without the requirement of a β-subunit, suggesting the existence of dimeric complexes composed by AKINβγ and one of the kinases in which this KIS/GBD domain could play the role of a β-subunit (Lumbreras et al., 2001). To test if the existence of the KIS/GBD in a γ-type subunit prevents the formation of trimeric complexes, two-hybrid, in vitro coimmunoprecipitation and in planta BiFC experiments were performed using AKINβγ, AKINβγI, and each domain of AKINβ-subunits (Fig. 3). Interactions have been detected between AKINβγ and each of the three β-subunits (AKINβ1, β2, and β3; Fig. 3B, lanes FL) in vitro. The β-galactosidase activity measured for the interaction involving AKINβγ was 2.3- and 11.2-fold higher with AKINβ2 than with AKINβ3 and β1, respectively. However, no interaction was detected with these proteins when using AKINβγI in two-hybrid experiments (data not shown). These results were confirmed by BiFC experiments performed between AKINβγ fused to the N-terminal domain of the yellow fluorescent protein (YFP; YN-AKINβγ) and the AKINβ-subunits fused with the C-terminal domain of the YFP (YC-AKINβ; Fig. 3A). All combinations led to a YFP signal showing a clear in planta interaction between YN-AKINβγ- and YC-AKINβ-subunits. Interestingly, the YFP signal is located both in the cytoplasm and the nucleus, as presented in Figure 3A, for a classical β-subunit (YN-AKINβγ/YC-AKINβ2 interaction) and for the plant-specific one (YN-AKINβγ/YC-AKINβ3 interaction). No YFP signal was detected when YN-AKINβγ and YC-AKINβ1/2/3 were independently infiltrated (data not shown). Moreover, as a negative control, the PAS1 protein (Faure et al., 1998) that does not interact with SnRK1 subunits was fused to YC and YN and cotransformed in Nicotiana benthamiana leaves with YN-AKINβγ and YC-AKINβ1/2/3, respectively. No YFP fluorescence was detected in any case (data not shown), confirming the relevance of the previously shown interactions.
Figure 3.
Protein-protein interactions between AKINβγ and the AKINβ-subunits. A, Subcellular localization of reconstructed YFP complexes determined in leaf epidermis of N. benthamiana. Left, YN-AKINβγ/YC-AKINβ2 interaction; and right, YN-AKINβγ/ YC-AKINβ3 interaction. Section 1, YFP fluorescence (green); section 2, chlorophyll autofluorescence (red); section 3, bright field; section 4, YFP/chlorophyll autofluorescence overlay. Scale bars correspond to 50 μm. B, Two-hybrid experiments between AKINβγ and AKINβ subdomains. Qualitative and quantitative β-galactosidase enzyme assays have been represented for the full-length proteins (FL). Activities were measured at least twice from six independent colonies grown with 2% Glc. C, AKINβγ in vitro protein binding assays. 35S-Met labeled proteins (*) were synthesized by in vitro transcription/translation system (Promega) and incubated alone or by pairs. After coimmunoprecipitation with c-Myc or HA antibodies, proteins were separated by 10% SDS-PAGE and exposed to an x-ray film.
To precisely determine the domains involved in these interactions, the sequences encoding SNF4, KIS/GBD, and CBS4 domains of AKINβγ and the N-terminal, KIS, and ASC domains of AKINβ1/2/3 were subcloned into pGADT7 and pGBKT7 vectors and used in similar experiments (Fig. 3B). The ASC domains of all AKINβ-subunits were sufficient to direct the interaction with the SNF4 domain of AKINβγ, while the KIS/GBD of AKINβγ does not interact with any AKINβ-subunits. No interaction was detected using other domains of AKINβγ or AKINβ1/2/3 (data not shown).
AKINβγ-Subunit Is Restricted to the Plant Kingdom
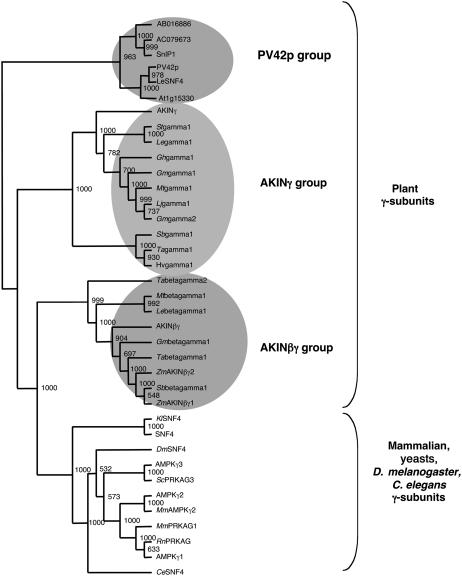
At present, the AKINβγ-type subunits have been described only in maize, Arabidopsis, and Medicago truncatula (Lumbreras et al., 2001; Buitink et al., 2004). To look for AKINβγ-related sequences in other organisms, the NCBI databases were screened using the AKINβγ sequence as a probe. The retrieved sequences from plants, mammals, yeast, D. melanogaster, and Caenorhabditis elegans were aligned using ClustalX (Thompson et al., 1997). The corresponding γ-related sequences were restricted to the SNF4 region and used to draw a phylogenetic tree (Fig. 4). The plant γ-subunits appear separated from other organisms and distributed into three subgroups corresponding to three γ-subunits previously published: PV42p from Phaseolus vulgaris (Abe et al., 1995) and AKINγ (Bouly et al., 1999) and AKINβγ from Arabidopsis. The group including AKINβγ appears closer to the nonplant group containing the mammalian, yeast, D. melanogaster, and C. elegans sequences than to the other plant γ-proteins. Interestingly, the proteins corresponding to the nine sequences grouped with AKINβγ present the same AKINβγ atypical structure with a KIS/GBD fused to an SNF4 domain. Moreover, proteins presenting similar structures have never been found in other groups, neither in the plant PV42p and AKINγ subgroups nor in nonplant organisms.
Figure 4.
Phylogenetic analysis of the γ-subunits from various organisms. Alignment of the SNF4 domain of γ-related proteins was created by ClustalX (Thompson et al., 1997) and bootstrapped 1,000 times. The tree was visualized by Treeview (Page, 1996). Bootstrap values >500 are shown to the right of each branch point. Numbers appearing in the plant PV42p group correspond to the accession numbers of the Arabidopsis proteins (AB016886, AC079673) or the AGI number (At1g15330). Sequences shown are human AMPKγ1, γ2, and γ3 (accession nos. U42412, AJ249976, and NM_017431), S. scrofa (ScPRKAG3, AF214520), R. norvegicus (RnAMPKγ, U42413), M. musculus (MmAMPKγ1, NM_016781; and MmAMPKγ2, BC015283), yeasts S. cerevisiae (SNF4, M30470) and K. lactis (KlSNF4, AJ277480), maize (ZmAKINβγ1, AF276085; and ZmAKINβγ2, AF276086), and Lycopersicon esculentum (LeSNF4, AF143742).
The large number of sequences analyzed in this work show that this type of protein is present in most plant organisms and emphasizes the hypothesis proposed by Lumbreras et al. (2001) that AKINβγ-type subunits could be specific to the plant kingdom.
AKINβγ Interacts with Two Hs1pro-1 Orthologs Implicated in Pathogen Resistance
To get information about the function of the AKINβγ-type subunits in plants, we decided to search for partners of this protein by performing a two-hybrid screen of a cDNA library from Arabidopsis 3-week-old rosettes using the full-length AKINβγ cDNA as bait. Thirty-two clones presenting a β-galactosidase activity higher than three units corresponding to the basal β-galactosidase activity level of BD-AKINβγ were further analyzed and sequenced. In this article, we present the characterization of 22 of these clones.
NCBI databases (nr and dbest) were screened with BLASTN and TBLASTX algorithms (Altschul et al., 1990) to find sequence similarities with the preys. Nine clones corresponded to two already-known members of the AKIN complex: AKINβ1 (one clone) and AKINβ2 (eight clones; Bouly et al., 1999). The clone corresponding to AKINβ1 is truncated to one-half of the ASC domain previously described by Bouly et al. (1999; position +236 after the first Met), thus lacking the Nterm, KIS domain, and part of the ASC domain.
The 13 other clones encode two Arabidopsis unknown proteins that present strong similarities with the Hs1pro-1 protein encoded by a nematode resistance gene isolated in sugar beet (Beta vulgaris; Cai et al., 1997). One clone, named AtHSPRO1, is assigned to BAC AL163832 on chromosome III (At3g55840). Analysis of the corresponding genomic sequence and of 13 overlapping expressed sequence tags (ESTs) found in the NCBI database suggests that this clone is full length. The 12 other clones found during the two-hybrid screen are assigned to BAC AF002019 on chromosome II (At2g40000). This cDNA, named AtHSPRO2, appears full length according to the analysis of the BAC and of the 93 available ESTs. AtHSPRO1 and AtHSPRO2 are the two Arabidopsis orthologs of the sugar beet Hs1pro-1 gene and no other related sequences have been characterized in the Arabidopsis genome. The location of these two genes in duplicated regions of chromosomes II and III could explain their high percentage of similarity (http://www.tigr.org/tdb/e2k1/ath1/Arabidopsis_genome_duplication.shtml). The two Arabidopsis predicted proteins AtHSPRO1 and 2 are 75% identical and 86% similar. Both of them share 59% identity and 78% similarity with the sugar beet Hs1pro-1 protein (Fig. 5). Moreover, three sequences from Glycine max, Pisum sativum, and Hordeum vulgare were available in the database. The alignment of the Hs1pro-1-related proteins found in Arabidopsis, G. max (AAG44839), and P. sativum (AAF67003) highlights an N-terminal extension compared to the Hs1pro-1 protein from sugar beet (Fig. 5). The two Arabidopsis proteins present imperfect LRR but no nucleotide-binding site (NBS) or kinase domain previously described for Hs1pro-1 (Cai et al., 1997; Fig. 5). The existence of a putative transmembrane domain in Hs1pro-1 has also been reported by Cai et al. (1997). Nevertheless, the corresponding regions in the two Arabidopsis proteins present several differences, and the Munich Information Center for Protein Sequences (MIPS) Arabidopsis database does not predict the presence of any transmembrane domain in these proteins.
Figure 5.
Alignment of the sugar beet Hs1pro-1 protein and the plant Hs1pro-1-related proteins available in the database. Alignments were realized using the ClustalX program (Altschul et al., 1990) and the Hs1pro-1 protein from sugar beet (U79733), the Hs1pro-1-like proteins from Glycine max (AAG44839) and Pisum sativum (AAF67003), and from the two Arabidopsis AtHSPRO1 and AtHSPRO2 proteins. Identical amino acids are visualized with an asterisk (*) and similar ones with a colon (:). The imperfect LRR repeats (red amino acids and regions delimited by red lines) and the transmembrane domain (blue) defined in the Hs1pro-1 protein are positioned.
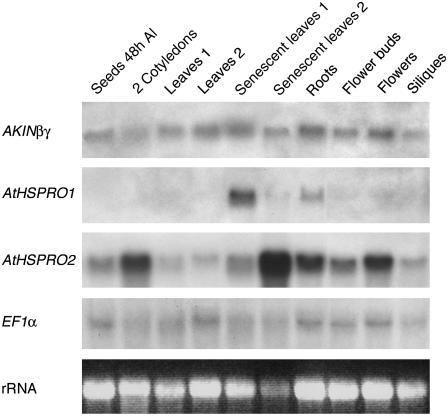
Northern-blot experiments were performed to analyze the expression patterns of AKINβγ and of the two isolated preys and to determine if they were expressed in the same organs or conditions (Fig. 6). AKINβγ gene is expressed constitutively in all the organs tested, while AtHSPRO1 and AtHSPRO2 appear to be differentially expressed. AtHSPRO2 mRNA is ubiquitous, high in cotyledons, roots, and flower organs, and increases dramatically during senescence (senescent leaves 1 and 2). At the contrary, the AtHSPRO1 messenger remains undetectable or at a very low level except in roots and in senescent leaves. These data are in accordance with the number of ESTs reported for these two proteins in the MIPS Arabidopsis database.
Figure 6.
Expression patterns of AKINβγ and AtHSPRO1/2 by northern-blot experiments. Seeds 48 h AI, Germinated seedlings 48 h after imbibition. Leaves 1 and 2, Leaves from 3- and 5-week-old rosettes. Senescent leaves 1, Leaves from mature rosettes before flowering. Senescent leaves 2, Yellow leaves from mature rosettes. Roots, Four-week-old roots grown under aeroponic conditions. Flowers, Open flowers. Siliques, One-centimeter green siliques. rRNA, EtBr visualization of ribosomic 28S. EF1α, Elongation Factor 1α. Hybridizations were performed at least twice on two independent membranes using the full-length cDNAs of AKINβγ and AtHSPRO1/2 as probes.
AKINβγ/AtHSPRO Interaction Is Located in the Cytoplasm and Occurs via the KIS/GBD
To confirm the putative interactions between AtHSPRO1/2 and AKIN complexes and to precisely determine the domains of interaction with AKINβγ, two-hybrid experiments and in vitro binding assays were performed using the full-length sequences of the preys and the AKINβγ subdomains described above cloned in pGADT7 and pGBKT7 vectors (Fig. 7). No interaction could be detected between these two proteins and AKINα, AKINβ, or AKINβγI either in two-hybrid experiments or in in vitro binding assays (data not shown). Concerning their interaction with AKINβγ, two-hybrid (Fig. 7B) and coimmunoprecipitation experiments (Fig. 7C) were confirmed by BiFC experiments performed between YN-AKINβγ and YC-HSPRO1 or YC-HSPRO2. Indeed, YFP signals were detected in the cytoplasm for both YN-AKINβγ/YC-HSPRO1 and YN-AKINβγ/YC-HSPRO2, confirming the in planta interaction between AKINβγ and HSPRO1/2 (Fig. 7A). Moreover, two-hybrid and in vitro binding assays show that the AtHSPRO1 and AtHSPRO2 proteins interact preferentially with the KIS/GBD domain of AKINβγ, while only a weak interaction remains with the SNF4 domain alone (Fig. 7, B and C).
Figure 7.
Protein-protein interaction between AtHSPRO1/2 and AKINβγ/βγKIS. A, Subcellular localization of reconstructed YFP complexes determined in leaf epidermis of N. benthamiana. Left, YN-AKINβγ/YC-AtHSPRO1 interaction; and right, YN-AKINβγ/YC-AtHSPRO2 interaction. Section 1, YFP fluorescence (green); section 2, chlorophyll autofluorescence (red); section 3, YFP/chlorophyll autofluorescence overlay. Scale bars correspond to 50 μm. B, Two-hybrid experiments between AKINβγ subdomains and AtHSPRO1/AtHSPRO2. Qualitative and quantitative β-galactosidase enzyme assays have been represented. Activities were measured at least twice from six independent colonies grown with 2% Glc. C, In vitro protein binding assays between AtHS1PRO1/AtHS1PRO2 and AKINβγ/βγKIS. AKINβγ proteins were synthesized by in vitro transcription/translation system (Promega) with 35S-Met labeled (*) and incubated alone or by pairs. AtHSPRO1 and AtHSPRO2 were synthesized without 35S-Met when used with the full-length AKINβγ due to their similar molecular masses. After coimmunoprecipitation with c-Myc or HA antibodies, proteins were separated by 10% SDS-PAGE and exposed to an x-ray film.
DISCUSSION
The isolation and characterization of ZmAKINβγ-1 and -2, two maize proteins interacting with the Arabidopsis AKINα2-subunit, corresponding to a γ-subunit fused in the N terminal to a truncated KIS domain, have been previously published by Lumbreras et al. (2001). Our work presents several breakthroughs concerning its Arabidopsis ortholog AKINβγ at the level of gene regulation, structure of the complex containing this unusual protein, function, and subcellular localization.
We have shown that the atypical KIS extension of AKINβγ is related to and corresponds exactly to a domain named GBD characterized in the mammalian AMPKβ1 (Hudson et al., 2003; Polekhina et al., 2003, 2005) and in the yeast GAL83 (Wiatrowski et al., 2004). To easily distinguish it from the classical KIS domain present in β-subunits, this domain has been named KIS/GBD in this article, even if it is unlikely to bind glycogen in plants. The interaction of this fragment with the AKINα2 kinase had previously led to the hypothesis of the existence of dimeric complexes (Lumbreras et al., 2001). Interestingly, our data show that AKINβγ interacts strongly also with the three Arabidopsis β-subunits (AKINβ1, β2, and the plant-specific AKINβ3 missing the KIS/GBD). This interaction occurs between the AKINβγ SNF4 region and the ASC domains of the AKINβ-subunits, while no interaction could be detected either with the KIS/GBD or the N-terminal domains. These data are in accordance with the binding of SNF4 to SIP1, SIP2, and GAL83 previously described in yeast (Jiang and Carlson, 1997) and of Arabidopsis AKINγ with AKINβ1/β2 (Bouly et al., 1999). Furthermore, the existence of trimeric complexes including AKINβγ is strengthened by the isolation of several AKINβ1/2 clones during the course of the two-hybrid screen using AKINβγ as bait.
BiFC experiments, an imaging technique recently adapted to the plant field (Walter et al., 2004; Bracha-Drori et al., 2004), allowed us to get the in planta confirmation of the interactions and also gave us the first data concerning the subcellular localization of SnRK1 subunits in plants. A clear localization of YN-AKINβγ/YC-AKINβ interactions was shown both in the cytoplasm and the nucleus. The nuclear localization of SnRK1 complex is particularly interesting because it is in accordance with the implication of the yeast SNF1 kinase at different levels of gene regulation as demonstrated in yeast (Hardie et al., 1998; Lo et al., 2001; Schuller, 2003).
Taken together, these data do not exclude the existence of dimeric complexes composed by AKINβγ and one of the two kinases proposed by Lumbreras et al. (2001), but strongly suggest that unusual trimeric complexes never described in the literature can also be formed by the association of one AKINα-, one AKINβ-, and one AKINβγ-subunit and located both in the cytoplasm and the nucleus.
The PCR-based cDNA isolation of two AKINβγ cDNAs, differing by the presence of the unspliced intron 10 in AKINβγI, highlighted the existence of alternative splicing events. Very few examples of alternative splicing have been reported yet in plants (Kazan, 2003), and in these cases the splicing is often regulated in a tissue-, environmental-, or organelle-specific manner (Lazar and Goodman, 2000; Kobayashi et al., 2001; Xu and Johnson, 2001; Shi et al., 2002). In our case, both mRNA seem to coexist in all the conditions tested but vary by their related levels. The existence of intron 10 alternative splicing produces a C-terminal end truncated putative protein of 394 amino acids that lacks mainly the fourth CBS domain (CBS4). CBS4 is the most conserved CBS domain found between plants, mammals, and yeast γ-subunits (Bradford et al., 2003). Our data have shown that this domain seems necessary for the interaction with α- and β-subunits. Yeast complementation, two-hybrid, and CoIP experiments indicate that AKINβγI would not be implicated in the formation of an active SnRK1 kinase complex. Further experimental supports will be necessary to test the simultaneous presence of both proteins in the plant and analyze the function of such a truncated βγ-protein. Alternative splicing could be one way of regulating these kinase complexes because splice variants have also been reported in mammals (Hardie, 2004).
To search for orthologs of AKINβγ in other organisms, a phylogenetic analysis of the sequences encoding all types of γ-subunits was performed. Plant γ-type subunits appear strictly separated from the mammal, yeast, and C. elegans γ-subunits and can be divided into three groups containing PV42P, AKINγ, or AKINβγ, respectively. The AKINβγ group is exclusively composed by plant proteins presenting the AKINβγ characteristics (SNF4 region fused in N terminus with a KIS/GBD), while such atypical subunits do not exist in yeasts, mammals, C. elegans, or D. melanogaster. Despite its unusual features, this group is the closest to the mammals and yeast. This observation is supported by the functional ability of three members of this group (ZmAKINβγ-1 and -2, AKINβγ) to complement the yeast snf4 mutant. LeSNF4 is the only protein belonging to one of the two other groups reported to complement the snf4 mutant (Bradford et al., 2003).
What could be the function of this plant-specific subunit with its unusual extension? Interestingly, when searching for AKINβγ orthologs in other organisms, more than 20 ESTs similar to AKINβγ were found in a sorghum (Sorghum bicolor) cDNA library prepared from mRNA extracted from a plant attacked by pathogens, suggesting that AKINβγ expression is regulated by plant-pathogen interaction.
In this work, a two-hybrid screen performed using AKINβγ as a bait led to the isolation of two types of cDNAs, AtHSPRO1 and AtHSPRO2, that encode proteins similar to the sugar beet Hs1pro-1 (Cai et al., 1997). The Hs1pro-1 protein has previously been shown to confer the resistance to the nematode Heterodora schachtii Schmidt on the basis of gene-for-gene relationship in sugar beet (Cai et al., 1997). Moreover, its mRNA expression is induced during nematode infection in sugar beet and Arabidopsis (Thurau et al., 2003). The absence of any NBS suggests that this protein does not belong to the NBS-LRR class of plant resistance genes but could be a member of another class of R genes (Jung et al., 1998). At present, nothing to our knowledge has been reported on the two Arabidopsis orthologs. An alignment between the sugar beet Hs1pro-1 and the predicted orthologs from several plants shows that the sugar beet Hs1pro-1 is smaller than its orthologs with fewer putative LRR repeats (Fig. 5). This difference in size can be explained either by the result of a divergence of these proteins during the evolution or by a wrong annotation of the sugar beet cDNA. Actually, the latter is more likely because northern-blot experiments performed by Cai et al. (1997) revealed a 1.6-kb mRNA that is close to the size of our two Arabidopsis full-length clones.
The expression patterns of the two AtHSPRO genes are very different and only slightly overlapping. AtHSPRO2 appears more expressed than AtHSPRO1 in most stages throughout Arabidopsis development. AtHSPRO1 and 2 mRNAs accumulate in the roots as previously shown for Hs1pro-1 in sugar beet (Cai et al., 1997). Interestingly, the mRNA level is dramatically increased during the two stages of senescence tested, with a higher level of AtHSPRO1 in the early senescent stage (leaf 3) and of AtHSPRO2 in the late senescent stage (senescent leaves) in the absence of pathogens. A similar observation had been reported previously for NIT2 and AtOSM34, two defense-related genes induced during leaf senescence in pathogen-free plants (Quirino et al., 1999). The induction of the two AtHSPRO genes putatively involved in plant defense response could be a component of the leaf senescence program.
Furthermore, an analysis of the microarray experiments available on the The Arabidopsis Information Resource site (www.arabidopsis.org) shows that the AtHSPRO2 mRNA is up-regulated by a factor comprised between 6 and 10 after inoculation by the bacteria Xanthomonas campestris pv campestris. Altogether, these data suggest that these two Arabidopsis Hs1pro-1 orthologs could be implicated in a more general resistance process and not only in response to a nematode attack. The interaction between the AKINβγ-subunit and these two Hs1pro-1-related proteins suggests that the SnRK1 complexes are implicated in plant-pathogen interactions. These results are in accordance with the data published by Hao et al. (2003). The authors have shown that the enhanced susceptibility of the transgenic plants expressing the geminivirus AL2 and L2 proteins is due to the interaction between these proteins and AKINα2, their interaction resulting in the inactivation of this kinase (Hao et al., 2003).
We have shown that the AtHSPRO1 and AtHSPRO2 proteins interact mainly with the AKINβγ KIS/GBD but not with the KIS subdomains of AKINβ1, β2, and β3 (data not shown), highlighting a specificity of interaction between these proteins. Recent data present evidence that the carbohydrate BD present in the dual-specificity protein phosphatase PTP-KIS (At3g52180) can bind starch (Kerk et al., 2006). This phosphatase, closely related to laforin, a mammalian glucan-binding protein phosphatase required for the metabolism of glycogen, binds to starch granules during the day and dissociates at night (Sokolov et al., 2006). Mutants of this dual-specificity protein phosphatase present a dramatic increase in their level of starch (Niittylä et al., 2006; Sokolov et al., 2006). Interestingly, the identified key conserved residues for this binding (W19, K39) are conserved in AKINβγ KIS/GBD. Because we have demonstrated here that this domain can mediate protein-protein interactions, more work will be necessary to understand if such a domain can mediate both protein-protein and carbohydrate-protein interactions.
Taken together, in vitro and in vivo results led us to propose that at least three different types of SnRK1 complexes could exist in plants: (1) dimeric complexes formed by AKINβγ and one of the two kinases as proposed by Lumbreras et al., (2001); (2) trimeric complexes composed by one AKINα-subunit, AKINβ1/β2-subunit, and AKINγ; and (3) other trimeric complexes composed by one AKINα, one AKINβ, and AKINβγ. The isolation of AtHSPRO1/2 proteins reinforces the latter hypothesis. In this case, the SNF4 domain of AKINβγ would interact with the AKINβ-subunits and with the kinase, leaving its KIS/GBD free for the interaction with AtHSPRO1/2. BiFC experiments show that this interaction is located exclusively in the cytoplasm, suggesting that the interaction with AtHSPRO proteins could retain the targeted complex in this compartment.
Altogether, the data presented in this article suggest that the SnRK1 heterotrimeric complex containing AKINβγ could be implicated in plant pathogen resistance through the interaction in the cytoplasm of the KIS/GBD domain of AKINβγ with AtHSPRO1/2 proteins.
MATERIALS AND METHODS
Plant Material
Wild-type Arabidopsis (Arabidopsis thaliana), ecotype Columbia, were grown in a culture room and were maintained in soil with a 15-h-light and 9-h-dark regime (long day). Relative humidity was 65% and temperature was 20°C/17°C during the light/dark cycle. Germinated seedlings (used for RNA extraction) were grown in the dark on a Whatman Number 1 paper imbibed by water, relative humidity was 65%, and temperature was 20°C.
Genomic Analysis of AKINβγ
The yeast (Saccharomyces cerevisiae) SNF4 (accession no. M30470) and mammalian AMPKγ1, γ2, and γ3 (U42412, AJ249976, and NM_017431) were used to screen the NCBI database (nr and dbest restricted to Arabidopsis) with the TBLASTN algorithm (Altschul et al., 1990). Three ESTs (AI999237, AV540740, and AV552460) were selected. The corresponding gene was assigned to the BAC clone AC000106 of chromosome I. NetPlantGene (Hebsgaard et al., 1996) and SIM4 (Florea et al., 1998) programs were used, respectively, to determine the intron/exon structure of the gene and align the different ESTs with the genomic sequence. Amplification of the 5′ and 3′ UTRs was realized by PCR experiments performed on an Arabidopsis shoot cDNA library as previously described by Bouly et al. (1999).
PCR Experiments
To determine the expression of the two AKINβγ mRNA containing (or not) the 10th intron, PCR experiments were performed on DNA of several cDNA libraries using three specific oligonucleotides. The downstream oligonucleotide (E9Forward) 5′-ACGCCTCTTTGGGTTCTGC-3′, localized in exon 9, was used with 5′-TATCATCAGAACACAAGGACG-3′ (I10Reverse) localized into the 10th intron for the amplification of a 175-bp fragment of the cDNA containing the 10th intron or with the oligonucleotide (E10/E11Reverse) 5′-GAGCAGTTCTATCACTTCGAGA-3′ localized on the exon10/intron10 junction in order to amplify the corresponding region of the AKINβγ cDNA spliced. cDNA libraries were previously described by Aubourg et al. (1999).
Northern-Blot Hybridization
Total Arabidopsis RNA were isolated in the middle of the light treatment from 48-h-old germinated seedlings (seeds 48 h after imbibition) and from green siliques according to Kay et al. (1987). Total RNA from 7-d-old germinated seedlings with two cotyledons (2 Cotyledons), leaves from 3- and 5-week-old rosettes (Leaves 1 and 2), leaves from mature rosettes before flowering (Senescent Leaves 1), yellowish mature leaves during flowering (Senescent Leaves 2), stems, floral buds, flowers, and 4-week-old roots grown under aeroponic conditions (Roots) were isolated according to Lessard et al. (1997). Samples have been taken in the middle of the light regime. RNAs (20 μg) were electrophoresed, blotted, and hybridized as previously described by Bouly et al. (1999). The probes have been obtained by PCR amplification using specific primers and correspond to the full-length AKINβγ, AtHSPRO1, AtHSPRO2, and EF1α cDNAs.
Protein-Protein Interaction Analysis by the Yeast Two-Hybrid System and in Vitro Binding Assays
AKINβγ full-length cDNA was cloned downstream of the Gal4 AD or BD in pGBT9 and pGAD424 vectors (CLONTECH) with specific oligonucleotides modified by 5′EcoRI and 3′BamHI restriction sites (5′ATGTTTGGTTCTAC3′ and 5′TCAAAGACCGAGCAG3′) to produce BD-AKINβγ and AD-AKINβγ. Specific oligonucleotides (5′-ATGGTTCCTGCTGGT-3′ and 5′-TCAAAGACCGAGCAG-3′, 5′-ATGTTTGGTTCTAC-3′ and 5′-TATTGTATTCACTAC-3′, 5′-ATGTTTGGTTCTAC-3′ and 5′-TACCTTCGAGAGTATATGTC-3′, and 5′-AGTGATATAACTGCTCTGGC-3′ and 5′-TCAAAGACCGAGCAGGAATTG-3′) modified with 5′EcoRI and 3′BamHI restriction sites were used to amplify AKINβγ SNF4107-598 domain, AKINβγ KIS/GBD1-106 domain, AKINβγ1-403 (truncated in the 10th intron), and AKINβγ424-598 (after intron 10 including the last CBS domain), respectively, and cloned into pGBKT7 and pGADT7 vectors (CLONTECH) to produce BD-βγ107-598/AD-βγ107-598, BD-βγ1-106/AD-βγ1-106, BD-βγ1-403/AD-βγ1-403, and BD-βγ424-498/AD-βγ424-498, respectively. All these AKINβγ constructs were tested against each α- and β-member of the AKIN complex as described by Bouly et al. (1999). The BD and AD constructs were used to transform the yeast strain Y190, and protein interactions were assayed using LacZ-filter lift and o-nitrophenyl-β-d-galactopyranoside assays and by monitoring growth on SD medium without Leu, Trp, and His (SD-LTH) containing 25 mm 3-aminotriazole (3-AT).
The vectors described above were used to produce 35S-Met-labeled proteins fused with the c-Myc or the HA epitope. These fusion proteins were prepared by in vitro transcription/translation using a TNTT7 Quick Transcription/Translation Lysate system (Promega). A total of 5 μL of each marked protein (or peptide) was incubated together into 400 μL of immunoprecipitation buffer (25 mm MOPS, 50 mm NaCl, 10% glycerol, 6 mg/mL bovine serum albumin, 1 mm EDTA, 0.5% Tween 20, 0.02% NaN3) at 4°C for 1 h. After 2 h of incubation at 4°C with 1 μL of c-Myc monoclonal antibody or 10 μL of HA-Tag polyclonal antibody (CLONTECH), the mixture was incubated with 6 mg of protein A-Sepharose (Sigma P3391) at 4°C for 1 h. The Sepharose was then washed eight times with 1 mL of the immunoprecipitation buffer. Proteins were then detached from the protein A-Sepharose in 20 μL of Laemmli buffer at 85°C for 5 min and separated in SDS-PAGE (15% acrylamide). The gel was dried and exposed to an x-ray film overnight at room temperature.
Two-Hybrid Screen Using AKINβγ as Bait
The yeast strain HF7C (Feilotter et al., 1994) carrying BD-AKINβγ bait was transformed with 500 μg of DNA from a pGAD424 cDNA library (AD-prey) from Arabidopsis 3-week-old rosettes (CLONTECH). The cells were then grown on SD-LTH (Durfee et al., 1993). A total of 6,000 transformants were streaked on SD-LTH plates in the presence of 25 mm 3-AT. A total of 286 transformants were grown on nitrocellulose filters (Whatman No. 5) and placed on SD-LTH plates containing 10, 25, or 50 mm 3-AT to check their LacZ+ phenotype by β-galactosidase assays. Yeast DNA was extracted according to Kaiser and Auer (1993) and used to transform bacterial strain MC1066. Bacteria were then streaked on minimum medium without Leu to select only the library AD plasmid. Each AD-prey isolated was used to cotransform the yeast strain Y190 (Harper et al., 1993) with BD-AKINβγ. β-Galactosidase measurement (as described previously by Bouly et al. [1999]) and a monitoring of growth on SD-LTH containing 25 or 50 mm 3-AT allowed a quantification of protein interaction and the selection of clones on the basis of high LacZ+ phenotypes and their capacity to grow on SD-LTH with a high concentration of 3-AT. After selection, the selected clones were sequenced and database screening was used to characterize the putative partners.
Finally, each AD-prey was cloned in the opposite vector pGBKT7 (CLONTECH) to produce BD-prey. After transformation of the yeast strain Y190, the β-galactosidase activity was measured.
BiFC and Confocal Microscopy
AKINβ2, AKINβ3, AKINβγ, AtHSPRO1, and AtHSPRO2 full-length cDNAs were cloned in the GATEWAY-compatible vector pDONR207 (Invitrogen) prior to being transferred in the BiFC GATEWAY-modified vector developed by F. Parcy (CEA-Grenoble, France). AKINβγ was fused to the N-terminal part of the YFP (YN) to produce 35S∷YN-AKINβγ. AKINβ2, AKINβ3, AtHSPRO1, and AtHSPRO2 were fused to the C-terminal part of the YFP (YC) to produce 35S∷YC-AKINβ2, 35S∷YC-AKINβ3, 35S∷YC-AtHSPRO1, and 35S∷YC-AtHSPRO2. Nicotiana benthamiana plants were grown in the greenhouse under 13 h light, 25°C day temperature, and 17°C night temperature for all the agroinfiltration experiments. Leaves were infiltrated (Rathjen et al., 1999) with an exponential phase culture Agrobacterium tumefaciens strain C58C1 resuspended to an OD600 of 0.5 with the infiltration medium (10 mm MES, pH 5.6, 10 mm MgCl2, 200 μm acetosyringone). For the coinfiltration, equal volumes of both cultures were mixed before agroinfiltration. Observations were performed 48 h after infiltration. Confocal microscopy was performed on an inverted Leica TCS-SP2-AOBS spectral confocal laser scanning microscope. Samples were excited with a 514-nm argon laser (50%) with an emission band of 510 to 550 nm for YFP detection and 640 to 700 nm for chlorophyll autofluorescence. Measurements of fluorescence emission spectra were carried out with an excitation at 488 nm (25%), and emission was detected between 500 and 600 nm at 5-nm intervals.
Eukaryotic SNF4/AMPKγ/SnRKγ-Subunit Analysis
The AKINγ sequence from Arabidopsis (CAB64720), Phaseolus vulgaris PV42p (AAA91175), yeast SNF4 (accession no. M30470), human AMPKγ1, γ2, and γ3 (U42412, AJ249976, and NM_017431), and Drosophila melanogaster (DmSNF4:AF094764) were used to search for overlapping ESTs and genomic sequences from different plant or nonplant organisms in the NCBI database (dbest and nr) using the TBLASTN algorithm (Altschul et al., 1990).
Sequences related to the γ-subunits from Caenorhabditis elegans (CeSNF4:CAC35836) were retrieved from the genomic sequence analysis using HMM (Krogh, 1997) and annotations of the BAC. Published mammals AMPKγ (Sus scrofa, ScPRKAG3: AF214520; Rattus norvegicus, RnAMPKγ: U42413; Mus musculus, MmAMPKγ1; NM_016781; and MmAMPKγ2; BC015283) and yeast Kluyveromyces lactis KlSNF4 (AJ277480) sequences were used. For each organism, overlapping sequences were used to create contigs and then translated. Full-length sequences were aligned using ClustalX (Thompson et al., 1997) and phylogenetic trees were generated, bootstrapped 1,000 times, and visualized by Treeview (Page, 1996). “Exclusions for positions with gaps” and “corrections for multiple sequences” were both set to “off.” The tree was outgrouped with PV42p and SnIP1 groups. Sequences related to SnIP1 correspond to a new group of SNF4-like protein isolated by Crawford et al. (2001). Names of the protein sequences deduced from EST contigs were given arbitrarily in bold, starting with the initials of the organism, “gamma” or “betagamma” for γ- or βγ-subunits, and one number when several sequences were found in the same organism. ESTs accession numbers used for each organism are listed in alphabetical order: Ghgamma1: AI730544, BF274631, AW187178; Gmgamma1: AW507983, AW348746, AI965571, AW350665, BG508480, BG041147, BI315858, BE474422, AI460404, AI495211, BF425409, BE661899, BE802388, AW100253, BE611594, AW755822; Gmgamma2: AW234285, BG726542, BF594973, BG790832, AW832024; Gmbetagamma1: BE329598, AW761384, BG045823, BI471330, AI938551, BF067362; Hvgamma1: BE413386, AL503884, BF620756; Legamma1: AW031797, AW930464, AW030495, AW931734, AW930111, AI896552, AW030567, AW221409, AW934655, BG132919, AW221379, AW616814, BE432801, AW034443, AW033795, AW033948, AW931052, AW931884, AI778239, BE435549, AI896954, BF052148, AW224492, BE441128, AW221367, AW221368, AW221366, BF051899, AI486513, AI485033, BE432466, AW222755, AW930016, BI209432, BI209432, BI207710, BG631518, BE433740; Lebetagamma1: AI896937, BE434764, AW933231, BE434544, AW033267, BG12450; Ljgamma1: AV410347, AV410863, AV412990, AV423982, AV420137, AV426298, AV428789; Mtgamma1: AA660841, AL284884, AL375470, AL375471, AL384885, AL388323, AW560279, AW573932, AW684384, AW685042, AW685140, AW686841, AW689677, AW773651, BE319326, BE942814, BF003163, BF003164, BF646632, BF649373, BG447936, BG589026, BG647811, BI266830; Mtbetagamma1: BG452241, AW774494; Stgamma1: BG590178, BE342824, BE341103, BG600892, BE342642, BG098543, BF890379; Sbgamma1: AW28645, BE363968, BE356406, AW564195, BE356340, BF421349, BE595667, BE563959, BF421265, BG465396, BG463490, BG464559, BG464134, BE594283, BG463409, BE363898, BE5945416; Sbbetagamma1: BG560098, BG241716, BF585763, AW744961, BI140637, BG412843, BG050314, BF481119, AW285543, AW285557, AW565509, AW565523, AW676933, AW745014, BE353014, BE598031, BE598304, BE598571, BE598914, BF421885, BF585679, BG050385, BG560098; Tagamma1: BE403487, BE426463, BG314395; Tabetagamma1: BG907542, BE489373, BI479562, BE415403, BE498573, BG605114, BE492292, BE445860, BG907543, BF293342; and Tabetagamma2: BE416528, BE417856, BE416027, BE417134, BE416979, BE416980.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ132632 (AKINβγ), DQ132633 (AKINβγI), DQ132634 (AtHSPRO1), and DQ132634 (AtHSPRO2).
Acknowledgments
We thank M. Schmidt (University of Pittsburgh) for the yeast mutants, M. Carlson (Columbia University) for the SNF1 and SNF4 two-hybrid constructs, N. Glab (University of Paris Sud, Orsay, France) for the pNSG2 vector, and F. Parcy (Commissariat à l'Energie Atomique, Grenoble, France) for the unpublished BiFC gateway-compatible vectors. We thank V. Geffroy for helpful discussions. We are grateful to I. Gy for the sequencing and her help with the phylogenetic analysis, J-P. Bares and G. Santé for growing the plants, and R. Boyer for the photographs.
This work was supported by the Ministère de l'Education Nationale et de la Recherche, France (to L.G., C.P., J.-P.B., and M.J.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Martine Thomas (martine.thomas@u-psud.fr).
References
- Abe H, Kamiya Y, Sakurai A (1995) cDNA clone encoding yeast SNF4-like protein from Phaseolus vulgaris. Plant Physiol 110: 336 [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Kreis M, Lecharny A (1999) The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res 27: 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A (1997) The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci 22: 12–13 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouly JP, Gissot L, Lessard P, Kreis M, Thomas M (1999) Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINalpha1, an SNF1-like protein kinase. Plant J 18: 541–550 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Downie AB, Gee OH, Alvarado V, Yang H, Dahal P (2003) Abscisic acid and gibberellin differentially regulate expression of genes of the SNF1-related kinase complex in tomato seeds. Plant Physiol 132: 1560–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitink J, Thomas M, Gissot L, Leprince O (2004) Starvation, osmotic stress and dessication tolerance lead to expression of different genes of the regulatory beta and gamma subunits of the SnRK1 complex in germinating seeds of Medicago truncatula. Plant Cell Environ 27: 55–67 [Google Scholar]
- Burwinkel B, Scott JW, Bührer C, van Landeghem FKH, Cox GF, Wilson CJ, Hardie DG, Kilimann MW (2005) Fatal congenital heart glycogenosis caused by a recurrent activating R531Q mutation in the γ2-subunit of AMP-activated protein kinase (PRKAG2), not by phosphorylase kinase deficiency. Am J Hum Genet 76: 1034–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Kleine M, Kifle S, Harloff HJ, Sandal NN, Marcker KA, Klein-Lankhorst RM, Salentijn EM, Lange W, Stiekema WJ, et al (1997) Positional cloning of a gene for nematode resistance in sugar beet. Science 275: 832–834 [DOI] [PubMed] [Google Scholar]
- Carlson M (1999) Glucose repression in yeast. Curr Opin Microbiol 2: 202–207 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Eng FJ, Carlson M (1989) Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol Cell Biol 9: 5045–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung PC, Salt IP, Davies SP, Hardie DG, Carling D (2000) Characterization of AMP-activated protein kinase gamma-subunit isoforms and their role in AMP binding. Biochem J 346: 659–669 [PMC free article] [PubMed] [Google Scholar]
- Crawford RM, Halford NG, Hardie DG (2001) Cloning of DNA encoding a catalytic subunit of SNF1-related protein kinase-1 (SnRK1-alpha1), and immunological analysis of multiple forms of the kinase, in spinach leaf. Plant Mol Biol 45: 731–741 [DOI] [PubMed] [Google Scholar]
- Daniel T, Carling D (2002) Functional analysis of mutations in the gamma 2 subunit of AMP-activated protein kinase associated with cardiac hypertrophy and Wolff-Parkinson-White syndrome. J Biol Chem 277: 51017–51024 [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7: 555–569 [DOI] [PubMed] [Google Scholar]
- Faure JD, Vittorioso P, Santoni V, Fraisier V, Prinsen E, Barlier I, Van Onckelen H, Caboche M, Bellini C (1998) The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development 125: 909–918 [DOI] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, Beach D (1994) Construction of an improved host strain for two hybrid screening. Nucleic Acids Res 22: 1502–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8: 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham-Skelton AP, Chilley P, Lumbreras V, Reignoux S, Fenton TR, Dahn CC, Pages M, Gatehouse JA (2002) A novel higher plant protein phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J 29: 705–715 [DOI] [PubMed] [Google Scholar]
- Gao G, Fernandez CS, Stapleton D, Auster AS, Widmer J, Dyck JR, Kemp BE, Witters LA (1996) Non-catalytic beta- and gamma-subunit isoforms of the 5′-AMP-activated protein kinase. J Biol Chem 271: 8675–8681 [DOI] [PubMed] [Google Scholar]
- Geigenberger P (2003) Regulation of sucrose to starch conversion in growing potato tubers. J Exp Bot 54: 457–465 [DOI] [PubMed] [Google Scholar]
- Gissot L, Polge C, Bouly JP, Lemaitre T, Kreis M, Thomas M (2004) AKINbeta3, a plant specific SnRK1 protein, is lacking domains present in yeast and mammals non-catalytic beta-subunits. Plant Mol Biol 56: 747–759 [DOI] [PubMed] [Google Scholar]
- Halford NG, Bouly JP, Thomas M (2000) SNF1-related protein kinases (SnRKs): regulators at the heart of the control of carbon metabolism and partitioning. In M Kreis, JC Walker, eds, Advances in Botanical Research. Academic Press, San Diego, pp 405–434
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 54: 467–475 [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Stapleton D, O'Donnell JB Jr, Kung JT, Dalal SR, Kemp BE, Witters LA (2001) An activating mutation in the gamma1 subunit of the AMP-activated protein kinase. FEBS Lett 500: 163–168 [DOI] [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM (2003) Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG (2004) The AMP-activated protein kinase pathway: new players upstream and downstream. J Cell Sci 117: 5479–5487 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M (1998) The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem 67: 821–855 [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 24: 3439–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K, Hong SP, Carlson M (2004) Pak1 protein kinase regulates activation and nuclear localisation of Snf1-Gal83 protein kinase. Mol Cell Biol 24: 8255–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg SM, Lee RH (1998) Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol Cell Biol 18: 4548–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ER, Pan DA, James J, Lucocq JM, Hawley SA, Green KA, Baba O, Terashima T, Hardie DG (2003) A novel domain in AMP-activated protein kinase causes glycogen storage bodies similar to those seen in hereditary cardiac arrhythmias. Curr Biol 13: 861–866 [DOI] [PubMed] [Google Scholar]
- Jiang R, Carlson M (1996) Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev 10: 3105–3115 [DOI] [PubMed] [Google Scholar]
- Jiang R, Carlson M (1997) The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol 17: 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Cai D, Kleine M (1998) Engineering nematode resistance in crop species. Trends Plant Sci 3: 266–271 [Google Scholar]
- Kaiser P, Auer B (1993) Rapid shuttle plasmid preparation from yeast cells by transfer to E. coli. Biotechniques 14: 552. [PubMed] [Google Scholar]
- Kazan K (2003) Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Plant Sci 8: 468–471 [DOI] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, Mc Pherson JD (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236: 1299–1302 [DOI] [PubMed] [Google Scholar]
- Kemp BE (2004) Bateman domains and adenosine derivatives form a binding contract. J Clin Invest 113: 182–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GBG (2006) A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J 46: 400–413 [DOI] [PubMed] [Google Scholar]
- Kleinow T, Bhalerao R, Breuer F, Umeda M, Salchert K, Koncz C (2000) Functional identification of an Arabidopsis Snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J 23: 115–122 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Dokiya Y, Sugita M (2001) Dual targeting of phage-type RNA polymerase to both mitochondria and plastids is due to alternative translation initiation in single transcripts. Biochem Biophys Res Commun 289: 1106–1113 [DOI] [PubMed] [Google Scholar]
- Krogh A (1997) Two methods for improving performance of an HMM and their application for gene finding. Proc Int Conf Intell Syst Mol Biol 5: 179–186 [PubMed] [Google Scholar]
- Lakatos L, Klein M, Hofgen R, Banfalvi Z (1999) Potato StubSNF1 interacts with StubGAL83: a plant protein kinase complex with yeast and mammalian counterparts. Plant J 17: 569–574 [DOI] [PubMed] [Google Scholar]
- Laurie S, McKibbin R, Halford NG (2003) Antisense SNF1-related (SnRK1) protein kinase gene represses transient activity of an α-amylase (α-Amy2) gene promoter in cultured wheat embryos. J Exp Bot 54: 739–747 [DOI] [PubMed] [Google Scholar]
- Lazar G, Goodman HM (2000) The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol Biol 42: 571–581 [DOI] [PubMed] [Google Scholar]
- Leclerc I, Kahn A, Doiron B (1998) The 5′-AMP-activated protein kinase inhibits the transcriptional stimulation by glucose in liver cells, acting through the glucose response complex. FEBS Lett 431: 180–184 [DOI] [PubMed] [Google Scholar]
- Lessard P, Decroocq V, Thomas M (1997) Isolation and analysis of messenger RNA from plant cells: cloning of cDNAs. In MS Clark, ed, Plant Molecular Biology. A Laboratory Manual. Springer-Verlag, Berlin, pp 154–201
- Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL (2001) Snf1: a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293: 1142–1146 [DOI] [PubMed] [Google Scholar]
- Lovas A, Bimbo A, Szabo L, Banfalvi Z (2003) Antisense repression of StubGAL83 affects root and tuber development in potato. Plant J 33: 139–147 [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Alba MM, Kleinow T, Koncz C, Pages M (2001) Domain fusion between SNF1-related kinase subunits during plant evolution. EMBO Rep 2: 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan D, Jeon JT, Looft C, Amarger V, Robic A, Thelander M, Rogel-Gaillard C, Paul S, Iannuccelli N, Rask L, et al (2000) A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science 288: 1248–1251 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Comparot-Moss S, Lue W-L, Messerli G, Trevisan M, Seymou MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen J, et al (2006) Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem 281: 11815–11818 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Polekhina G, Feil SC, Gupta A, O'Donnell P, Stapleton D, Parker MW (2005) Crystallization of the glycogen-binding domain of the AMP-activated protein kinase β subunit and preliminary X-ray analysis. Acta Crystallograph Sect F Struct Biol Cryst Commun 61: 39–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polekhina G, Gupta A, Michell BJ, van Denderen B, Murthy S, Feil SC, Jennings IG, Campbell DJ, Witters LA, Parker MW, et al (2003) AMPK beta subunit targets metabolic stress sensing to glycogen. Curr Biol 13: 867–871 [DOI] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford NG (1998) Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J 14: 195–202 [Google Scholar]
- Quirino B, Normanly FJ, Amasino RM (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40: 267–278 [DOI] [PubMed] [Google Scholar]
- Rathjen JP, Chang JH, Staskawicz BJ, Michelmore RW (1999) Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J 18: 3232–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43: 139–160 [DOI] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT (2006) SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA 103: 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG (2004) CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest 113: 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xiong L, Stevenson B, Lu T, Zhu JK (2002) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov LN, Dominguez-Solis JR, Allary A-L, Buchanan BB, Luan S (2006) A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA 103: 9732–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton D, Gao G, Michell BJ, Widmer J, Mitchelhill K, Teh T, House CM, Witters LA, Kemp BE (1994) Mammalian 5′-AMP-activated protein kinase non-catalytic subunits are homologs of proteins that interact with yeast Snf1 protein kinase. J Biol Chem 269: 29343–29346 [PubMed] [Google Scholar]
- Stapleton D, Mitchelhill KI, Gao G, Widmer J, Michell BJ, The T, House CM, Fernandez CS, Cox T, Witters LA, et al (1996) Mammalian AMP-activated protein kinase subfamily. J Biol Chem 271: 611–614 [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG (1999) Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander M, Olson MA, Ronne H (2004) Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J 23: 1900–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurau T, Kifle S, Jung C, Cai D (2003) The promoter of the nematode resistance gene Hs1pro-1 activates a nematode-responsive and feeding site-specific gene expression in sugar beet (Beta vulgaris L.) and Arabidopsis thaliana. Plant Mol Biol 52: 643–660 [DOI] [PubMed] [Google Scholar]
- Vincent O, Carlson M (1999) Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J 18: 6672–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Townley R, Kuchin S, Carlson M (2001) Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev 15: 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Warden SM, Richardson C, O'Donnell J Jr, Stapleton D, Kemp BE, Witters LA (2001) Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem J 354: 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiatrowski HA, Van Denderen BJ, Berkey CD, Kemp BE, Stapleton D, Carlson M (2004) Mutations in the gal83 glycogen-binding domain activate the snf1/gal83 kinase pathway by a glycogen-independent mechanism. Mol Cell Biol 24: 352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Azzout-Marniche D, Foretz M, Stein SC, Lemarchand P, Ferre P, Foufelle F, Carling D (2000) Characterization of the role of AMP-activated protein kinase in the regulation of glucose-activated gene expression using constitutively active and dominant negative forms of the kinase. Mol Cell Biol 20: 6704–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Johnson CH (2001) A clock- and light-regulated gene that links the circadian oscillator to LHCB gene expression. Plant Cell 13: 1411–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Jiang R, Carlson M (1994) A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J 13: 5878–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hubbard EJ, Carlson M (1992) A protein kinase substrate identified by the two-hybrid system. Science 257: 680–682 [DOI] [PubMed] [Google Scholar]
- Yoshida EN, Benkel BF, Fong Y, Hickey DA (1999) Sequence and phylogenetic analysis of the SNF4/AMPK gamma subunit gene from Drosophila melanogaster. Genome 42: 1077–1087 [DOI] [PubMed] [Google Scholar]