Abstract
In Arabidopsis thaliana, the Salt Overly Sensitive 2 (SOS2) gene is required for intracellular Na+ and K+ homeostasis. Mutations in SOS2 cause Na+ and K+ imbalance and render plants more sensitive toward growth inhibition by high Na+ and low K+ environments. We isolated the SOS2 gene through positional cloning. SOS2 is predicted to encode a serine/threonine type protein kinase with an N-terminal catalytic domain similar to that of the yeast SNF1 kinase. Sequence analyses of sos2 mutant alleles reveal that both the N-terminal catalytic domain and the C-terminal regulatory domain of SOS2 are functionally essential. The steady-state level of SOS2 transcript is up-regulated by salt stress in the root. Autophosphorylation assays show that SOS2 is an active protein kinase. In the recessive sos2-5 allele, a conserved glycine residue in the kinase catalytic domain is changed to glutamate. This mutation abolishes SOS2 autophosphorylation, indicating that SOS2 protein kinase activity is required for salt tolerance.
Control of intracellular ion homeostasis is essential for all cellular organisms. Most cells maintain relatively high K+ and low Na+ concentrations in the cytosol. In plants, this is achieved through coordinated regulation of transporters for H+, K+, and Na+. At the plasma membrane, a family of P-type H+-ATPases serves as the primary pump that generates a protonmotive force driving the active transport of other solutes, including K+ and Na+ (1). Several plant K+ channels and transporters have been molecularly characterized. The inward rectifying K+ channel AKT1 is essential for root K+ uptake in Arabidopsis thaliana (2, 3). Expression characteristics indicate that the KAT1 channel is involved in K+ influx in Arabidopsis guard cells (4, 5). Recently, an outward rectifying K+ channel has been shown to be essential for unloading K+ into the Arabidopsis root xylem (6). The wheat HKT1 gene product functions as a high-affinity K+ transporter (7). In addition, a family of KUP genes exists in Arabidopsis. At least one of them, KUP1, encodes a protein that can function as a dual-affinity K+ transporter (8, 9). Na+ enters plant cells passively, presumably through K+ transport systems (10). Unlike animals or fungi, plants do not seem to possess Na+/K+-ATPases or Na+-ATPases. Na+ efflux is achieved through the activities of Na+/H+ antiporters on the plasma membrane. Much of the Na+ that enters the cell is compartmentalized into the vacuole through the action of vacuolar Na+/H+ antiporters (11, 12). The driving force for the vacuolar transporters is the protonmotive force created by vacuolar V-type H+-ATPases and the H+-pyrophosphatase (1, 13). Although there has been great progress in the characterization of K+ and Na+ transporters in plants, little is currently known about their regulation.
In the trophic chain, plant roots play pivotal roles by taking up mineral nutrients from soil solutions. Plant roots experience constant fluctuations in soil environments. A frequent variant in the soil solution is Na+ concentration (14). Na+ is not an essential ion for most plants. In fact, the growth of the majority of plants, glycophytes, is inhibited by the presence of high concentrations of soil Na+. External Na+ causes K+ deficiency by inhibiting K+ uptake into plant cells (15). Na+ accumulation within the cell is toxic to many cytosolic enzymes. In contrast, many cellular enzymes are activated by K+, which is the most abundant cation in the cytoplasm. Certain cytoplasmic enzymes are especially prone to Na+ inhibition when K+ concentration is reduced (16). Therefore, maintaining intracellular K+ and Na+ homeostasis to preserve a high K+/Na+ ratio is important for all cells and especially critical for plant cells.
A family of Arabidopsis sos (salt overly sensitive) mutants defective in the regulation of intracellular Na+ and K+ homeostasis was recently characterized (15, 17, 18). The sos mutants are specifically hypersensitive to inhibition by high concentrations of external Na+ or Li+ (17, 18). In response to high Na+ challenge, the sos2 and sos3 mutants accumulate more Na+ and retain less K+ than wild-type plants (18). The mutants are also unable to grow when the external K+ concentration is very low (17, 18). These phenotypes suggest that the mutant plants are defective in the regulation of K+ and Na+ transport (18). The SOS3 gene was recently cloned and shown to encode an EF hand-type calcium-binding protein that shares significant sequence similarities with animal neuronal calcium sensors and the yeast calcineurin B subunit (19). In yeast, calcineurin is a central component in the signaling pathway that regulates Na+ and K+ homeostasis (20, 21). Loss-of-function mutations in calcineurin B cause increased sensitivity of yeast cells to Na+ or Li+ stress.
We report here the positional cloning of the SOS2 locus. SOS2 is predicted to encode a serine/threonine type protein kinase with an N-terminal catalytic domain highly similar to those of yeast SNF1 and mammalian AMPK kinases. Sequence analyses of several sos2 mutant alleles point to a functional requirement of both the N-terminal catalytic domain and the C-terminal regulatory domain of SOS2. SOS2 is expressed in both the root and shoot. In the root, SOS2 mRNA is up-regulated by salt stress. Autophosphorylation assays demonstrate that SOS2 is an active protein kinase. Furthermore, a mutation that abolishes SOS2 autophosphorylation renders plants hypersensitive to salt stress, indicating that SOS2 protein kinase activity is necessary for salt tolerance. This demonstrates that a protein kinase is essential for intracellular Na+ and K+ homeostasis and plant salt tolerance.
Materials and Methods
Genetic and Physical Mapping.
Genetic mapping with restriction fragment length polymorphism and PCR-based markers was as described (19). Construction of yeast artificial chromosome (YAC) and bacterial artificial chromosome (BAC) clone contigs (19) was partly based on information available at http://www.nucleus.cshl.org/protarab/chrom5map.htm and http://www.kazusa.or.jp/kaos.
Nucleic Acid Analysis.
For sequence determination, DNA was amplified from wild-type plants and sos2 mutants by PCR. Nine sos2 mutant alleles are known (18). All of the alleles were analyzed except sos2-4 and sos2-9 because viable seeds were not available. To avoid errors resulting from PCR, the products of five independent PCRs were pooled and sequenced. Reverse transcription–PCR was carried out on mRNA isolated from 2-wk-old Arabidopsis seedlings. Salt stress treatment, RNA extraction, and Northern blot analysis were carried out as described by Ishitani et al. (22).
Protein Expression.
To produce bacterially expressed recombinant proteins, the coding region of SOS2, SOS2(K40N), and SOS2(G197E) cDNAs were amplified by PCR with primers harboring restriction sites, cloned in frame into BamHI–EcoRI of pGEX-2TK (Amersham Pharmacia), and transformed into Escherichia coli BL21 DE3 cells (Amersham Pharmacia). Mutations K40N and G197E in the SOS2 protein were created by site-directed mutagenesis. For glutathione S-transferase (GST)-SOS2(K40N), primer pairs 5′-GCGGATCCATGACAAAGAAAATGAGAAGAGTGGGC and 5′-ATTGTACTCT- TAGCCATAATGTTGATGGCT were used for the first PCR, and 5′-GCGAATTCTTAAGTTGGGATCAAAACGTGA- TTGTTCTG and 5′-GTGATAATGTAGCCATCAACATTATGGCTA were used for the second PCR. For the mutant protein GST-SOS2(G197E), primer pairs 5′-GCGGATCCAT- GACAAAGAAAATGAGAAGAGTGGGC and 5′-ATATAACGAAAAGAATAACCTCGCAAGACC were used for the first reaction and 5′-GCTGATATTTGGTCTTGCGAGGTTATTCTT and 5′-GCGAATTCTTAAGTTGGGATCAAAACGTGATTGTTCTG were used for the second reaction. The final amplification was done with 5′-GCGGATCCATGACAAAGAAAATGAGAAGAGTGGGC and 5′-GCGAATTCTTAAGTTGGGATCAAAACGTGATTGTTCTG on both templates. The final constructs were confirmed by sequencing. E. coli cultures were induced with 0.5 mM isopropyl β-d-thiogalactoside, and recombinant proteins were affinity-purified from bacterial lysates with glutathione-Sepharose beads (Amersham Pharmacia).
Kinase Assay.
GST-fusion proteins were incubated in kinase buffer [20 mM Tris⋅HCl (pH 8.0)/5 mM MgCl2/1 mM CaCl2/1 mM DTT]. The kinase reaction was started by adding [γ-32P]ATP and was transferred to 30°C for 30 min. The reaction was stopped by adding 4× SDS-sample buffer and analyzed by SDS/PAGE and autoradiography.
Results
Positional Cloning of SOS2.
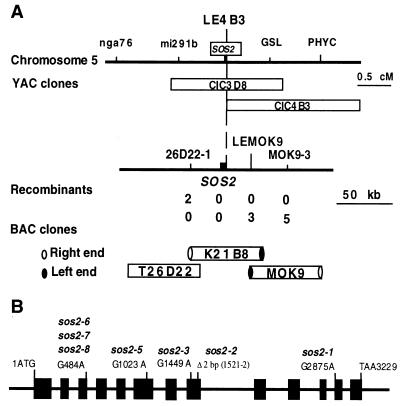
The SOS2 gene was mapped by crossing the sos2-2/sos2-2 mutant, which is in the Columbia ecotype, to the SOS2/SOS2 Landsberg ecotype. On the basis of analysis of 1230 recombinant chromosomes, the SOS2 locus was previously mapped to chromosome V, between molecular markers nga76 and PHYC (18). Further genetic mapping using the recombination crossover points narrowed SOS2 to a region between the restriction fragment length polymorphism markers mi291b and GSL (Fig. 1A). A YAC contig covering this region was assembled. The left end of YAC CIC4B3 (LE4B3) was found to be tightly linked to SOS2 because no recombination occurred (Fig. 1A). A contig of BAC clones centered around LE4B3 was assembled. Simple sequence length polymorphism markers 26D22-1 and MOK9-3 and single nucleotide polymorphism marker LEMOK9 were developed based on sequence information of the respective BAC clones. Genetic mapping using these markers delimited the SOS2 locus to a 60-kb region of K21B8. Sequence analysis revealed that a candidate gene within this region carries a 2-bp deletion in the sos2-2 allele, which was generated by fast neutron irradiation (Fig. 1B). Further sequence analyses revealed that other sos2 alleles all carry mutations in this gene (Fig. 1B). Each mutation causes a change in amino acid sequence in the predicted ORF. We therefore conclude that this candidate is the SOS2 gene.
Figure 1.
Positional cloning of the SOS2 gene. (A) Physical mapping of SOS2. Genetic mapping delimited SOS2 to a region in the BAC clone K21B8. The SOS2 gene was identified by sequencing candidate genes in this region from sos2 mutant and wild-type plants. (B) Structure of SOS2 and position of sos2 mutations. Positions are relative to the initiation codon. Filled boxes indicate the ORF, and lines between boxes indicate introns.
SOS2 Encodes a Protein Kinase.
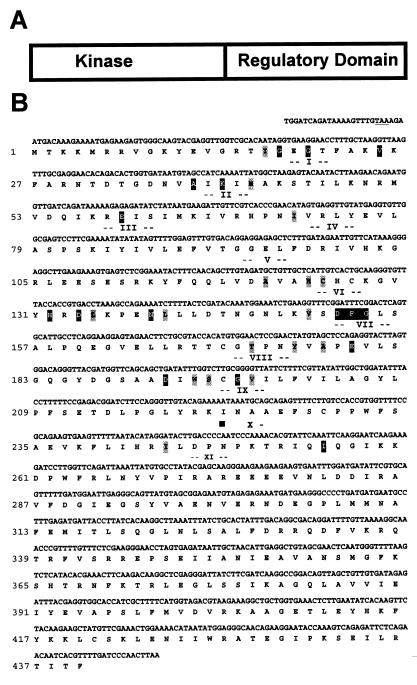
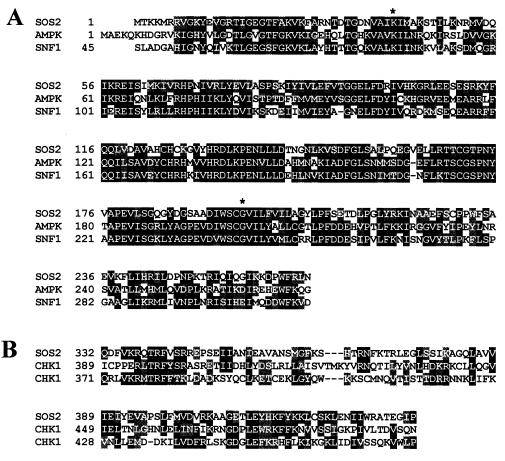
The transcribed sequence of the SOS2 gene was determined by sequencing cDNAs obtained by reverse transcription–PCR. Comparison with the genomic sequence showed that the SOS2 gene contains 13 exons and 12 introns (Fig. 1B). SOS2 is predicted to encode a protein of 446 amino acids with an estimated molecular mass of 51 kDa (Fig. 2). Database searches revealed that the deduced amino acid sequence of SOS2 has similarity with various serine/threonine protein kinases. The putative kinase catalytic domain of SOS2 resides in the N-terminal portion of the protein (Fig. 2A) and contains the 11 subdomains common to protein kinases (25). The putative catalytic domain sequence is most similar to the yeast SNF1 and mammalian AMPK kinases (23, 24) (Fig. 3A). The sos2-5, sos2-6, sos2-7, and sos2-8 mutations are predicted to disrupt the kinase catalytic domain. In the sos2-5 allele, Gly-197, which corresponds to one of the invariant amino acid residues of subdomain IX of protein kinases (25, 26), is changed to a negatively charged glutamic acid residue. sos2-6, sos2-7, and sos2-8 are identical mutations that disrupt the donor site of an intron splice junction (Fig. 1B), resulting in mal-splicing, premature termination, and a truncated polypeptide of 130 amino acid residues.
Figure 2.
SOS2 encodes a putative serine/threonine protein kinase. (A) Diagrammatic representation of SOS2 structure. (B) SOS2 cDNA sequence and the conceptual translational product of its longest ORF (GenBank accession number AF237670). Underlined is a stop codon (TAA) at −6 to −4 that precedes the ATG in-frame. Numbers I–XI indicate kinase subdomains as defined by Hanks et al. (25), with invariant and nearly invariant amino acid residues highlighted in black and gray, respectively.
Figure 3.
Amino acid alignments. (A) Alignment of putative kinase catalytic domain of SOS2 with Saccharomyces cerevisiae SNF1 (23) and human AMPK kinases (24). Amino acid residues identical in at least two proteins are highlighted in black and conservative substitutions in gray. Mutations that abolish SOS2 autophosphorylation (see Fig. 4) are indicated; first * is K40N, and second * is G197E, which corresponds to the sos2-5 allele. (B) Alignment of the C-terminal portion of SOS2 with the regulatory domains of Schizosaccharomyces pombe (yCHK1) and human CHK1 (hCHK1) kinases (27).
The C-terminal putative regulatory domain of SOS2 is relatively unique. Part of this domain of SOS2 shows low sequence homology with the regulatory domains of DNA replication checkpoint kinase CHK1 from yeast and humans (27–29) (Fig. 3B). Analysis of the other sos2 mutant alleles revealed that the mutations disrupt only the putative regulatory domain, leaving the catalytic domain intact. This suggests an essential function of the putative regulatory domain in plant Na+ tolerance. In the sos2-1 mutant allele, a single base pair substitution at the acceptor site of an intron splicing junction results in the addition of 29 amino acid residues between Glu-390 and Ile-391, presumably disrupting the function of the putative regulatory domain. The sos2-2 mutation that was created by fast neutron bombardment (18) has a 2-bp deletion that causes frameshift and premature termination, resulting in a truncated polypeptide of 287 amino acids. In the sos2-3 mutant allele, a single nucleotide substitution creates a stop codon that truncates the protein at Pro-262.
Protein Kinase Activity Is Required for SOS2 Function.
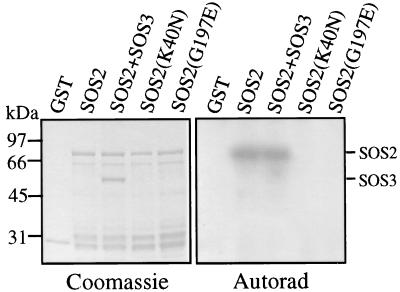
To determine whether SOS2 encodes a functional protein kinase, the SOS2 ORF was cloned into pGEX-2TK and expressed in bacteria as a C-terminal fusion protein to the bacterial GST. GST-SOS2 was purified from bacterial lysate by affinity chromatography with glutathione-Sepharose beads and shown to have the expected molecular mass of 78 kDa (Fig. 4). Incubation of the recombinant protein with [γ-32P]ATP in an in vitro kinase assay produced a strong phosphorylation signal that is likely the result of SOS2 autophosphorylation (Fig. 4, lanes 2 and 3). Lys-40 of SOS2 corresponds to a highly conserved residue in subdomain II (Fig. 2) that is required for activity in most protein kinases (25). To verify that the phosphorylation signal was because of SOS2 autophosphorylation, Lys-40 was changed to Asn by means of site-directed mutagenesis and the resulting mutant GST-SOS2(K40N) subjected to autophosphorylation assays. The Lys-40 to Asn mutation abolished the autophosphorylation of SOS2 (Fig. 4, lane 4).
Figure 4.
Autophosphorylation of SOS2 kinase. GST, GST-SOS2, GST-SOS2 plus GST-SOS3, and mutated kinases GST-SOS2(K40N) and GST-SOS2(G197E) were expressed in E. coli, purified from bacterial lysates by means of glutathione-Sepharose chromatography, incubated with [γ-32P]ATP in kinase buffer, electrophoresed on SDS/polyacrylamide gel, and Coomassie stained (Left), and exposed to x-ray film (Right).
In the sos2-5 mutant allele, the highly conserved Gly-197 is changed to Glu. We expressed the sos2-5 allele in bacteria, and the resulting mutant protein GST-SOS2(G197E) was examined for kinase activity. Like the Lys-40Asn mutation, the sos2-5 mutation also abolished the autophosphorylation activity of SOS2 (Fig. 4, lane 5). Because sos2-5 is a recessive mutation (18), the results show that kinase activity is required for SOS2 function in plant salt tolerance.
SOS2 kinase apparently has a very specific substrate requirement because none of the commonly used protein kinase substrates, such as histone H1, myelin basic protein, and casein, was phosphorylated by SOS2 (data not shown). SOS3 was not phosphorylated by SOS2, nor did it appear to affect SOS2 autophosphorylation in vitro (Fig. 4, lane 3). We have recently found several synthetic serine- or threonine-containing peptides that can be readily phosphorylated by SOS2 (30). In addition, phosphorylation of the peptides by SOS2 depended on the presence of both SOS3 and Ca2+ (30).
SOS2 Expression in the Root Is Up-Regulated by Salt Stress.
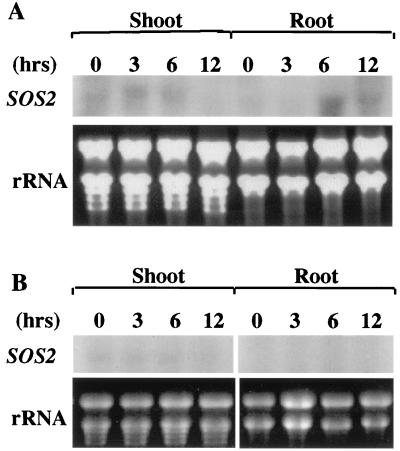
To analyze SOS2 expression under salt stress, 10-day-old Arabidopsis seedlings in agar plates were pulled out of agar medium and placed on filter papers soaked with 200 mM NaCl in Murashige and Skoog nutrient solution for 3, 6, or 12 h. Control plants were treated in the same manner, except no NaCl was added to the nutrient solution. After the treatment, roots and shoots were separated at the base of hypocotyls. Total RNA was extracted from control and NaCl-treated tissues. Northern blot analysis using SOS2 cDNA as a probe detected a single transcript of approximately 1.5 kb, the expected size of SOS2 mRNA (Fig. 5). In the root, a very low level of SOS2 mRNA was present in the sample before salt treatment (0 h). After 6 or 12 h of NaCl treatment, increased levels of SOS2 mRNA were detected (Fig. 5A). In the shoot, slight up-regulation of SOS2 transcript was found after 3 or 6 h of NaCl treatment. However, 12 h of NaCl treatment appeared to decrease SOS2 expression in the shoot (Fig. 5A). In the control treatments, only very low levels of SOS2 transcript were detected, and no up-regulation could be seen throughout the time course (Fig. 5B). Overall, the steady-state level of SOS2 transcript was very low, and it took approximately a week of x-ray exposure to obtain the signals shown, whereas a few hours were enough for most other stress-induced genes.
Figure 5.
Regulation of SOS2 expression by salt stress. Plants were treated with 200 mM NaCl (A) or with nutrient solution as a control (B) for the indicated time periods. Total RNA were extracted from roots and shoots, and subjected to Northern blot analysis with 32P-labeled SOS2 cDNA as probe. Thirty-five micrograms of total RNA was loaded in each lane. Ethidium bromide-stained rRNA bands were used as controls for equal loading.
Discussion
SOS2 is a major salt tolerance locus in A. thaliana (18). Mutations in the SOS2 gene drastically reduce plant tolerance to high Na+ stress and to low K+ stress. Based on mutant characterization, we have postulated previously that SOS2 might encode a regulatory protein that controls the expression and/or activities of certain K+ and Na+ transporters (18). In the present study, we have isolated the SOS2 gene through positional cloning. Indeed, SOS2 encodes a regulatory protein, a protein kinase. Protein phosphorylation is a frequent theme of cellular signal transduction, and its involvement in plant ion homeostasis and salt tolerance has been hypothesized (31). Our results provide direct evidence that protein phosphorylation is involved in Na+ and K+ homeostasis and plant salt tolerance. Future identification of protein substrate(s) that are phosphorylated by the SOS2 kinase will help understand how plant salt tolerance is regulated by protein phosphorylation. Candidate physiological substrates of SOS2 might include K+ and Na+ transporters and/or transcription factor(s) that control their expression.
The similar phenotypes of sos2 and sos3 mutants suggested that SOS2 and SOS3 may function in the same regulatory pathway (18). We tested and found that SOS3 is not phosphorylated by SOS2, nor did it affect SOS2 autophosphorylation (Fig. 4). Nevertheless, we have discovered that SOS2 physically interacts with SOS3, and SOS2 phosphorylation of peptide substrates is activated by SOS3 in a calcium-dependent manner (30).
SOS2 transcript is present in both roots and shoots. This is consistent with the observation that both the root and the shoot of sos2 mutant plants are hypersensitive to NaCl stress (18). SOS2 expression in the root appears to be up-regulated by NaCl stress. The significance of this up-regulation is unclear. There is certainly a very low level of expression in the root even without stress treatment, which could be detected by very long exposures in the Northern blot analysis or by reverse transcription–PCR (data not shown). The expression of SOS2 in the absence of stress is consistent with its role in primary signal transduction leading to salt adaptation. The slight up-regulation of SOS2 transcript may be important to maintain a sufficient level of SOS2 protein during salt stress. Like SOS2, SOS3 expression level is also very low (J.-K.Z., unpublished results). This probably reflects that SOS3 and SOS2 play regulatory roles that do not necessarily require abundant expression.
SOS2 encodes a protein kinase with a catalytic domain at the N terminus and a regulatory domain at the C terminus. The kinase catalytic domain is essential for SOS2 function. The sos2-5 mutation causes a single amino acid substitution within the catalytic domain that abolishes kinase autophosphorylation, resulting in the loss of SOS2 function and therefore increased sensitivity to salt stress. The regulatory domain also appears to be essential for SOS2 function because mutations that truncate (sos2-2 and sos2-3) or disrupt this domain (sos2-1) render plants hypersensitive to high Na+ and low K+ stresses. The catalytic domain of SOS2 is highly similar to the catalytic domains of SNF1/AMPK kinases (Fig. 3A). SNF1/AMPK kinases function to protect cells against nutritional or environmental stresses that deplete cellular ATP by regulating both metabolism and gene expression (23, 24). Although the catalytic domain of SOS2 is very similar to those of yeast SNF1 and mammalian AMPK kinases, SOS2 clearly is not a plant homolog of SNF1/AMPK. This is because true plant SNF1/AMPK kinases, such as SnRK1, share substantial sequence similarity with yeast SNF1 and mammalian AMPK at their C-terminal regulatory domain in addition to very high similarity at the N-terminal catalytic domain (32). Part of the regulatory domain of SOS2 is similar to the DNA repair and replication checkpoint kinase CHK1 (Fig. 3B), which is required for cell cycle arrest in response to DNA damage (27–29). The sequence similarity with CHK1 kinase is interesting because sos2 mutants show cell cycle defect at the root meristem in the presence of Na+ stress (J.-K.Z., unpublished data).
Although several protein kinases were previously reported to play roles in plant stress responses, none of them functions in ion homeostasis (33–36). The AtDBF2 protein kinase was identified by its ability to increase not only salt tolerance but also osmotic, heat and cold stress tolerance when overexpressed in Saccharomyces cerevisiae or in cultured tobacco cells (33). The mitogen-activated protein kinase MKK4 from alfalfa was shown to be activated by cold and drought but not by salt stress (34). Ectopic expression of a calcium-dependent protein kinase in maize protoplasts activates the expression of cold- and abscisic acid-responsive genes (36). The transcript levels of several protein kinases were shown to be up-regulated by various stresses, including touch, cold, and osmotic stress (35); however, their functions remain unknown. In contrast to these previously reported protein kinases that are involved in either general stress responses or in osmotic and cold stress responses, the SOS2 kinase has specific roles in plant adaptation to high Na+ and low K+ stresses (18).
Acknowledgments
We thank Becky Stevenson and Jing Ma for excellent technical assistance, and Dr. Xiaoyin Lin of The Institute for Genome Research for annotation of the BAC clone K21B8. This work was supported by National Institutes of Health Grant R01GM59138 and U.S. Department of Agriculture-National Research Initiative Grant 9801270.
Abbreviations
- SOS
salt overly sensitive
- YAC
yeast artificial chromosome
- BAC
bacterial artificial chromosome
- GST
glutathione S-transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF237670).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060034197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060034197
References
- 1.Sze H, Li X, Palmgren M G. Plant Cell. 1999;11:677–689. doi: 10.1105/tpc.11.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sentenac H, Bonneaud N, Minet M, Lacroute F, Salmonm J-M, Gaymard F, Grignon C. Science. 1992;256:663–665. doi: 10.1126/science.1585180. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch R E, Lewis B D, Spadling E P, Sussman M R. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- 4.Anderson J A, Huprikar S S, Kochian L V, Lucas W J, Gaber R F. Proc Natl Acad Sci USA. 1992;89:3736–3740. doi: 10.1073/pnas.89.9.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura R L, Mckendree W L, Jr, Hirsch R E, Sedbrook J C, Gaber R, Sussman M R. Plant Physiol. 1995;109:371–374. doi: 10.1104/pp.109.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferriere N, Thibaud J-B, Sentenac H. Cell. 1998;94:647–655. doi: 10.1016/s0092-8674(00)81606-2. [DOI] [PubMed] [Google Scholar]
- 7.Rubio F, Gassmann W, Schroeder J I. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- 8.Kim E J, Kwak J M, Uozumi N, Schroeder J I. Plant Cell. 1998;10:51–62. doi: 10.1105/tpc.10.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu H-H, Luan S. Plant Cell. 1998;10:63–73. doi: 10.1105/tpc.10.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder J I, Ward J M, Gassmann W. Annu Rev Biophys Biomol Struct. 1994;23:441–471. doi: 10.1146/annurev.bb.23.060194.002301. [DOI] [PubMed] [Google Scholar]
- 11.Gaxiola R A, Rao R, Sherman A, Grisafi P, Alper S L, Fink G R. Proc Natl Acad Sci USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apse M P, Aharon G S, Snedden W A, Blumwald E. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 13.Rea P A, Poole R J. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:157–180. [Google Scholar]
- 14.Epstein E, Norlyn J D, Rush D W, Kingsbury R W, Kelley D B, Cunningham G A, Wrona A F. Science. 1980;210:399–404. doi: 10.1126/science.210.4468.399. [DOI] [PubMed] [Google Scholar]
- 15.Wu S-J, Ding L, Zhu J-K. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murguia J R, Belles J M, Serrano R. Science. 1995;267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Zhu J-K. Proc Natl Acad Sci USA. 1997;94:14960–14964. doi: 10.1073/pnas.94.26.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J-K, Liu J, Xiong L. Plant Cell. 1998;10:1181–1192. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Zhu J-K. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T Y, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo J M. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- 22.Ishitani M, Xiong L, Stevenson B, Zhu J-K. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celenza J L, Carlson M. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 24.Mitchelhill K I, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters L A, Kemp B E. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 25.Hanks S K, Quinn A M, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 26.Hanks S K, Hunter T. In: The Protein Kinase Facts Book. Hardie D, Hanks S, editors. Vol. 1. London: Academic; 1995. pp. 7–47. [Google Scholar]
- 27.Walworth N, Davey S, Beach D. Nature (London) 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 28.Walworth N C, Bernards R. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 29.Boddy M N, Furnari B, Mondesert O, Russell P. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 30.Halfter, U., Ishitani, M. & Zhu, J.-K. (March 21, 2000) Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040577697. http://www.pnas.org/cgi/doi/10.1073/pnas.040577697
- 31.Niu X, Bressan R A, Hasegawa P M, Pardo J M. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halford N G, Hardie D G. Plant Mol Biol. 1998;37:735–748. doi: 10.1023/a:1006024231305. [DOI] [PubMed] [Google Scholar]
- 33.Lee J H, Van Montagu M, Verbruggen N. Proc Natl Acad Sci USA. 1999;96:5873–5877. doi: 10.1073/pnas.96.10.5873. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Jonak C, Kiegerl S, Lighterink W, Barker P J, Huskisson N S, Hirt H. Proc Natl Acad Sci USA. 1996;93:11274–11279. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. Proc Natl Acad Sci USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheen J. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]