Abstract
The chlorophyll a/b, chlorophyll a/c, and chlorophyll a/a light-harvesting proteins are part of an extended gene family that also includes the transiently expressed stress proteins, the Elips (early light-induced proteins). Four Elip homologue proteins, encoded by single-copy nuclear genes, have been identified in the Arabidopsis thaliana database. These proteins were divided into two groups according to the expression pattern under light-stress conditions and the predicted secondary structure. Group one included two members of the Elip family with three predicted transmembrane helices and a gene expression strictly related to light stress. Group two included two proteins, the Seps (stress-enhanced proteins), which possessed two predicted transmembrane segments. The transcripts of Sep1 and Sep2 were present under low light conditions, but their level increased 4- to 10-fold during illumination of plants with high-intensity light. Preliminary data indicated that the induced transcripts were translated in vivo. Other physiological stress conditions, such as cold, heat, desiccation, salt, wounding, or oxidative stress, did not significantly influence the expression of Sep genes. In vitro import of radioactively labeled precursors of Seps into isolated chloroplasts confirmed the thylakoid membrane localization of these proteins. Considering the predicted protein structure and homology to other pigment-antenna proteins, the two-helix Seps might represent an evolutionary missing link between the one- and three-helix antenna proteins present in pro- and eukaryota.
Keywords: ancestors, chloroplast, pigment proteins
Photosynthetic eukaryotes are traditionally divided into three major groups, largely on the basis of their light-harvesting antenna systems. The Chlorophytes (green algae and higher plants) have chlorophyll a/b antennas, the Chromophytes have chlorophyll a/c antennas, and the Rhodophytes (red algae) have only chlorophyll a and use phycobilisomes as the major photosystem (PS) II antenna (1, 2). The primary function of photosynthetic light-harvesting complexes is the absorption of light and the transfer of the excitation energy to the photochemical reaction centers. Because of their high sequence homology and similar structure and function, all of the eukaryotic light-harvesting antenna proteins are considered part of an extended gene family.
The chlorophyll a/b-binding (Cab) family in Arabidopsis thaliana contains at least 30 different members, associated with PSI or PSII (3). During the last few years, the Cab gene family was extended by the distant relatives, the PSII-S protein (4–6), the one-helix protein, the OHP (3), and the early light-induced proteins, the Elips (7, 8). The discovery of the cyanobacterial Hlip (high light-induced proteins) and the SCPs (small Cab-like proteins), clearly sharing the same conserved residues with the eukaryotic Cab proteins (9, 10), supported the idea that the prokaryotic Hlips and SCPs and the eukaryotic antenna proteins had a common ancestor (1, 11).
The higher plant Elips are nuclear-encoded chloroplast proteins (12, 13) localized in the nonappressed regions of the thylakoid membranes (14). In contrast to the typical Cab family members, which are constitutive structural components of PSI and PSII, Elips accumulated only transiently in the thylakoid membranes in substoichiometric amounts. Induction of Elips during greening of etiolated pea seedlings (12, 13) in mature green plants exposed to light stress (8) or during acclimation of plants to increased light intensities (15) was reported. Functions other than light harvesting were proposed for this group of proteins in higher plants (8, 16).
In this work, four proteins containing an Elip consensus sequence were identified and characterized in A. thaliana. Two of these proteins represented recently cloned Elip1 (17) and Elip2 (18), and two others belonged to a group of the stress-enhanced proteins, the Seps. The secondary structure of Seps and their homology to the Cab proteins make them strong candidates for the two-helix ancestor of the eukaryotic antenna systems.
Materials and Methods
Growth of Plant and Stress Conditions.
A. thaliana L. cv. Columbia were grown in a growth chamber on soil at 25°C at a light intensity of 100 μmol m−2 s−1 under short-day conditions.
Light-stress treatment was performed on mature leaves, detached from 4- to 5-wk-old plants, floated on water, and exposed to a light intensity of 2.500 μmol m−2 s−1 for 3 hr. For cold or heat stress, detached leaves floated on water were transferred for 2 hr to incubators set at 4°C or 42°C, respectively. Wounding stress was obtained by cutting the leaves into 5-mm2 small segments, which were then incubated for 2 hr on water at room temperature at a light intensity of 10 μmol m−2 s−1. Desiccation stress was performed on leaves dehydrated on Whatman 3MM paper at room temperature at a light intensity of 10 μmol m−2 s−1. After 2 hr of incubation, the relative water content of leaves was reduced to 50%. Leaves subjected to high salt or oxidative stress were submerged for 2 hr in 400 mM NaCl or in 2% H2O2 solutions, respectively. UV-A treatment was performed by illumination of detached leaves with a UV lamp (366 nm) at a light intensity of 25 μmol m−2 s−1 for 2 hr. Plant material was collected and either immediately used for extractions or frozen in liquid nitrogen and stored at −70°C for further preparations.
Gene Cloning, Sequencing, and Data Analysis.
Arabidopsis EST clones of Elip1 (clone ID 174P5T7, accession no. TC8652), Elip2 (clone ID VCVCD09, accession no. Z26549), Sep1 (clone ID 235A5T7, accession no. N65188), and Sep2 (clone ID 212I19T7, accession no. P 19105) were obtained from the Arabidopsis Biological Resource Center at Ohio State University, Columbus, OH, and the identity of each clone was verified by sequencing of both cDNA strands (CyberGene, Stockholm, Sweden).
The EST clone of Elip1 contained a full-length cDNA insert of 873 bp with a coding region of 588 bp (195 amino acids). The EST clone of Elip2 was truncated at the 5′-end and a full-length cDNA clone of Elip2 was amplified by PCR as described (18). The full-length clone of Elip2 contained a 674-bp cDNA insert with a coding region of 582 bp (193 amino acids). The EST clones of Sep1 and Sep2 contained the full-length cDNA inserts. The Sep1 clone contained a 691-bp cDNA insert with a coding region of 438 bp (146 amino acids) and the Sep1 clone, an insert of 862 bp with a coding region of 606 bp (202 amino acids). The cDNA sequence data of the Sep1 and Sep2 are available in GenBank (accession nos. AF133716 and AF133717, respectively).
Similarity searches were done by using the advanced blast program (19). Alignment of amino acid sequences was performed by using clustalw (Des Higgins, European Bioinformatics Institute, U.K.) with manual correction of gaps, and an Elip consensus sequence was obtained by using the multalin program (20). An Elip consensus sequence was constructed on the basis of Elip or Elip-related amino acid sequences in Pisum sativum (21), Hordeum vulgare (13), Glycine max (22), Helianthus annuus (23), and Craterostigma plantagineum (24), the red algae, such as Porphyra purpurea (25) and Cyanidium caldarium (26), the green alga Dunaliella bardawil (27), and the cyanobacteria Synechocystis (9).
Localization prediction and determination of the processing site were analyzed by the programs psort (28) and chlorop Ver. 1.1 (29). Secondary structure prediction was performed by using the predator (30) and gorIV (31) programs. The transmembrane regions were predicted by using the dense alignment surface method (32), sosui (33), and tmhmm (34). The hydropathy plot was raised according to ref. 35. Protein pattern and motif predictions were performed by using sequence motif search (30) and protein motif fingerprint databases (36).
Southern and Northern Blot Analysis.
Genomic DNA was extracted from leaves by using a DNA extraction kit (Qiagen, Chatsworth, CA). DNA (5 μg per reaction) was digested by the restriction enzymes and DNA fragments were separated in 0.8% agarose gel and transferred to Hybond-N+ membrane (Amersham) according to the manufacturer's protocol. The hybridization was performed according to ref. 37. The signal on the filter was analyzed by using a Phosphorimager BAS1000 (Fujifilm, Fuji).
Total RNA was extracted from control- or light stress-treated leaves by using an RNeasy mini kit (Qiagen) according to the manufacturer's protocol. After separation of 5 μg RNA in 1.2% agarose gel, RNA was transferred to Hybond-N+ membrane before hybridization, according to ref. 37.
The cDNA probe was labeled with α-32P-dCTP by using a megaprime DNA labeling kit (Amersham Pharmacia).
In Vitro Transcription, Translation, and Import.
Plasmids containing inserts of the Elip1, Sep1, Sep2 (Bluescript, Lambda-PRL2), and Elip2 (pGEM-TEasy) cDNA clones were linearized with the restriction enzymes, NotI (for Elip1 and Sep2), SacII (for Elip2), or SmaI (for Sep1), and used for in vitro transcription with T7 (Elip1, Sep2) or SP6 (Sep1, Elip2) polymerases. In vitro translation was performed in a wheat germ lysate (Boehringer Mannheim) in the presence of [35S]methionine (1,220 Ci/mmol, Amersham Pharmacia).
In vitro imports into isolated intact pea chloroplasts were performed according to ref. 14. Proteins were separated by SDS/PAGE according to ref. 38, and the gels were exposed to x-ray film (Cronex, Sterling Diagnostic Imaging, New York) or Phosphorimager plates (Fujifilm).
Results
Isolation of Two Light Stress-Regulated Genes from A. thaliana.
Conserved domains of Elips or Elip-related proteins from higher plants, red algae, green algae, and the cyanobacteria were used for the construction of an Elip amino acid consensus sequence “ERINGRL(A)AMI(V)GF.” A blast search for related sequences in the database of expressed sequence tags (ESTs) of anonymous Arabidopsis cDNA clones (39) revealed four clones with a significant sequence homology to the Elip consensus sequence.
The EST clone 174P5T7 was identical to cDNA encoding Elip (accession no. gb/U89014) described in ref. 17 and is designated here as Elip1. The EST clone VCVCD09 encoded a protein with 78% identity and 89% similarity to Elip1 and is designated here as Elip2 (Fig. 1). The EST clones 235A5T7 and 212I19T7 encoded a group of proteins designated Sep1 and Sep2, respectively.
Figure 1.
Amino acid sequence alignment of Elips and Seps. The sequences were aligned manually with the assistance of the multiple alignment program clustalw. Identical amino acids are shown on a gray background. Gaps introduced for the alignment are indicated by dashes.
Alignment of the deduced amino acid sequences of Sep1 and Sep2 revealed that these proteins share only 14% identity and 32% similarity in the entire sequences (Fig. 1). However, the middle regions and the C-terminal tails of these two proteins were much more conserved than their N terminus. Comparison of amino acid sequences of Elips and Seps showed (Fig. 1) that these proteins share several conserved amino acids. The overall sequence similarity between the Elips and the Sep1 or the Sep2 was 26% and 33%, respectively.
Chloroplast Localization and Predicted Thylakoid Membrane Topology of Seps.
Predictions of the protein secondary structure and subcellular localization suggested that Seps are polytopic integral membrane proteins localized in the chloroplast. Hydropathy plots of the Sep1 and Sep2 revealed that they possessed two transmembrane helices of 19–23 aa (Fig. 2A). Furthermore, Sep1 and Sep2 significantly differed from one another in the length of the mature proteins, which was 103 aa for Sep1 and 181 aa for Sep2 (Fig. 2B). This difference was mainly the result of the lengths of the hydrophilic C- and N-terminal regions, which were much longer for the Sep2 than for the Sep1 (Fig. 2B).
Figure 2.
Predicted secondary structure of Elips and Seps. (A) Hydropathy plots of the translated cDNA sequences of Elips and Seps; (B) schematic representation of the predicted secondary structure of Elips and Seps. The predicted length of transmembrane domains, stromal and luminal connectors, and transit peptides are shown. Numbers indicate the amount of amino acids (aa) in each domain.
Hydropathy plots of the Elip1 and Elip2 in Arabidopsis confirmed that these proteins contained three transmembrane helices of 19–23 aa each (Fig. 2 A and B), which is in agreement with the predictions made for Elips from pea and barley (7). The N-terminal and the C-terminal hydrophilic regions of Elip1 and Elip2, as well as linkers between the helices, were similar in length for these two proteins (Fig. 2B).
As all organelle polypeptides of a nuclear origin, the Seps were expected to be synthesized as precursor proteins with cleavable transit peptides. Two possible N-terminal cleavage sites, one between 73–74 (CS-score 3.1) and a second between 43–44 (CS-score 1.45) amino acids, were predicted for Sep1 by using the chlorop Ver. 1.1 program (29). Because the chloroplast transit peptides of 14 analyzed members of Elip family were predicted to be shorter than 50 aa, we assumed that the latter value was correct for Sep1 (Fig. 2B). The Sep2 was not recognized by the chlorop program as a chloroplast protein, thus the prediction of the N-terminal signal peptide was based only on the psort analysis (28). A transit peptide with a cleavage site between 21–22 aa was predicted for Sep 2 (Fig. 2B). This cleavage site could be aligned manually with the logoplot constructed from the 62 sequences used in the development of a new version of the chlorop program (29). A predicted discrete cleavage site was between 41–44 aa for Elip1 and Elip2 (Fig. 2B).
To verify the predicted chloroplast localization of Seps, these proteins were transcribed and translated in vitro in the presence of [35S]methionine, and the radioactively labeled precursors were used for in vitro import into isolated intact pea chloroplasts. As a control, the Elip1 and Elip2 transcripts were translated and imported under the same experimental conditions. The results (Fig. 3) showed that both Elips and Seps were imported into the chloroplasts and inserted into the thylakoid membranes. No traces of these proteins were detected in the stromal fraction. During the import assay, the 15.6-kDa and 24.5-kDa precursors of the Sep1 and Sep2 were processed to 9-kDa and 21-kDa mature products, respectively (Fig. 3). The Elip1 and Elip2 precursors with apparent molecular masses of 25.5 kDa and 21.5 kDa, respectively, were processed to mature products of 19.5 kDa and 16 kDa (Fig. 3).
Figure 3.
Thylakoid membrane localization of Elips and Seps. In vitro-transcribed and -translated Elip and Sep precursors (lane 1) were imported into intact pea chloroplasts before their separation into the thylakoid membranes (lane 2) and the soluble stromal fraction (lane 3). Autoradiogram of proteins from both chloroplast fractions separated by SDS/PAGE is shown. pElip and pSep, precursors of Elip and Sep.
The predicted thylakoid membrane topology of Seps revealed (not shown) that both Seps represent most likely type III membrane proteins, with the N and the C terminus located in the stromal compartment. In agreement with data reported for pea and barley Elips (7, 13), Elip1 and Elip2 in Arabidopsis are predicted to be type II membrane proteins with the N terminus on the stromal and the C terminus on the lumenal side of the membrane.
Homology of Seps with the Elips and Cab Proteins in Arabidopsis.
Comparison of amino acid sequences of Seps revealed that these proteins show a very high homology with the Cab gene family members, especially in the first transmembrane helix.
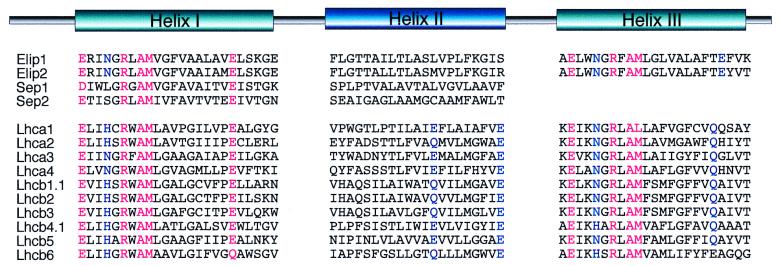
The Cab gene family of higher plants consists of chlorophyll a/b-binding proteins associated with the PSI (Lhca1–4 gene products) or PS II (Lhcb1–6 gene products). Recently, this family was extended by 12 additional members identified in the Arabidopsis EST databank (3). The availability of the array of Cab (3) and Elip (17, 18) sequences in Arabidopsis allowed us to compare helices I and II of Seps with the corresponding domains of Cab and Elip family members (Fig. 4).
Figure 4.
Comparison of the transmembrane domains of Elips, Seps and Cab proteins from Arabidopsis. Highly conserved amino acid residues common for the Elips, Seps, and Cabs are marked in red; additional residues with important structural functions discussed in text are marked in blue. The sequences of Cab proteins from Arabidopsis are according to ref. 3.
The first helix of Seps contains a number of amino acid residues, which are strikingly conserved for the whole Cab gene family members (11), like an Arg (R) or two highly conserved Glu (E) residues, of which the first is replaced by an Asp (D) in Sep1 (Fig. 4). Whereas the first Glu (E) residue was reported to serve a dual function in chlorophyll binding and in locking together the two transmembrane helices of Lhcb1 (11, 40), the role of a second Glu (E) is not yet determined. Because the latter Glu (E) is highly conserved in Cab proteins, it is expected that it must play an important structural role (11).
Helices I and III of Elips in Arabidopsis share the Glu (E), Arg (R), and Asn (N) residues involved in binding the four core chlorophylls a in Lhcb1 (11, 40). The second Glu (E) residue present in the C-terminal region of helix III (Fig. 4) seems to be conserved for all known eukaryotic and prokaryotic Elips or algal Fcps (fucoxanthin chlorophyll a/c-binding proteins), but not for Cabs (11).
Furthermore, helix I of Seps and both helices I and III of Elips and Cab proteins contained a conserved LAM(GAM)/FAM motif for Seps and Elips and a WAM(FAM)/LAM motif for Cab proteins (Fig. 4).
Helix II of Seps, Elips, and Cab proteins is highly variable in sequence and differs significantly for all three groups of proteins. The amino acid composition of helix II in Sep1 and Sep2 showed 19% identity and 62% similarity between these two proteins (Fig. 4), but there was no significant homology between helix II of Seps and Elips or Seps and Cab proteins. For Elip1 and Elip2 in Arabidopsis, this region shows 86% identity and 95% similarity between these two proteins. Sequence alignment of the Arabidopsis Elips with other Elip or Elip-related proteins from soybean (22), pea (21), barley (13), common sunflower (23), or Craterostigma (24) revealed between 38–62% identity and 81–95% similarity in the helix II region.
Genomic Organization of Elips and Seps.
To investigate the copy number of the Sep1, Sep2, Elip1, and Elip2 genes in Arabidopsis, genomic DNA was digested by the restriction endonucleases and hybridized to the Sep or Elip cDNAs. Independently of the stringency of hybridization (results of a low stringency hybridization are not shown), a single band was detected for all four genes tested (Fig. 5). This indicated that the Sep1, Sep2, Elip1, and Elip2 genes exist at a single locus, and closely related nucleotide sequences are not present in the Arabidopsis genome.
Figure 5.
Genomic DNA gel blot analysis of Elips and Seps. Five micrograms of Arabidopsis genomic DNA were digested with the restriction enzymes: (1) BamHI, (2) EcoRI, or (3) HindIII, and the blot was hybridized with the complete Elip1, Elip2, Sep2, and Sep2 coding regions derived from the EST cDNA clones. The sizes of the fragments are marked.
Recently, the genomic sequences of Sep and Elips were localized on the Arabidopsis chromosomes. The Sep1 gene was localized on chromosome IV (accession nos. AL035521 and AL021961) and consisted of four exons interrupted by three introns, and the Sep2 gene was localized on chromosome II (accession no. AC007019) and was composed of two exons separated by a single intron. The positions of the introns were not conserved for Seps.
The Elip1 gene was localized on chromosome III (accession no. AB022223), and the Elip2 gene was found on chromosome IV (accession no. Z97336). The coding region of Elip1 and Elip2 genes had a similar genomic structure, comprising three exons and two introns. In contrast to the Sep genes, the positioning of the introns was conserved between Elip1 and Elip2 genes, and they were placed 12 bp downstream the transit peptide cleavage site and 48 bp upstream the first transmembrane helix of these two proteins. The number of introns reported for the Cab gene family was between one (Lhcb2, Lhcb4, and Lhcb6) and five (Lhcb5), and the Lhcb1 gene contained no introns (41).
Regulation of Sep Gene Expression by Light Stress.
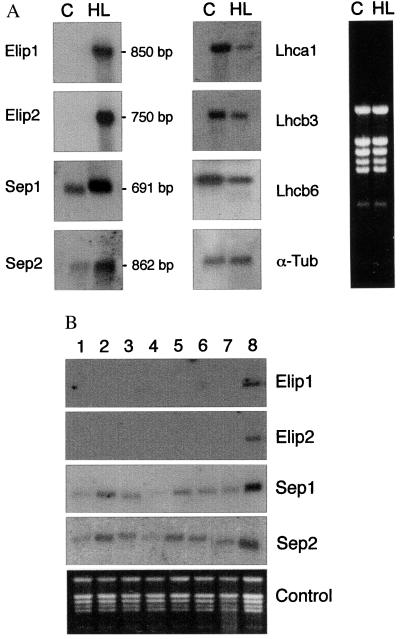
To analyze the gene expression of Seps, total RNA was isolated from detached leaves of Arabidopsis exposed to low light (control) or high light conditions and used for hybridization with the Sep-specific cDNA probes. For comparison, the expression of Elip1 and Elip2 genes was assayed under the same experimental conditions. The results revealed (Fig. 6A) that the transcripts of Sep1 (691 bp) and Sep2 (862 bp) were detected under low light conditions, but their level increased significantly during light stress (Fig. 6A Left). A 4- to 10-fold increase in the transcript levels was measured for Sep2 and Sep1, respectively. Differently from Seps, the transcripts for Elip1 (850 bp) and Elip2 (750 bp) were not detected in control plants kept at low light conditions but were specifically induced by light stress (Fig. 6A Left). The amounts of Sep and Elip transcripts increased with the time of high light exposure, but their levels declined rapidly after transferring of leaves from high- to low-intensity light (not shown). The half-lifetime of Sep and Elip mRNAs under low light conditions was calculated to be less than 1 hr (not shown). Furthermore, the accumulation of Sep and Elip transcripts was light intensity dependent between 100 and 2,000 μmol m−2 s−1 (not shown). These results are in agreement with the previously published results for pea or barley Elips (8, 42).
Figure 6.
Regulation of Elip and Sep gene expression by stress. (A) Arabidopsis leaves were either directly used for isolation of total RNA (C, control) or exposed to light stress for 3 hr before RNA extraction (HL, high light). Northern blot hybridization was performed by using 32P-labeled cDNA fragments of the Elip1, Elip2, Sep1, Sep2 clones and selected members of the Cab gene family, such as Lhca1, Lhcb3, and Lhcb6. The α-tubulin gene (clone ID 32C11T7) was hybridized as an internal control. (B) Arabidopsis leaves were exposed for 2 hr to various stress conditions: (1) control, (2) cold stress, (3) heat shock, (4) wounding, (5) salt stress, (6) desiccation, (7) oxidative stress, and (8) UV-A illumination. Isolated total RNA was used for Northern blot hybridization with the radioactively labeled inserts of the Elip and Sep cDNA clones. The rRNA pattern in the gel, visualized by staining with ethidium bromide, is shown in A (Right) and B (Lower).
In contrast to the light-stress up-regulation of Seps and Elips transcripts, the transcript levels for other members of the Cab gene family, such as the Lhca1, Lhcb3, or Lhcb6, were down-regulated under light-stress conditions (Fig. 6A Middle). The transcript level of the α-tubulin, assayed as a control, did not change significantly under either illumination condition tested (Fig. 6A Middle).
The regulation of Sep gene expression by other stresses, such as cold, heat, wounding, salt, desiccation, oxidative stress, or exposure to UV-A radiation, was also investigated. The expression of Sep genes was significantly up-regulated by UV-A illumination (Fig. 6B). In contrast, the level of the Sep1 and Sep2 transcripts was much lower in leaves exposed to mechanical wounding than in control leaves.
It was reported before that the induction of pea Elip (16) and the prokaryotic Hlip (9) was triggered by blue and UV-A radiation. The Elip1 and Elip2 genes in Arabidopsis were also induced by UV-A illumination, and other stresses were not effective in the activation of Elip genes (Fig. 6B).
The binding of mRNAs to polyribosomes is assumed to be indicative of active protein synthesis. Assay of mRNA distribution between cytoplasmic and polysomal fractions revealed (not shown) that under light-stress conditions, 95% of Sep and Elip transcripts were associated with the polysomes, and only 5% were present as free mRNAs. Furthermore, the incorporation of Sep and Elip transcripts into polysomes increased with increasing light intensity. In vivo labeling studies demonstrated that the proteins with apparent molecular masses similar to those calculated for Elips and Seps accumulated in the thylakoid membranes of Arabidopsis under light-stress conditions (not shown). However, the identity of these proteins still requires confirmation.
Discussion
The superfamily of the chlorophyll-binding proteins in eukaryota includes several members synthesized in response to environmental stresses, which do not appear to play a role in light harvesting (8, 24, 27). In this work, two groups of the Cab-related light stress-regulated genes were isolated and characterized from A. thaliana. Group one was represented by two members belonging to the previously described Elip family (7, 13), and group two consisted of two proteins, the Seps.
Despite a sequence homology between Seps, Elips, and Cab proteins, the predicted secondary structure of Sep1 and Sep2 significantly differed from those reported for Elips and Cab proteins. It was shown that the Cab polypeptides fold into three membrane-spanning helices, where helices I and III are held together by reciprocal ion pairs involving an Arg (R) on one helix and a Glu (E) on the another (11, 40). A similar three-helix structure was predicted for the Elip1 and Elip2 from Arabidopsis and suggested earlier for pea and barley Elips (7, 13). Interestingly, Sep1 and Sep2 contain only two hydrophobic transmembrane segments, and the conserved Arg (R) and Glu (E)/Asp (D) residues are present only in the first conserved helix. Because two transmembrane helices are required to make the reciprocal ion pair, we expect that in vivo these proteins might form homodimers. A similar homodimer structure was proposed for one-helix Hlip from cyanobacteria (9, 11). The pattern of small residues essential for close packing of the two helices of Cab proteins (11) and the conserved Met (M) that interlocks with them is present in helix I of Seps (Fig. 4), supporting the concept of their homodimeric structure.
Recent purification of Elips from light-stressed pea leaves confirmed the theoretical expectations that these proteins bind chlorophylls (43). The predicted amino acid sequences implied that also the Seps are potential chlorophyll-binding proteins. One putative chlorophyll-binding domain, an Asp (D) or a Glu (E), was localized in helix I of Seps1 or Sep2, respectively (Fig. 4). However, to provide a fifth ligand to the Mg atom of chlorophyll, the negative charges of these amino acids have to be neutralized by an ionic bridge to an Arg (R) side chain, as was reported for Lhcb1 (40). This suggests that the Sep1 or Sep2 dimers could bind two chlorophyll a molecules.
It is assumed that the light-harvesting proteins of all photosynthetic eukaryotes and prokaryotes have a common origin (1). It has previously been proposed (11, 41) that the three-helix members of the Cabs, Fcps, or Elips originated from a four-helix protein that in turn arose from a two-helix protein as the result of internal gene duplication and fusion. This hypothesis was based on the discovery of the Cab-related PSII-S protein with four transmembrane helices, where helices I and III and II and IV were clearly related (5, 6). It was suggested that similar gene duplication, followed by deletion of the fourth helix, has given rise to the three-helix ancestors of the eukaryotic antenna proteins and Elips. Furthermore, it was speculated that the two-helix ancestor might derive from the fusion of a one-helix Hlip-like gene with a gene for another one-helix transmembrane protein of unknown origin. Considering the high degree of homology between the transmembrane domain of Hlip and the first helix of Sep1 or Sep2 (71% and 79% similarity, respectively) and the first helix of Cab proteins and Seps (up to 30% identity and 50% similarity), the two-helix Seps might represent an evolutionary missing link between one- and three-helix antenna proteins. However, there is no obvious relatedness between the second helix of the eukaryotic Seps and Elip or Cab proteins. Furthermore, the second helix of Sep1 and Sep2 has no significant homology to any known protein from pro- and eukaryota. A similar situation was reported for all Cab proteins, where the second helix differs between the closely related Lhca and Lhcb. Thus, it is possible that the middle helices of Cab family members and their distant relatives have diverged during evolution to the point where similarity can no longer be discerned.
It was suggested that the early ancestors of the antenna proteins in prokaryota were part of the family of generalized light stress-response proteins localized in the plasma membranes (11). In agreement with this concept, the gene expression of Seps was positively regulated by light stress at the transcriptional and/or posttranscriptional levels. Preliminary data indicated that the accumulation of Sep transcripts under light-stress conditions was followed by their active translation and accumulation of the corresponding proteins in the thylakoid membranes of Arabidopsis (not shown). In contrast to the Elip genes, which were expressed only under light-stress conditions, low levels of Sep transcripts were present also under moderate light conditions. The regulation of Elip and Sep gene expression by light stress seems to be very specific for these two groups of proteins, because other physiological stresses such as cold, heat, wounding, desiccation, salt, or oxidative stress did not promote accumulation of Elip and Sep transcripts. This indicates that the accumulation of these transcripts in UV-A-exposed leaves was triggered by specific UV light receptor(s) and not by photooxidative damage itself.
Single-copy nuclear genes, spread over chromosomes II, III, or IV, encoded the Seps and Elips in Arabidopsis. In this respect, genomic organization of Elips in Arabidopsis resembled that present in tobacco and pea (21), where a single-copy Elip gene was reported. In barley (13) or spinach (15), multigene Elip families have been described. The nucleotide sequences of the coding regions show 60% identity for the Elip1 and Elip2 and 46% identity for the Sep1 and Sep2.
It was shown that the Elip relatives in red algae are encoded by plastid chromosomes (25, 26). The scattered distribution of Elip and Sep genes in the Arabidopsis genome indicates that the translocation of these genes from chloroplast to nuclear genome occurred individually. Furthermore, the identical position of the introns in the Elip1 and Elip2 genes and the overall sequence homology suggest that these two genes might evolve through a subsequent duplication event.
The accumulation of the Sep transcripts and their translation products under light-stress conditions suggest that similarly to Elips, the physiological function(s) of Seps is likely not light harvesting. It was proposed that higher plant Elips might be involved in a transient binding of pigments during biogenesis and/or turnover of the chlorophyll-binding proteins in the thylakoid membranes (8). A similar chlorophyll-storage function has been proposed for the IsiA, a cyanobacterial chlorophyll a-binding protein induced by iron starvation (44). However, the physiological significance of Seps in Arabidopsis is not yet understood and requires further investigation.
Acknowledgments
We thank Drs. Patrick Dessi and Klaas van Wijk for comments on the manuscript. We gratefully acknowledge the Arabidopsis Biological Resource Center for providing the cDNA clones. This work was supported by a research grant from the Swedish Natural Science Research Council, the Swedish Strategic Fund, and the Carl Tryggers Foundation.
Abbreviations
- Cab
chlorophyll a/b-binding proteins, Elip, early light-induced proteins
- Lhca
Lhcb, light-harvesting chlorophyll a/b-binding proteins of PSI or II, respectively
- PS
photosystem
- Sep
stress-enhanced proteins
- EST
expressed sequence tag
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF133716 and AF133717).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050391397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050391397
References
- 1.Wolfe R G, Cunningham F X, Durnford D, Green B R, Gantt E. Nature (London) 1994;367:566–568. [Google Scholar]
- 2.Durnford D G, Deane J A, Tan S, McFadden G I, Gantt E, Green B R. J Mol Evol. 1999;48:59–68. doi: 10.1007/pl00006445. [DOI] [PubMed] [Google Scholar]
- 3.Jansson S. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- 4.Funk C, Schröder W, Napiwotzki A, Tjus S, Renger G, Andersson B. Biochemistry. 1995;34:11133–11141. doi: 10.1021/bi00035a019. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Sandusky P, Bowlby N R, Aebersold R, Green B R, Vlahakis S, Yokum C F, Pichersky E. FEBS Lett. 1992;314:67–71. doi: 10.1016/0014-5793(92)81463-v. [DOI] [PubMed] [Google Scholar]
- 6.Wedel N, Klein R, Ljungberg U, Andersson B, Herrmann R G. FEBS Lett. 1992;314:61–66. doi: 10.1016/0014-5793(92)81462-u. [DOI] [PubMed] [Google Scholar]
- 7.Green B R, Pichersky E, Kloppstech K. Trends Biochem Sci. 1991;16:181–186. doi: 10.1016/0968-0004(91)90072-4. [DOI] [PubMed] [Google Scholar]
- 8.Adamska I, Ohad I, Kloppstech K. Proc Natl Acad Sci USA. 1992;89:2610–2613. doi: 10.1073/pnas.89.7.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miroshnichenko-Dolganov N A, Bhaya D, Grossmann A R. Proc Natl Acad Sci USA. 1995;92:636–640. doi: 10.1073/pnas.92.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funk C, Vermaas W. Biochemistry. 1999;38:9397–9404. doi: 10.1021/bi990545+. [DOI] [PubMed] [Google Scholar]
- 11.Green B R, Kühlbrandt W. Photosynth Res. 1995;44:139–148. doi: 10.1007/BF00018304. [DOI] [PubMed] [Google Scholar]
- 12.Meyer G, Kloppstech K. Eur J Biochem. 1984;138:201–207. doi: 10.1111/j.1432-1033.1984.tb07900.x. [DOI] [PubMed] [Google Scholar]
- 13.Grimm B, Kruse E, Kloppstech K. Plant Mol Biol. 1989;13:583–593. doi: 10.1007/BF00027318. [DOI] [PubMed] [Google Scholar]
- 14.Adamska I, Kloppstech K. Plant Mol Biol. 1991;16:209–223. doi: 10.1007/BF00020553. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl M, Funk C, Webster J, Bingsmark S, Adamska I, Andersson B. Photosynth Res. 1997;54:227–236. [Google Scholar]
- 16.Adamska I, Kloppstech K, Ohad I. J Biol Chem. 1993;268:5438–5444. [PubMed] [Google Scholar]
- 17.Moscovici-Kadouri S, Chamovitz D A. Plant Physiol. 1997;115:1287–1290. [Google Scholar]
- 18.Heddad M, Adamska I. In: Photosynthesis: Mechanisms and Effects. Garab G, editor. Dordrecht, The Netherlands: Kluwer; 1998. pp. 389–392. [Google Scholar]
- 19.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;24:217–221. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgenstern B, Dress A, Werner T. Proc Natl Acad Sci USA. 1996;93:12098–12103. doi: 10.1073/pnas.93.22.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolanus W, Scharnhorst C, Kühne U, Herzfeld F. Mol Gen Genet. 1987;209:234–239. doi: 10.1007/BF00329648. [DOI] [PubMed] [Google Scholar]
- 22.Yamagata H, Bowler C. Biosci Biotechnol Biochem. 1996;61:2143–2144. doi: 10.1271/bbb.61.2143. [DOI] [PubMed] [Google Scholar]
- 23.Ouvrard O, Cellier F, Ferrare K, Tousch D, Lamaze T, Dupuis J M, Casse-Delbart F. Plant Mol Biol. 1996;31:819–829. doi: 10.1007/BF00019469. [DOI] [PubMed] [Google Scholar]
- 24.Bartels D, Hanke C, Schneider K, Michel D, Salamini F. EMBO J. 1992;11:2771–2778. doi: 10.1002/j.1460-2075.1992.tb05344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reith M E, Munholland J. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- 26.Kessler U, Maid U, Zetsche K. Plant Mol Biol. 1992;18:777–780. doi: 10.1007/BF00020019. [DOI] [PubMed] [Google Scholar]
- 27.Lers A, Levy H, Zamir A. J Biol Chem. 1991;266:13698–13705. [PubMed] [Google Scholar]
- 28.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emanuelsson O, Nielsen H, von Heijne G. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frishman D, Argos P. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 31.Garnier J, Gibrat J F, Robson B. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 32.Cserzo L, Wallin L, Simon G, von Heijne G, Elofsson A. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 33.Hirokawa S, Boon-Chieng S, Mitaku S. BioInformatics. 1998;4:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 34.Sonnhammer E L L, von Heijne G, Krogh A. In: Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Menlo Park, CA: AAAI; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 35.Kyle J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 36.Attwood T K, Findlay J B C. Protein Eng. 1994;7:195–203. doi: 10.1093/protein/7.2.195. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 1–5.78. –5.79. [Google Scholar]
- 38.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Newman T, Bruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kühlbrandt W, Wang D N, Fujiyoshi Y. Nature (London) 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 41.Green B R, Pichersky E. Photosynth Res. 1994;39:149–162. doi: 10.1007/BF00029382. [DOI] [PubMed] [Google Scholar]
- 42.Pötter E, Kloppstech K. Eur J Biochem. 1993;214:779–786. doi: 10.1111/j.1432-1033.1993.tb17980.x. [DOI] [PubMed] [Google Scholar]
- 43.Adamska I, Roobol-Bóza M, Lindahl M, Andersson B. Eur J Biochem. 1999;260:453–460. doi: 10.1046/j.1432-1327.1999.00178.x. [DOI] [PubMed] [Google Scholar]
- 44.La Roche J, van der Staay G W M, Partensky F, Ducret A, Aebersold R, Li R, Golden S S, Hiller R G, Wrench P M, Larkum A W D, et al. Proc Natl Acad Sci USA. 1996;93:15244–15248. doi: 10.1073/pnas.93.26.15244. [DOI] [PMC free article] [PubMed] [Google Scholar]