Abstract
The β protein of bacteriophage λ acts in homologous genetic recombination by catalyzing the annealing of complementary single-stranded DNA produced by the λ exonuclease. It has been shown that the β protein binds to the products of the annealing reaction more tightly than to the initial substrates. We find that β protein exists in three structural states. In the absence of DNA, β protein forms inactive rings with ≈12 subunits. The active form of the β protein in the presence of oligonucleotides or single-stranded DNA is a ring, composed of ≈15–18 subunits. The double-stranded products of the annealing reaction catalyzed by the rings are bound by β protein in a left-handed helical structure, which protects the products from nucleolytic degradation. These observations suggest structural homology for a family of proteins, including the phage P22 erf, the bacterial RecT, and the eukaryotic Rad52 proteins, all of which are involved in homologous recombination.
The bacterial RecA and homologous eukaryotic Rad51 proteins play a central role in recombination and repair, and they have been the most intensively studied enzymes in homologous genetic recombination (reviewed in refs. 1–3). Proteins of the universal RecA family, which promote the recognition of homology in duplex DNA by a single strand from a DNA homolog, are central to a pathway of recombination named single strand invasion, after the underlying mechanism. It has become clear, however, that other pathways of homologous recombination exist that may employ totally different mechanisms. In particular, there is a second apparently universal pathway, the single strand annealing pathway, named after its underlying mechanism (4). In Escherichia coli this pathway is governed by proteins of the λ Red system, β protein and a 5′ exonuclease (Redα). Although an atomic-resolution crystal structure now exists for the exonuclease (5), very little information has been obtained about physical properties of β protein. Nevertheless, genetic and biochemical data provide some insight into the function of β.
β is capable of annealing complementary DNA strands (6, 7), and the annealing of two parental chromosomes cut at nonallelic sites was shown to be the major mode of Red recombination in recA mutant cells (8). Recombination by invasion of the intermediates produced by Redαβ at the site of a double-strand break was proposed (9, 10). The cooperative action of λ exonuclease and β was used to explain the Red-mediated double strand break repair (11). No pairing of single-stranded DNA (ssDNA) with double-stranded DNA (dsDNA) in the presence of β was detected (7), whereas β was shown to drive a strand displacement only when the two substrates have complementary single-stranded regions (12). β protein binds to ssDNA (7), and the minimum length of ssDNA required for stable binding was reported to be between 28 and 36 nucleotides (13). β protein was inefficient in protecting dsDNA from the action of DNase I (7), suggesting a poor binding of β to dsDNA, which was later confirmed (14). However, partially duplex substrates, and duplex products produced by the action of β protein, formed stronger associations with β than did ssDNA (14).
In this paper we describe three quaternary states of the β protein: two ring-like structures and a helical filament. The bacteriophage P22 erf (essential recombination function) protein was shown to be a functional homolog of the β protein of bacteriophage λ (15), and it was previously demonstrated that erf also forms rings (16). The E. coli RecT protein is involved in a RecA-independent pathway of recombination (17) and forms rings in the absence of DNA and helical filaments on ssDNA (18). The eukaryotic Rad52 protein is essential in both Rad51-dependent and -independent pathways, the latter including single strand annealing (19–21), and assembles into rings in the presence of ssDNA (22). On the basis of these functional and structural similarities, we suggest that β protein is a member of a family of recombination proteins found in phage, bacteria, and eukaryotes.
MATERIALS AND METHODS
β Protein.
The protein was prepared as described by Karakousis et al. (14).
DNA Substrates.
λ phage, ssM13 mp18, and dsM13 mp18 were purchased from New England Biolabs. Calf thymus DNA and φX174 (replicative form) were obtained from Sigma. φX174 fragments with blunt ends: 10 μg of φX174 DNA was digested with 50 units of HpaI (10 units/μl, Promega) in 60 μl of reaction mixture, containing commercially provided buffer for 1 hr at 37°C. DNA was then loaded on a low-melting-point agarose gel and subjected to electrophoresis. The resulting 412-, 1,264-, and 3,710-bp bands were excised and extracted. Denatured DNA (the DNA fragments from the latter experiment and calf thymus DNA): DNA was placed in a boiling water bath for 2 min and 15 sec and then quickly put on ice. dsDNA with 3′ single-stranded tails of increasing lengths: 25 μg of φX174 DNA was cut with 26 units of PstI (13 units/μl, IBI) in a 60-μl reaction mixture, containing commercially provided buffer, after 1-hr incubation at 37°C. Twelve microliters from the reaction mixture were diluted in 105.4 μl of 20 mM potassium phosphate buffer, pH 6.0, and further digested with 13 units of [λ] exonuclease (5 units/μl, Boehringer Mannheim) for 20 sec, 40 sec, and 5 min at 37°C. DNA was loaded on a low-melting-point agarose gel, electrophoresed, excised, and extracted. dsDNA with a blunt end and a 4-base protrusion: 12.5 μg of φX174 DNA was digested with 13 units of PstI in a 30-μl reaction mixture, containing commercially provided buffer, after 1-hr incubation at 37°C. NaCl was added to a final concentration of 150 mM, and 60 units of XhoI (10 units/μl, Promega) were added. The reaction mixture was incubated for another hour at 37°C. DNA was loaded on a low-melting-point agarose gel, electrophoresed, excised, and extracted.
Small Ring Preparation.
β protein (0.8 μM), 10 mM MgCl2, and 20 mM potassium phosphate buffer (pH 6.0) were incubated for 10 min at 37°C.
Large Ring Preparation.
β protein (0.8 μM), 180 μM 30-mer (a generous gift of Smita Patel, Ohio State University), 10 mM MgCl2, and 20 mM potassium phosphate buffer (pH 6.0) were incubated for 10 min at 37°C. Either 9 or 14.5 μM β protein and 90 or 120 μM heat-denatured calf thymus DNA, 10 mM MgCl2, and 20 mM potassium phosphate buffer (pH 6.0) were incubated for 10 or 25 min at 37°C. Also, 0.9 μM β protein, 91 μM ssM13 (New England Biolabs), 10 mM MgCl2, and 20 mM potassium phosphate buffer (pH 6.0) were incubated for 10 min at 37°C.
Filament Preparation.
Thirty micromolar dsDNA with 3′ single-stranded tails was incubated with 3.6 μM β protein, 10 mM MgCl2, and 20 mM potassium phosphate buffer (pH 6) for 10 min at 37°C. Also, 1.6 μM β protein, 40 μM heat-denatured 1,264-base φX174 DNA fragment, 10 mM MgCl2, and 20 mM potassium phosphate buffer (pH 6) were incubated for 30 min at 37°C.
Electron Microscopy and Image Analysis.
Samples (5 μl) were placed on carbon-coated glow discharged grids and blotted after 30 sec. Grids were then stained with 2% (wt/vol) uranyl acetate for ≈40 sec, blotted dry, and examined with a JEOL 1200 EXII electron microscope at magnification of 30,000× and accelerating voltage of 80 kV. Samples for low-angle rotary shadowing were prepared as described (23). Micrographs were scanned using a sampling that corresponded to 4 Å/pixel. Single particle image analysis employed the spider software package (24).
Rings formed in the absence of a polynucleotide.
Using a reference-free alignment procedure (25), we generated an average of 1,373 small rings. This average (data not shown) displayed a weak 12-fold symmetry, so all images were then ranked according to their 12-fold rotational power (26) and the top 900 images were aligned and averaged (Fig. 1d). The new average displays a clear 12-fold rotational symmetry. To test the reliability of this method, the 1,373 rings were also ranked according to their 10-, 11-, 13-, and 14-fold rotational powers. Averages were then created containing from 300 to 900 of the top images for each rotational symmetry-based sorting. All of these averages were quite poor, with the exception of an average of 600 of the top 11-fold rings. This average, although not as good as the average of the 900 top 12-fold images (Fig. 1d), did suggest 11 subunits.
Figure 1.
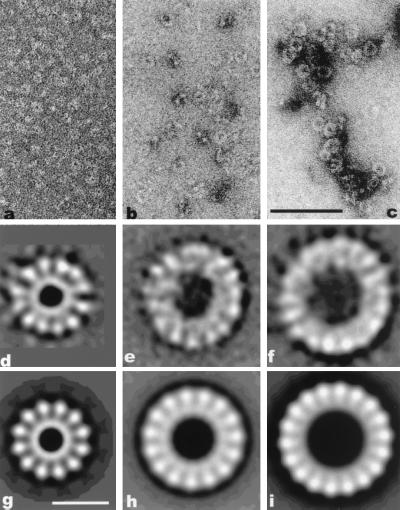
(a–c) Electron micrographs of β protein forming small rings in the presence of MgCl2 (a), oligonucleotides (b), or ssM13 (c). (Scale bar = 1,000 Å.) (d–i) Image averages. (d) Average of 900 small rings showing 12 subunits. (e) Average showing ≈15 subunits from the 300 large rings with the strongest 15-fold rotational power. The rings were formed in the presence of a 30-mer oligonucleotide. (f) Average of 600 large rings with 18 subunits assembled after incubation with heat-denatured calf thymus DNA. (g) Twelve-fold symmetrized image from d. (h) Fifteen-fold symmetrized image from e. (i) Eighteen-fold symmetrized image from f. (Scale bar = 100 Å.)
Rings formed in the presence of a 30-base oligonucleotide.
The averages of 969 images displayed rings with 15 or 16 subunits, hence the images were arranged according to their strongest 15 and 16 rotational powers. The top 200 to 750 rings were selected for each rotational power, aligned, and averaged. Both groups of rings gave averages with 15- or 16-fold symmetry with uneven resolution along the circumference, 180–184 Å in diameter with a central hole of ≈70 Å.
Rings formed in the presence of long ssDNA.
An average of 1,942 rings, assembled in the presence of ssDNA, showed a ring composed of ≈17–19 subunits (not shown). The aligned images were then ranked according to their 17-, 18-, and 19-fold rotational power, and the top 300 to 1,000 were selected for each symmetry. These subsets were then independently aligned, and the rotational power spectrum was computed for each image and averaged. Only the averaged power spectrum from the images selected for 18-fold symmetry showed a distinct peak. Therefore, rings were selected for the strongest 18-fold power, and an average of the 600 rings with the strongest 18-fold rotational power is shown in Fig. 1f. It can be seen that this average is somewhat asymmetric, with subunits on one side resolved more distinctly than those on the opposite side. This asymmetry would arise from disorder, where the alignment procedure can bring subunits on one side into register by rotational and translational shifts, but not on the other side at the same time.
Helical filaments.
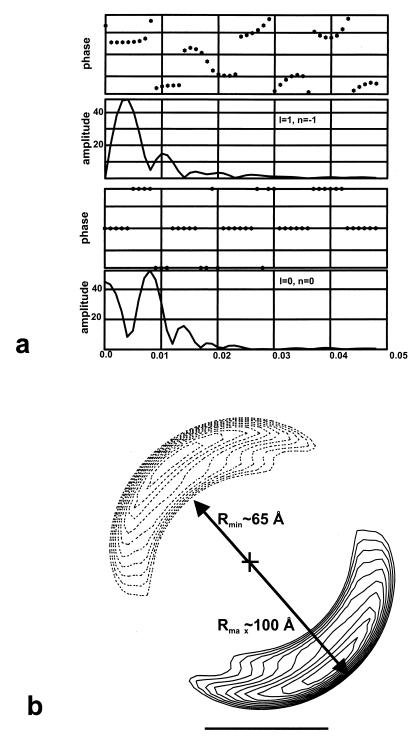
Filament images were straightened by using a spline algorithm (27). Fourier transforms of individual filaments did not reveal features arising from the individual subunits, but rather showed only the diffraction associated with a continuous helical strand. Because the strand cannot be actually continuous, but must arise from individual β subunits, the inability to see the subunit spacing is most likely caused by a large degree of disorder. We therefore created a simulated fiber pattern by averaging the Fourier transforms of 59 filaments, each having a mean helical pitch near 125 Å, in an attempt to enhance any weak features. This transform (Fig. 2f) shows only the single layer line (at ≈1/125 Å−1) arising from the left-handed one-start helix. Treating the filament as a continuous helix, we extracted the equator and the 1/125 Å−1 layer line from each of these 59 filaments, and averaged them together, searching for the axial translation and polarity (up versus down) that would bring each into best register with all of the others. After seven cycles of averaging, 34 filaments were found that averaged together quite well, with a clear polarity (average up–down phase residuals of 30 versus 50). The averaged layer lines from these filaments are shown in Fig. 4a, and a reconstruction generated from these layer lines is shown in Fig. 4b.
Figure 2.
(a–e) Electron micrographs of β protein forming helical filaments on dsDNA. (a) Compressed helix with a pitch less than 115 Å. (b) Helix with a pitch ranging from 115 to 135 Å. (c) Extended helix with a pitch greater than 135 Å. (d) Rotary-shadowed filament, showing left-handed protein helix. (e) Uncoiling of the filaments (arrows). (Scale bar = 700 Å.) (f) Fourier transform (simulated fiber pattern) generated by averaging the intensities from transforms of 59 individual β-DNA filaments. Only the single layer line at 1/125 Å−1 is seen. (g) Pitch distribution of the β-DNA filaments.
Figure 4.
(a) Averaged layer lines from 34 filaments. (b) Cross section of the helical reconstruction generated from the layer lines in a. The reconstruction is shown by two slices, one at z = 0 Å (solid lines) and one at z = −62.5 Å (dotted lines), half of a helical turn away. The outer contour shown encloses a volume of 700,000 Å3 for one helical turn, and was chosen so as to yield a thickness (between Rmin and Rmax shown) of about 35 Å. This was the approximate thickness of the strands seen in regions (Fig. 2e) where the filaments unravel. The cross sections show that the helix consists of a single compact nucleoprotein strand, as shown in the model of Fig. 5b.
RESULTS
Small Rings.
When β protein alone, in the absence of a polynucleotide, was examined by electron microscopy, large numbers of rings were seen (Fig. 1a). Mg2+ was essential for the assembly of these rings. The signal-to-noise ratio in the raw images was insufficient to reliably see internal structure in these rings, so image averaging was employed. The results are consistent with either a polymorphism in the number of subunits (11 or 12), or rings with a fixed symmetry (12 subunits), but with a large degree of disorder.
The averaged ring (Fig. 1d) is about 145 Å in diameter, with a central hole ≈35 Å in diameter. Surrounding the central hole there is a continuous ring of density, with 12 knob-like projections extending out from it. A 12-fold symmetrized image of this average is shown in Fig. 1g. Since a monomer of the β protein has a molecular weight of 25,814 (28), it would be about 20 Å in radius, assuming that it is perfectly spherical (and assuming a partial specific volume for proteins of 0.75 cm3/g). Thus, the size of the observed ring is consistent with containing ≈12 subunits of the β protein, and the possibility that two β subunits are contained in each “spoke,” or that two spokes are part of the same β subunit, appears unlikely.
Large Rings.
In the presence of ssDNA or oligonucleotides, β protein formed large rings as well as small rings. Unlike the small rings, these large rings were not seen at all in the absence of DNA. The addition of 58-fold excess of ssM13 over β resulted in the formation of numerous large rings, and the absence of any small rings. This observation suggests that the small rings convert directly or indirectly (by disassembly and reassembly) into large rings.
An oligonucleotide containing 30 bases was adequate to produce many larger rings when it was incubated with the β protein (Fig. 1b). Image analysis was used to examine the structure of these rings, and the results suggested that these rings were composed of 15 or 16 subunits. A raw average of 300 of these rings is shown in Fig. 1e, while a 15-fold symmetrized version of this average is shown in Fig. 1h. The averaged ring is ≈185 Å in diameter, with a central hole of about 75 Å.
In the presence of ssDNA, the rings formed were larger than those seen with an oligonucleotide, but their structure appears similar (Fig. 1c). An average of these larger rings is ≈210 Å in diameter (Fig. 1f), with a central hole ≈100 Å in diameter. An 18-fold symmetrized version of this average is shown in Fig. 1i. The size of the individual subunits within the average is comparable to that seen in the small rings (Fig. 1g), so we conclude that each subunit corresponds to a single β protein.
Mg2+ and long ssDNA both stabilized the formation of the large rings. In the absence of Mg2+ and the presence of an oligonucleotide, no large rings were observed. However, in the presence of a ssDNA (1.2 kb) and the absence of Mg2+, large rings were observed, but many were incomplete and the circular ring structure was disrupted. DNA ends are not required for ring formation, as circular ssDNA molecules were effective as substrates. However, circular dsDNA failed to induce the formation of the large β rings.
Helical Filaments.
In the presence of DNA, filaments of β protein were also found (Fig. 2 a–c). These filaments are helical (as shown by further analysis below), have a diameter similar to that of the large rings (≈200 Å), and have a variable pitch.
Only short helices (up to ≈750 Å) were observed with noncomplementary ssDNA. The length of the filaments increased up to 4 times when heat-denatured complementary strands (a 3,710-base fragment from φX174) were introduced as substrates. The length of the filaments was the longest (≈6,000 Å) when λ DNA (linear dsDNA, 48.5 kbp) was used. Filament formation was enhanced by the presence of single-stranded overhangs. Linearized φX174 dsDNA treated with λ exonuclease for 20 sec, 40 sec, and 5 min gave rise respectively to ≈3, 7, and 10 times more β filaments than dsDNA with a blunt end and a 4-base protrusion. DNA ends were required, as covalently closed circular dsDNA failed to induce β polymerization into filaments.
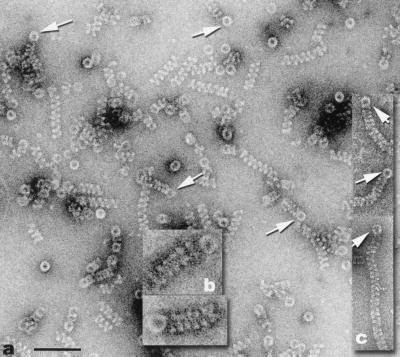
β large rings and filaments had different times of appearance during the course of DNA renaturation. Heat denatured 3.7-kb φX174 fragment was mixed with β and incubated for 10 sec and 30 sec at 37°C. Only large rings were observed after 10-sec incubation, whereas large rings and filaments were detected after 30-sec incubation. Filaments originating from rings were very often observed under DNA annealing conditions (Fig. 3 a and b). This association did not seem coincidental because of the high frequency of occurrence. When a heat-denatured 1,264-base φX174 DNA fragment was incubated with β, many large rings were attached to filaments of less than 9 turns (the maximum number of helical turns observed was 12–13). The average of 31 such rings had a ≈216-Å diameter, which suggested a structure of ≈18 subunits (see above). Rings and filaments were joined frequently on dsDNA with single-stranded tails (Fig. 3c). We can exclude the possibility that the orthogonal orientation of the ring to the filament axis is due to adsorption to the grid or specimen preparation for negative stain electron microscopy, since the same relationship was observed in frozen–hydrated specimens (data not shown).
Figure 3.
(a and b) Electron micrographs showing the association of helical filaments and large rings (arrows), after β protein was incubated with MgCl2 and heat-denatured 1,264-base φX174 fragment. (c) Filaments originating from large rings when partially duplex φX174 DNA was incubated with β and MgCl2. (Scale bar for a and c = 1000 Å. The magnification of b is 2× that of a and c.)
The length of the filaments was not related to the length of the single-stranded overhangs, suggesting that the filaments form mainly on the dsDNA. The polarity of the ssDNA tails was not important, as DNA with both 3′ (dsDNA digested with λ exonuclease) and 5′ single-stranded tails (λ phage cohesive ends) were adequate substrates for nucleation of the filaments.
In the presence of dsDNA, the formation of helices did not require Mg2+. The same was not true for filaments formed when complementary ssDNA was the starting substrate. The incubation of β protein with heat-denatured DNA and Mg2+ resulted in the formation of many large rings and filaments of all lengths. Without Mg2+ in this reaction only a very few short filaments and many abnormal, incomplete rings were observed.
The helical filaments were left-handed as determined by examining rotary shadowed specimens (Fig. 2d) and existed in three states: the majority had a medium pitch ranging from 115 to 135 Å, but compressed helices, with a pitch as low as 75 Å, and extended helices with a pitch as high as 205 Å were also encountered (Fig. 2 a–c and g). Sometimes, portions of the helices were fully uncoiled (Fig. 2e). In an effort to see whether the polymorphism and unraveling were due to specimen preparation for negative-stain electron microscopy, images of unstained, frozen–hydrated specimens were obtained by cryo-electron microscopy. These images (data not shown) were very similar to those obtained by negative staining, suggesting that specimen preparation was not responsible for the range of structures observed in Fig. 2.
A three-dimensional reconstruction (Fig. 4b) was generated from an average of 34 filaments, each having a pitch of ≈125 Å. The helical image analysis and three-dimensional reconstruction provide no direct evidence for the number of β subunits per turn of the left-handed helix, so other methods were used. An analysis of 27 short unraveled segments of the helical filaments (e.g., Fig. 2e) showed a weak periodicity of about 30 Å. The path length along the 125-Å pitch helix at the center of the density (r = 83 Å) is about 530 Å for one turn. Since it is reasonable to assume that the spacing of subunits is similar within the helix and the unraveled strand, this periodicity within the unraveled segments would correspond to about 18 subunits per turn within the helix. The volume enclosed by the reconstruction shown in Fig. 4b corresponds to that expected from about 20 β subunits.
The images provide no direct evidence for the position of the DNA within the β helix, but the possibility that the DNA runs linearly along the axis of the helix can be excluded by the density distribution shown in Fig. 4b. If this were the case, there would be no contact between the protein and the DNA. The observed density distribution suggests that the radius of the DNA will be somewhere between about 65 and 100 Å. The longest filaments on a 3.7-kb double-stranded fragment of φX174 DNA contained ≈25 helical turns. Assuming that the DNA was not stretched, this would correspond to a DNA path length of 503 Å per turn [3.4 Å × (3,700/25)]. With a 1,264-base double-stranded fragment of φX174 DNA, the longest β filaments formed had ≈12 helical turns, corresponding to a path length of 340 Å per turn. Similarly, with a 412-bp φX174 dsDNA fragment, the longest filaments formed contained ≈4 helical turns, corresponding to a path length of 350 Å per turn. Because the path length per 125-Å pitch helical turn, lp, is given by lp = [(2πr)2 + 1252]1/2, these values for the path length correspond to radii of 77 Å, 50 Å, and 52 Å, respectively. The disparity between the first value and the second two may arise from the fact that when longer dsDNA substrates were used, the β filaments did not continue to grow in length. For example, when intact λ DNA was used (48.5 kbp) filaments longer than 50 turns were never observed. Similarly, when calf thymus dsDNA was used, the longest filaments were of comparable length. This suggests that the value of ≈50-Å radius is the best estimate, and the DNA is coiling at the inner radius of the β filament. This would also be consistent with data showing a partial protection of the dsDNA by β protein (7), making it unlikely that the DNA is exposed on the outside of the filament.
DISCUSSION
Three quaternary states of β protein were discovered: small rings, large rings, and helical filaments. Evidence suggests that the small rings (≈145 Å in diameter) are inert and inactive, whereas the large rings (≈185–210 Å in diameter) are the active form of β in the initial catalysis of annealing. With 30-base oligonucleotides, β protein assembled into 15- or 16-subunit rings. The behavior of β in the presence of oligonucleotides is consistent with the results of Mythili et al. (13), who reported that 28–36 nucleotides was the minimum length required for a stable binding of β to DNA. Thus, the form of β detected in this binding assay was the large ring (15 or 16 subunits), while the 12-subunit ring did not bind to ssDNA.
In the presence of complementary ssDNA and Mg2+, β formed numerous large rings and filaments. The two forms appeared to be related, as a ring was frequently found at one of the helical ends. The large rings bind to one DNA strand and initiate its annealing to the complementary strand. Once initiated, the annealing can proceed spontaneously, followed by the formation of a ring-nucleated filament on the duplex DNA. The fact that under reannealing conditions rings are the first β structure to form on the ssDNA, followed by filament assembly as the renaturation progresses, supports our hypothesis that rings initiate the filaments rather than terminate them.
Evidence for the possible mechanism of β-catalyzed DNA renaturation is provided by the preferential binding of the two quaternary structures to different substrates: large rings to ssDNA and filaments to dsDNA. We have shown that filament formation on dsDNA requires the presence of an end, since no filaments are formed on circular dsDNA. Large rings and filaments are formed on partially duplex DNA, and there is reduced filament formation on blunt-ended dsDNA. Because large rings will form on the ssDNA tails present in the partially duplex DNA, there is a higher likelihood for the filaments to nucleate adjacent to the large rings than on the opposite end of the duplex. When the entire DNA fragment is renatured the large rings may detach from the dsDNA, as full-length filaments (12–13 helical turns on 1,264-bp DNA fragment) associated with a ring are never detected.
We have assumed that the main role of the large β rings is to catalyze the initiation of reannealing, involving the recognition of complementarity between two strands, which is the rate-limiting step (29). In this model, the extension of reannealing will proceed rapidly and spontaneously, and the β filament forms on dsDNA as it spontaneously anneals. A possible active role for filaments is that they displace any additional β rings bound to both complementary strands.
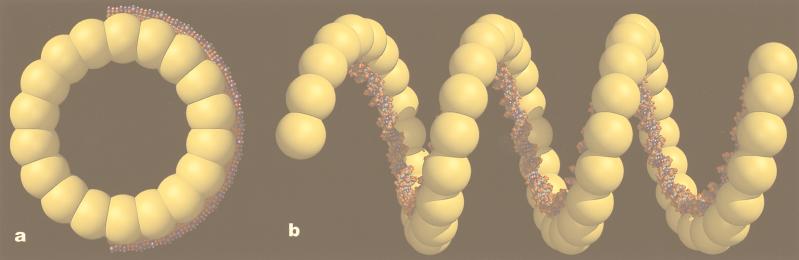
On the basis of our observations, we have generated a model for the relationship between β protein and DNA (Fig. 5). The annealing activity of the large rings (Fig. 5a) is very likely to follow the similar model proposed for the P22 erf protein (16), in which the ssDNA is wrapped around the outside of the rings, exposing the bases. The facts that longer DNA stabilizes rings formed in the absence of Mg2+ and produces larger rings than those formed by oligonucleotides are consistent with such a mode of binding. If the interaction of the ssDNA with the large ring were local (at the level of a single subunit), we would not expect to see either of these phenomena, because the oligonucleotide would bind in the same manner as the ssDNA. However, if there is a wrapping of the ssDNA around the outside of the ring, a long ssDNA would be binding many subunits, whereas a short oligonucleotide would be binding only a few subunits.
Figure 5.
Models for the complex of the large β ring and ssDNA (a) and the β helix formed on dsDNA (b). The ssDNA is believed to wrap around the outside of the 18-subunit rings, as proposed for phage P22 erf protein rings (16). The exposure of the bases would provide the catalytic role played by these rings in DNA annealing. The helical filaments are formed by β protein on preformed dsDNA, or on dsDNA products of the β-catalyzed annealing reaction. The β filament induces a left-handed supercoil in the DNA, with about one supercoil turn for every 100 bp.
One of the barriers to DNA annealing is the secondary structure that random ssDNA will form, so a catalytic role will be provided by the protein if it can maintain the bases in an exposed conformation. In the model presented here the DNA backbone is bound to the β ring, which would provide protection against nucleolytic degradation, in agreement with the observed protection from exo- and endonucleolytic attack provided to oligonucleotides by β (14).
We envision that the annealing reaction proceeds via the interaction of a single β–ssDNA ring complex with a complementary naked ssDNA, but experiments are needed to directly test this. After the initiation of annealing, the two single strands zipper together into a dsDNA that is supercoiled within a helical filament formed by β protein (Fig. 5b). A model has been generated assuming that there is no stretching of the DNA, which would lead to about one left-handed supercoil turn per 100 bp. Interestingly, this is not that different from the amount of supercoiling found in the eukaryotic nucleosome, which is 1.75 left-handed supercoils per 146 bp (or 1 left-handed supercoil per 83 bp) (30).
There are two possibilities for the origin of this supercoiling. One is that the β protein imposes it on the DNA by the formation of a helical β “scaffold.” The other is that the local interaction of β with dsDNA results in a change in the twist of the DNA, from about 10.5 to 9.7 bp per turn, so that a left-handed supercoiling is induced by the DNA to relax this change in twist. The latter possibility would be likely only if the β protein formed a very flexible structure, but the existence of the unraveled strand of the β–dsDNA complex (Fig. 2e) is consistent with such a very flexible structure. However, we have no direct information about any local change in twist of the DNA, so this possibility remains speculative.
The uncoiling of the β–dsDNA helical filament into a linear strand suggests a large conformational flexibility for this structure. Similar helix unraveling has been seen for a nucleocapsid protein–RNA complex from an animal virus (31) and for bacterial pili (32). Because the large ring has a diameter similar to that of the filament, and it is likely that there are ≈18 subunits of β in both the ring and one turn of the filament, it is possible that the subunit–subunit interactions are similar in both. This might arise if the ring and filament are two stable forms of the same linear polymer of β and DNA (present in the unraveled regions of the helix), with the ring being a β–ssDNA complex and the filament being a β–dsDNA complex.
The erf protein is the bacteriophage P22 analog of β protein functioning in homologous recombination (33). erf anneals complementary ssDNA strands (34) by means of ring-like oligomers which were suggested to bind DNA around their outer surface (16). erf rings resemble, both functionally and structurally, the large β rings which we also envision to wrap ssDNA externally (Fig. 5a).
RecT is a member of the RecE recombination pathway of E. coli, which is analogous to the Red pathway of the λ phage. RecT promotes homologous pairing and strand exchange (35, 36), and it carries out double-strand break repair (37). It exists in multiple forms, but only one of them, a helical filament, binds to DNA (18). Even though β protein and RecT are functionally and to some extent structurally similar, their DNA-binding modes are quite different. The RecT rings do not require DNA, whereas ssDNA is essential for the large β rings. On the other hand, RecT filaments form on ssDNA whereas β filaments are generally associated with dsDNA. Further research is needed to sort out the functions of each RecT quaternary state.
Rad52 is another protein with annealing activity (19, 20) that is involved in DNA recombination and repair (38). It has been demonstrated biochemically that human Rad52 self-associates (39), and yeast Rad52 assembles into rings as shown by electron microscopy (22). Rad52 forms complexes with both ssDNA and dsDNA, whereas Rad52 rings bind only to ssDNA (22).
Functional similarities between β on the one hand and E. coli RecT and P22 erf, on the other, have been previously suggested (15, 35), and it has been shown that both RecT (18) and erf (16) form rings. Our observation that the active form of β is a ring is particularly interesting, given the role of eukaryotic Rad52 in the single strand annealing pathway of recombination (4, 40) and the recent demonstrations that Rad52 from both Saccharomyces cerevisiae (22) and humans (41) forms rings. This finding suggests that while sequence homology among these different proteins is insignificant, they are likely to be structural and functional homologs.
Acknowledgments
We thank Jack Griffith for initial observations that motivated this project. This research was supported by National Institutes of Health Grant GM35269 and Human Frontier Science Program Grants RG335 (to E.H.E.) and GM33504 (to C.M.R.).
ABBREVIATIONS
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
References
- 1.West S C. Philos Trans R Soc London B. 1995;347:21–25. doi: 10.1098/rstb.1995.0004. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roca A I, Cox M M. Prog Nucleic Acids Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 4.Haber J E. BioEssays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 5.Kovall R, Matthews B W. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 6.Kmiec E, Holloman W. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 7.Muniyappa K, Radding C M. J Biol Chem. 1986;261:7472–7478. [PubMed] [Google Scholar]
- 8.Stahl M M, Thomason L, Poteete A R, Tarkowski T, Kuzminov A, Stahl F W. Genetics. 1997;147:961–977. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stahl F W, Kobayashi I, Stahl M M. J Mol Biol. 1985;181:199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- 10.Thaler D S, Stahl M M, Stahl F W. J Mol Biol. 1987;195:75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi N, Kobayashi I. Proc Natl Acad Sci USA. 1990;87:2790–2794. doi: 10.1073/pnas.87.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Karakousis G, Chiu S, Reddy G, Radding C. J Mol Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 13.Mythili E, Kumar K, Muniyappa K. Gene. 1996;182:81–87. doi: 10.1016/s0378-1119(96)00518-5. [DOI] [PubMed] [Google Scholar]
- 14.Karakousis G, Ye N, Li Z, Chiu S, Reddy G, Radding C. J Mol Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 15.Poteete A R, Fenton A C. Virology. 1984;134:161–167. doi: 10.1016/0042-6822(84)90281-2. [DOI] [PubMed] [Google Scholar]
- 16.Poteete A, Sauer R, Hendrix R. J Mol Biol. 1983;171:401–418. doi: 10.1016/0022-2836(83)90037-2. [DOI] [PubMed] [Google Scholar]
- 17.Noirot P, Kolodner R D. J Biol Chem. 1998;273:12274–12280. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- 18.Thresher R J, Makhov A M, Hall S D, Kolodner R, Griffith J D. J Mol Biol. 1995;254:364–371. doi: 10.1006/jmbi.1995.0623. [DOI] [PubMed] [Google Scholar]
- 19.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy G, Golub E I, Radding C M. Mutat Res. 1997;377:53–59. doi: 10.1016/s0027-5107(97)00057-2. [DOI] [PubMed] [Google Scholar]
- 21.Sugiyama T, New J H, Kowalczykowski S C. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 23.Egelman E H, Stasiak A. J Mol Biol. 1988;200:329–349. doi: 10.1016/0022-2836(88)90245-8. [DOI] [PubMed] [Google Scholar]
- 24.Frank J, Shimkin B, Dowse H. Ultramicroscopy. 1981;6:343–358. [Google Scholar]
- 25.Penczek P, Radermacher M, Frank J. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 26.Yu X, Jezewska M J, Bujalowski W, Egelman E H. J Mol Biol. 1996;259:7–14. doi: 10.1006/jmbi.1996.0297. [DOI] [PubMed] [Google Scholar]
- 27.Egelman E H. Ultramicroscopy. 1986;19:367–373. doi: 10.1016/0304-3991(86)90096-3. [DOI] [PubMed] [Google Scholar]
- 28.Sanger F, Coulson A R, Hong G F, Hill D F, Petersen G B. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 29.Wetmur J G. Annu Rev Biophys Biomol Struct. 1998;5:337–361. [Google Scholar]
- 30.Van Holde K E. Chromatin. New York: Springer; 1988. [Google Scholar]
- 31.Egelman E H, Wu S S, Amrein M, Portner A, Murti G. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullitt E, Makowski L. Nature (London) 1995;373:164–167. doi: 10.1038/373164a0. [DOI] [PubMed] [Google Scholar]
- 33.Poteete A. Virology. 1982;119:422–429. doi: 10.1016/0042-6822(82)90101-5. [DOI] [PubMed] [Google Scholar]
- 34.Poteete A, Fenton A. J Mol Biol. 1983;163:257–275. doi: 10.1016/0022-2836(83)90006-2. [DOI] [PubMed] [Google Scholar]
- 35.Hall S D, Kane M F, Kolodner R D. J Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall S, Kolodner R. Proc Natl Acad Sci USA. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusano K, Takahashi N K, Yoshikura H, Kobayashi I. Gene. 1994;138:17–25. doi: 10.1016/0378-1119(94)90778-1. [DOI] [PubMed] [Google Scholar]
- 38.Benson F E, Baumann P, West S C. Nature (London) 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 39.Shen Z, Peterson S R, Comeaux J C, Zastrow D, Moyzis R K, Bradbury E M, Chen D J. Mutat Res. 1996;364:81–89. doi: 10.1016/0921-8777(96)00025-0. [DOI] [PubMed] [Google Scholar]
- 40.Baumann P, West S C. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 41.Van Dyck E, Hajibagheri N M A, Stasiak A, West S C. J Mol Biol. 1998;284:1027–1038. doi: 10.1006/jmbi.1998.2203. [DOI] [PubMed] [Google Scholar]