Abstract
The binding of the cell surface molecule CD58 (formerly lymphocyte function-associated antigen 3) to its ligand, CD2, significantly increases the sensitivity of antigen recognition by T cells. This was the first heterophilic cell adhesion interaction to be discovered and is now an important paradigm for analyzing the structural basis of cell–cell recognition. The crystal structure of a CD2-binding chimeric form of CD58, solved to 1.8-Å resolution, reveals that the ligand binding domain of CD58 has the expected Ig superfamily V-set topology and shares several of the hitherto unique structural features of CD2, consistent with previous speculation that the genes encoding these molecules arose via duplication of a common precursor. Nevertheless, evidence for considerable divergence of CD2 and CD58 is also implicit in the structures. Mutations that disrupt CD2 binding map to the highly acidic surface of the AGFCC′C′′ β-sheet of CD58, which, unexpectedly, lacks marked shape complementarity to the equivalent, rather more basic CD58-binding face of human CD2. The specificity of the very weak interactions of proteins mediating cell–cell recognition may often derive largely from electrostatic complementarity, with shape matching at the protein–protein interface being less exact than for interactions that combine specificity with high affinity, such as those involving antibodies.
Antibodies to CD2 and CD58 (lymphocyte function-associated antigen 3) were among the first found to block the functions of human T lymphocytes in in vitro assays (reviewed in ref. 1). The cognate ligand-receptor relationship of CD2 and CD58, the first heterophilic protein interaction to be identified at the cell surface, was directly established when purified CD2 was shown to bind to cellular CD58 (2). CD58 is expressed in both haemopoietic and nonhaemopoietic lineages, including the endothelium (3). The binding of CD2 to CD58 is known to significantly enhance the efficiency of antigen recognition in vitro (4), and studies of CD2-deficient mice indicate that the interaction of CD2 with the murine ligand CD48 influences both positive selection and T cell activation (5).
The sequencing of cDNAs encoding CD58 revealed that, like CD2, the extracellular region of CD58 consists of single V-set and C2-set Ig superfamily (IgSF) domains (reviewed in ref. 6). Along with CD48 and several other molecules, CD2 and CD58 belong to a subset of the IgSF that is likely to have arisen via the duplication of an ancestral gene encoding a homophilic cell–cell recognition molecule (6).
Studies of the interaction of CD2 with its ligands have been informative with respect to the mechanism(s) of protein–protein recognition at the cell surface (reviewed in ref. 7). The crystal structures of soluble forms of rat (8) and human (9) CD2 [soluble CD2 (sCD2)] provided the initial views of the complete extracellular regions of cell adhesion molecules. This work drew attention to charged residues clustered at the ligand binding site of CD2 and to the unusual flatness of the binding site, two structural features that distinguish this surface from all other well characterized sites of protein–protein recognition. The affinities of the interactions of CD2 and its ligands in the solution phase, and at the cell surface, have been analyzed in detail and have been shown to be very low and characterized by extremely fast off-rates [koff (in solution) >5 s−1; reviewed in ref. 7]. It has been proposed that protein interactions at the cell surface may generally need to have very low affinities to facilitate highly dynamic, reversible cell–cell contacts and because the generation of an appropriate intracellular response, when binding initiates signaling, may require very brief interactions (7, 10).
A link between the unusual structural properties of the ligand binding face of CD2 and its capacity to effect weak specific binding at the cell surface was implied by the finding that electrostatically favorable, energetically neutral interactions involving the charged residues clustered in the ligand binding site appear to ensure that ligand recognition by CD2 is both weak and specific (11). The analysis of the crystal structure of the CD2-binding domain of CD58, described here, strengthens this view.
MATERIALS AND METHODS
Expression of Chimeric CD58.
Expression and crystallization methods are described in detail elsewhere (T. D. Butters, L.M.S., K.H., S.I., D.I.S., E.Y.J., and S.J.D., unpublished work). In brief, in vitro mutagenesis was used to replace the sequence encoding the C2-set domain of a soluble form of CD58 with the analogous rat CD2 domain 2 sequence to encode a chimeric soluble form of CD58 (cCD58) starting with the native CD58 N-terminal sequence “FSQQ… ,” continuing to the CD58 domain 1/rat CD2 domain 2 junctional sequence “… . LYVL/EMVS… . ,” and ending with the C-terminal sequence “… CPEK” of domain 2 of rat sCD2 (ref. 8 and references therein). The construct was expressed in Lec3.2.8.1 cells by using the glutamine synthetase-based gene expression system as described (12) and in the presence of 0.5 mM N-butyldeoxynojirimycin (a kind gift of R. A. Dwek of the Glycobiology Institute, Oxford). The deglycosylated, affinity-purified protein was crystallized by using the Crystal Screen and Crystal Screen 2 crystallization kits (Hampton Research, Riverside, CA). Data were collected from crystals grown in the presence of the precipitants ammonium sulfate (unbuffered) or sodium chloride [buffered with 0.1M sodium acetate (pH 4.6)].
CD2 and Antibody Binding.
All binding experiments were performed at 25°C on a BIAcore2000 (BIAcore AB, Stevenage, Herts, United Kingdom) essentially as described (13). For immobilizing cCD58, purified monoclonal anti-CD2 domain 2 antibody, OX54, at 20–60 μg/ml in 10 mM sodium acetate (pH 5.0) was covalently coupled to a research grade CM5 sensor chip (BIAcore AB) via primary amine groups by using the Amine Coupling Kit (BIAcore AB). Soluble CD58 (sCD58) was coupled by thiol coupling as described (13).
Structure Determination and Analysis.
Diffraction data were collected in-house at 20°C and on BM14 (λ = 0.898 or 0.946 Å) of the European Synchrotron Radiation Facility (Grenoble, France) at 100 K by using, respectively, 18- and 34.5-cm MAR-Research (Hamburg, Germany) detectors (Table 1). Data were processed and scaled by using denzo and scalepack (14). The structure was determined by molecular replacement using amore (15), the coordinates of rat sCD2 (Protein Data Bank code 1HNG), and the in-house x-ray data set (all observed data for 15- to 3.0-Å resolution). An unambiguous solution was found for a single molecule that, after rigid body refinement, gave an Rcryst of 37.4%. Polyalanine was then substituted for the domain 1 sequence. Further refinement was carried out in xplor (16) with manual rebuilding in o (17). Two-domain rigid-body refinement resulted in a relative reorientation of the domains. After simulated annealing and positional and overall B factor refinement, 2Fo − Fc and Fo − Fc electron density maps showed bias-free density and allowed the correct sequence to be substituted into domain 1. Subsequent refinement was carried out against the high resolution x-ray data set, with bulk solvent correction, by using all observed data but with 5% of the data set aside for Rfree cross-validation. The refined atomic model comprises residues 1 to 171 of the chimera, 3 N-linked GlcNAc carbohydrates, and 197 ordered water molecules. Of the nonglycine residues, none lie in energetically disallowed regions of the Ramachandran plot, according to procheck (18). Further statistics for the final model are given in Table 1.
Table 1.
Data collection and refinement statistics
| Crystal space group | P3221 | |
| Cell parameters, Å | a = 118.1, b = 118.1, c = 52.1 (ambient temperature) | |
| Cell parameters, Å | a = 116.4, b = 116.4, c = 51.4 (cryocooled) | |
| Data processing | In house | European Synchrotron Radiation Facility |
| Maximum resolution | 3 Å | 1.8 Å |
| No. of observations | 44,303 | 187,375 |
| Unique reflections | 8,532 | 37,412 |
| Completeness (percent) | 99.7 | 100.0 (100.0)* |
| I/σ | 10.2 | 15.9 (2.3) |
| Rmerge (percent)† | 17.9 | 9.0 (42.3) |
| Refinement | ||
| Data range, Å | 15–1.8 | |
| Reflections (F > 0) | 35,955 (4,095) | |
| Completeness (percent) | 96.3 (88.8) | |
| Reflections in Rfree set | 1,785 | |
| Nonhydrogen atoms | 1,650 (protein, 1411; H2O, 197; sugar, 42) | |
| rms Δ bond lengths, Ň | 0.011 | |
| rms Δ bond angles, degrees | 1.601 | |
| rms Δ B-factors for bonded atoms, Ň | 2.2 | |
| Mean B factor, protein/sugar/H2O, Ň | 36.9/78.3/48.5 | |
| Mean B factor, main/sidechain, ņ | 34.5/39.1 | |
| Rfree (percent) | 24.34 (34.52) | |
| Rcryst (percent) | 20.18 (33.47) | |
Values in parenthesis correspond to the highest resolution shell (1.86–1.8 Å for data processing and 1.88–1.8 Å for refinement).
Rmerge = Σ|I − 〈I〉|/Σ〈I〉; Rcryst = Σ∥Fobs| − |Fcalc∥/Σ|Fobs|; Rfree is as for Rcryst, but calculated for a test set comprising reflections not used in refinement.
Root mean squared deviations (rms Δ) in bond length and angles are given from ideal values.
Structural superpositions were performed by using shp (19). Figs. 2, 3, and 5 were produced by using bobscript (20), raster 3d (21), and volumes (R. Esnouf, personal communication). grasp (22) was used for the electrostatic analysis and surface display in Fig. 4.
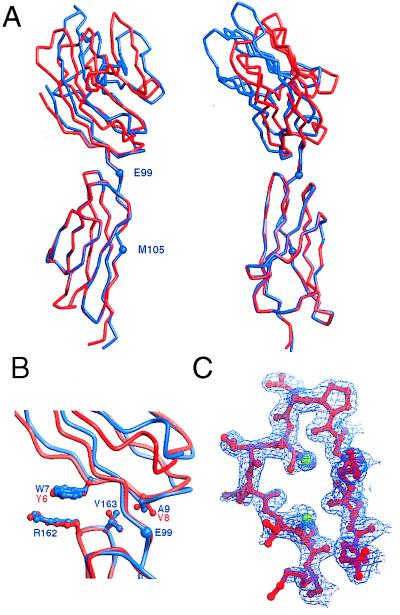
Figure 2.
The structure of cCD58. (A) Two orthogonal views of schematic α-carbon representations of cCD58 (red) and rat sCD2 (blue), superimposed on domain 2 of each molecule. Residues E99 and M105 at the boundaries of the linker region of rat CD2 domain 2 are marked with blue spheres. (B) The interdomain region is shown enlarged with the sidechains of the key nonlinker interface residues of cCD58 (red) and CD2 (blue) drawn as ball-and-stick models. (C) Portion of the 2Fo − Fc electron density map (final model phases) contoured at 1σ in the region of the FG loop of domain 1 of CD58. Running from right to left, residues 76–87 are shown in ball and stick representation and are colored red. The green spheres represent modeled water.
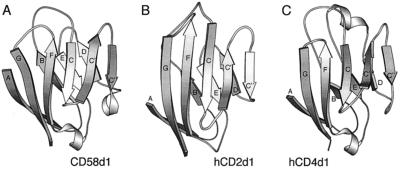
Figure 3.
Illustrations of the folds of domain 1 of CD58 (A), human CD2 (B), and CD4 (C). The domains are shown in the same orientation as defined by the pairwise superpositions referred to in the text. The coordinates for CD4 domain 1 were obtained from the Protein Data Bank.
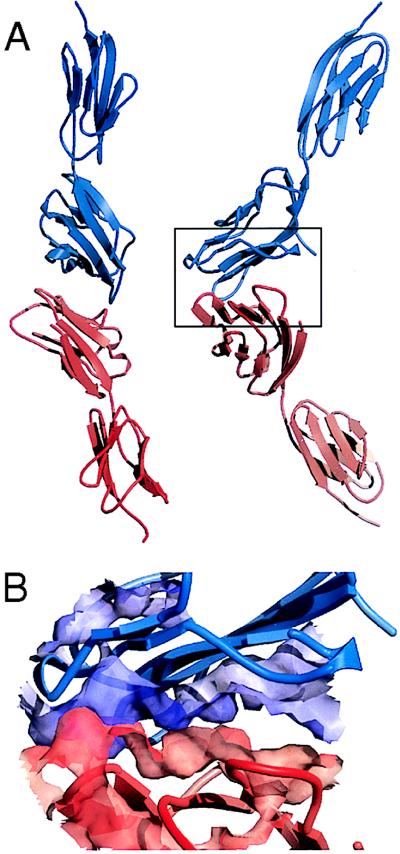
Figure 5.
Lack of shape complementarity in the presumptive complex of CD58 and CD2. (A) Two orthogonal views of cCD58 (red) and human sCD2 (blue) docked according to the orthogonal homodimeric human sCD2 crystal lattice contacts (9). (B) The region boxed in A is enlarged, and the solvent-accessible molecular surfaces of central sections of the interacting β-sheets of CD58 and CD2 are depicted semitransparently.
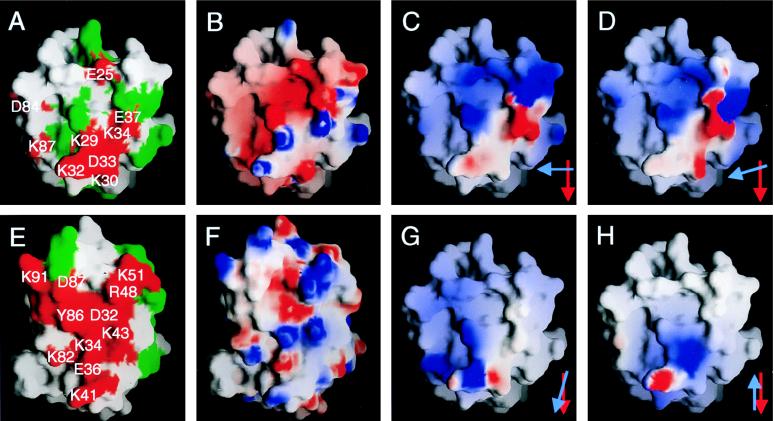
Figure 4.
Properties of the ligand binding faces of CD58 (A–D, G, and H) and human CD2 (E and F) viewed as in Fig. 3 A and B, respectively. In A and E, the grasp (22) surfaces of residues whose mutation disrupts or has no effect on binding are colored red and are labeled or are colored green, respectively (only a subset of the mutated human CD2 residues are labeled in E for clarity). In B and F, the electrostatic potential calculated at neutral pH is shown projected onto the grasp surfaces of the two domains; blue represents positive potential, white represents neutral, and red represents negative potential contoured at ±8.5 kT. In C, D, G, and H, the electrostatic potential of the ligand binding surface of human CD2, contoured at ±2.5 kT after docking with CD58, is shown projected onto the grasp surface of CD58 domain 1. The models used to dock the proteins are the homodimeric human sCD2 (9) (C) and rat sCD2 [molecule 2 of the asymmetric unit (8) (D)] crystal lattice contacts, the homodimeric interaction of CD8 monomers (27) (G), and the putative, membrane-spanning homodimeric interaction of P0 monomers (28) (H). Red and blue arrows in the lower right hand corners of C, D, G, and H indicate the relative orientations of the C β-strands of the domain 1 AGFCC′C′′ sheets of CD58 and CD2, respectively, in each of the four model complexes. In C and D, the projected electrostatic surface of CD2 is highly complementary to that of CD58 shown in B.
RESULTS
Expression and Functional Analysis of a Crystallizable Form of CD58.
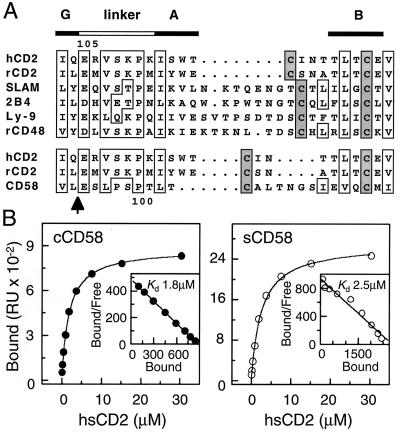
Attempts to crystallize a secreted form of wild-type CD58 truncated immediately before the transmembrane domain, either alone or in complex with human sCD2, were unsuccessful. Sequence alignments suggested that extended AB loops that could, in principle, interfere with the formation of reproducible crystal contacts might be present in domain 2 of each of the non-CD2 members of the CD2 subset (Fig. 1A). The same alignments also revealed the high degree of “linker” conservation in the CD2 subset of the IgSF, raising the possibility that the ligand binding domains of the CD2 subset molecules might be interchangeable. A crystallizable chimeric form of CD58 (cCD58), consisting of domain 1 of CD58 and domain 2 of rat sCD2, was therefore produced. This protein bound to human sCD2 with wild-type affinity (Fig. 1B) and to three CD2-blocking anti-CD58 antibodies (TS2/9, 1A2, and 1A3) (ref. 13 and references therein) with a stoichiometry approaching 1:1 (data not shown), indicating that cCD58 folded correctly.
Figure 1.
Sequence alignments and surface plasmon resonance analysis of the affinity of cCD58 for CD2. (A) Interdomain sequence alignments for the CD2-subset of the IgSF: human CD2 (hCD2 European Molecular Biology Laboratory accession number M16445), rat CD2 (rCD2; X05111), SLAM (U33017), 2B4 (L19057), Ly-9 (M84412), rat CD48 (rCD48; M37766), and CD58 (Y00636). The arrow indicates the position in the CD58 sequence at which the sequence for domain 2 was substituted with that of rat sCD2 to generate cCD58. (B) Human sCD2 was injected over BIAcore2000 sensor surfaces to which cCD58 (Left), sCD58 (Right), or a control protein (OX54; not shown) had been immobilized. The background response observed with injection of sCD2 over the control flow-cell was subtracted to give the amount of sCD2 bound. The lines drawn are a fit of the Langmuir 1:1 binding isotherm to the data. The insets show Scatchard plots of the data with linear fits. Both approaches gave the same Kd values.
Structure Determination and Overall Structure of cCD58.
The chimeric protein readily formed crystals yielding diffraction data to a resolution limit of 1.8 Å (Table 1). The overall structure of cCD58, solved by molecular replacement by using the crystal structure of rat sCD2 (8) as a search model, resembles that of sCD2 (Fig. 2A). The structure of domain 2 is not significantly different from that of domain 2 of rat sCD2 (rms deviation of 0.4 Å for superposition of 77 equivalent Cα atoms), and the conformation of the interdomain linker region (rCD2 residues 99–105) is also essentially fully conserved (Fig. 2A). The main interdomain contacts in rat CD2 are mediated by residues in β-strand A and the AB loop of domain 1 (W7GAL), which pack against residues R162 and V163 in the FG loop of domain 2. These interactions are duplicated in the chimera by CD58 domain 1 residues Y6GVV (Fig. 2B). The distinctive lattice contacts present in the rat and human sCD2 crystals (8, 9), which involved the ligand binding, AGFCC′C′′ face of all molecules in both unit cells, are absent from the crystals of cCD58, possibly because of the large net negative electrostatic potential of the AGFCC′C′′ face of cCD58.
CD58 Domain 1 Structure.
Domain 1 of CD58 has standard IgSF V-set AGFCC′C′′:DEB domain topology (Fig. 3A). The locations of the three N-glycosylation sequons in domain 1 of CD58 at N12 (β-strand B), N66 (β-strand E) and N81 (FG loop) indicate that the ligand binding AGFCC′C′′ face of CD58 is largely free of glycosylation.
Existing genetic, functional, and sequence data suggested that the genes encoding CD58 and CD2 arose by duplication, implying that these molecules would prove to be structurally similar (reviewed in ref. 6). Consistent with this view, the structure of the core of domain 1 of CD58 is very similar to that of CD2 and, also like CD2, it lacks the canonical disulfide bond between β-strands B and E. Several other distinguishing features of CD2 also are conserved in the CD58 structure, including the distinctive orientation of the C′′ strand, the general lack of twist of the AGFCC′C′′ β-sheet, and the shortened DE loop (Fig. 3 A and B). However, there are also clear structural differences. In particular, the structure of domain 1 of CD58 differs markedly from that of CD2 in the region of the FG loop, which is truncated in CD58 and lacks the characteristic β-bulge of the NH2-terminal portion of CD2 β-strand G (Fig. 2C).
Automated structure comparisons using dali (23) selected CD4 (1cdy) followed by human CD2 (1hnf), myelin P0 (1neu), and a murine Ig Fv fragment (1 mfa) as the four structures most similar to CD58 domain 1 in the Protein Data Bank. Similarities between CD58 and CD4 include the conformations of the BC and EF loops and the length of the C′C′′ loop (Fig. 3 A and C). Pairwise comparisons of CD58 domain 1 with human and rat CD2 domain 1 (rms differences of 1.0 and 1.3 Å for 79 and 83 equivalent residues, respectively, superimposed by using shp) and with other IgSF domains, including CD4 (rms difference = 1.3 Å for 86 equivalent residues), P0 (rms difference = 1.3 Å for 78 equivalent residues), and Fab NEW Vλ (rms difference = 1.2 Å for 68 equivalent residues), are consistent with the similarities detected with the dali analysis. However, detailed inspection of these superpositions suggests that the automated procedure recognized largely fortuitous similarities between CD58, CD4, P0, and the Fv, which arise from the adoption of canonical conformations by loops of similar or equivalent length (24) and do not reflect the evolutionary history of these molecules (data not shown). Overall, these comparisons indicate that CD2 and CD58 have diverged considerably, as might have been expected given the very low level of conservation of the sequences of the extracellular domains of these molecules (15% amino acid identity).
The Ligand Binding Face of CD58.
The substitution of exposed charged residues in the C (E25, K29, K30), C′ (E37), and G (K87) β-strands and in the CC′ (K32, D33, K34) and FG (D84) loops of domain 1 of CD58 has been shown to substantially reduce CD2 binding by CD58 in various assays (refs. 25 and 26; Fig. 4A). The analogous region of CD2 binds CD58 [Fig. 4E; discussed by Bodian et al. (9)].
Thus defined, the ligand binding, AGFCC′C′′ β-sheet surface of CD58 has two important structural features. First, although the surfaces of the AGFCC′C′′ β-sheets of rat and human CD2 are both flat, the equivalent surface of CD58 is characterized by two relatively large depressions separated by a “ridge” formed, principally, by K29 and E78 (Fig. 4A). The average distance of surface atoms from the least-squares-plane defining the AGFCC′C′′ face is 1.6–1.8 Å for rat and human CD2 and ≈3 Å for CD58. The depressions in the AGFCC′C′′ face of CD58 arise through the repositioning of side-chains centered on E25 in β-strand C (equivalent to D32 in human CD2), the substitution of E78 and P80 in CD58 for the equivalent β-strand F residues S84 and Y86 in human CD2, and differences in the length and conformation of the FG loop. As a result, there appears to be no marked shape complementarity in the ligand binding sites of CD2 and CD58. Second, and as anticipated (25), the ligand binding surface of CD58 is highly populated with charged residues: 16 of the 22 charged residues of domain 1 are surface-exposed on the AGFCC′C′′ β-sheet, and, of these, 10 are acidic. This acidic binding site exhibits overall electrostatic complementarity with the more basic ligand-binding site of CD2 (Fig. 4 B and F).
Implications for the Topology of CD2-CD58 Interactions.
The likely dependence of the interaction of CD2 and CD58 on electrostatic contacts, based on the compositions of the two binding sites, allows reconsideration of the topology of binding. On docking the CD2 and CD58 V-set domains in an orientation based on homodimeric, orthogonal lattice contacts observed in crystals of human sCD2 (Fig. 5A), all but three (K30, K87, and D84) of the residues implicated in the mutational analyses of CD58 (25, 26) can, by selecting alternative sidechain rotamer conformations, be brought within binding-distance of oppositely charged residues in the AGFCC′C′′ β-sheet of CD2. The inclusion of K87 in the interface only requires slight adjustment of the sCD2 lattice model, as noted (25), and the sidechain of K30 lies beneath the CC′ loop, where it could affect CD2 binding indirectly. Thus, only D84, for which the mutagenesis data are, in any case, discordant (25, 26), lies well outside the region of contact and seems unlikely to have a role in binding.
Without any alteration of the unliganded sidechain conformations, the human sCD2 crystal lattice contact model generates five contacts between oppositely charged residues [K34-D31, E37-R48, E39-R48, E42-K51, and E78-K34 (with the CD58 residue given first, followed by the paired CD2 residue)] and one unfavorable interaction of residues with the same charge (R44-R48). In contrast, docking CD2 and CD58 according to the homodimeric interactions of CD8α V-set domains (27) or antiparallel, homodimeric lattice contacts seen in crystals of P0 (28) generates only one complementary interaction (E74-K41) or a single, unfavorable contact (K32-K34), respectively. The electrostatic potential of the AGFCC′C′′ face of CD2, docked according to each model, is mapped onto the grasp surface of the AGFCC′C′′ face of CD58 in Fig. 4 C, D, G, and H, for comparison with the native electrostatic potential of CD58 (Fig. 4B). The family of models based on the sCD2 crystal lattice contacts all exhibit a substantially higher degree of electrostatic complementarity than the CD8- or P0-based models. Moreover, the total protein surface areas buried in the interactions modeled on the CD8 and P0 homodimers are only 68 and 49% of that buried in the complex modeled on the sCD2 lattice contacts (1,638 Å2). Overall, these analyses suggest that the topology of the CD2–CD58 interaction is most likely to resemble the homodimeric interaction seen in the crystal lattices of rat and human sCD2 (8, 9), in accordance with complementary mutagenesis of rat CD2 and CD48 (29).
DISCUSSION
Cell–cell contact involves the simultaneous engagement of large arrays of cell surface molecules whose rotational and translational freedom are severely limited by membrane anchorage. Previous structural studies of CD2 suggest that the constraints of membrane attachment are, to some extent, offset by the somewhat flexible, highly conserved interdomain region that ensures that the ligand binding AGFCC′C′′ surface of domain 1 is maximally exposed at the “top” of the molecule (8, 9). The compatibility of the heterologous V-set and C2-set domains apparent in the structure of cCD58, along with the high degree of conservation of the linker sequence, suggests that the interdomain region is a functionally important part of the molecule. The overall organization, dimensions, and flexibility of the extracellular region of native CD58 and, very probably, the remainder of the CD2 subset are therefore likely to be similar to those of CD2 and cCD58.
Detailed kinetic studies have implied that protein interactions at the cell surface are required to be very weak to accommodate the multivalent nature of cell–cell recognition (reviewed in ref. 7). The question then arises as to how specificity and low affinity are simultaneously achieved in these interactions. For rat CD2 binding to CD48, it has been proposed that the interactions of charged residues clustered in each binding face allow recognition to be both weak and specific because of the requirement that electrostatic complementarity compensates for the removal, on binding, of water interacting with the charged residues (11). This argument can probably be extended from the binding of rat CD2 to CD48 to the interactions of human CD2 and CD58, given the unusually high degree of clustering of charged residues in the ligand binding sites of both sets of molecules (ref. 29 and Fig. 4). This mode of recognition is clearly distinct from that seen in the well characterized interactions of other proteins, such as antibodies and proteases, which bind their ligands 104- to 105-fold more strongly, and where specificity appears to depend on a good fit of rather hydrophobic interacting surfaces. In addition to ensuring their specificity, such precise shape complementarity allows the interactions of these molecules to be highly energetically favorable by maximizing the contribution of van der Waal’s contacts (30).
In the absence of the structure of the complex it is impossible to establish the precise nature of the interaction of CD2 with CD58. The ligand binding surfaces have each been well characterized by mutagenesis, however, and this clearly indicates that the binding interface is formed by the exposed residues on the domain 1 AGFCC′C′′ β-sheets of both molecules. The clustering of charged residues at these two surfaces implies that electrostatic contacts have a prominent role in the binding of CD2 to CD58. With regard to the topology of binding, therefore, the most likely arrangement will be that which generates the highest degree of electrostatic complementarity, buries the largest number of residues implicated by mutagenesis as being at the interface, and also occludes the largest overall surface area. Although crystal symmetry contacts must be interpreted cautiously, according to these criteria, the orthogonal, homodimeric lattice contacts observed in crystals of rat and human sCD2 currently provide a better model of the complex than either the homodimeric interactions of CD8α V-set domains or antiparallel homodimeric lattice contacts seen in crystals of P0. Significantly, this arrangement is also strongly supported by complementary mutagenesis of rat CD2 and CD48 (29).
In the uncomplexed state, the ligand binding surfaces of human CD2 and CD58 exhibit little shape complementarity. Although the ligand binding surfaces of rat and human CD2 are unusually flat, the degree of nonplanarity of the ligand binding face of CD58 is comparable to that of other well characterized sites of protein–protein interactions. Regardless of the topology of binding, without major structural rearrangements, it is difficult to envisage how the depressions between the F and G and the C and C′ β-strands of CD58 domain 1 could be filled by the flat AGFCC′C′′ β-sheet of domain 1 of CD2. The cavities that remain after docking CD2 and CD58 in the orthogonal packing mode seen in crystals of human sCD2 are illustrated in Fig. 5B. Although the complex formed in vivo may achieve somewhat better packing than seen in this presumptive interface, it seems likely that poor complementarity will in part be responsible for the low affinities of CD2 for its ligands. Consistent with this, the enthalpic component of the free energy for CD2 binding to CD48 is between one-third and one-sixth of the enthalpies measured for the formation of four well characterized antibody–protein antigen complexes, each of which interacts over a comparable surface area (31), implying that substantially less contact occurs between the interacting surfaces of CD2 and CD48 than for the antibody complexes (J. E. Ladbury, R. O’Brien, P.A.v.d.M., and S.J.D., unpublished work). The absence of detailed shape complementarity is a prominent feature of the low affinity interactions of human and mouse T cell receptors with peptide-MHC class I antigens, as revealed by the first crystal structures of complexes of these molecules (32, 33). Shape mismatches between the interacting surfaces may frequently contribute to the generally low affinities of protein complexes involved in cell–cell interactions. For such interactions, electrostatic complementarity is likely to determine the specificity of binding.
Acknowledgments
The authors thank K. Starr, and A. N. Barclay and M. H. Brown, for insightful comments on the experiments and manuscript, respectively. Also thanked are R. Bryan, K. Measures, and R. Esnouf for providing computing facilities and programs, E. Garman, A. May, and A. Sharma for assistance during data collection, and S. Lee for help with figure preparation. The authors are grateful to the staff at BM14 and the European Molecular Biology Laboratory outstation at the European Synchrotron Radiation Facility (Grenoble, France). The Oxford Centre for Molecular Sciences is supported by the Biotechnology and Biological Sciences Research Council, the Engineering and Physical Sciences Research Council, and the Medical Research Council. S.I. and D.I.S. are members of the Tsukuba Advanced Research Alliance project of Tsukuba University, Tsukuba, Japan. S.I. is supported by the Human Frontier Science Program, E.Y.J. is supported by the Royal Society, P.A.v.d.M. and D.I.S. are supported by the Medical Research Council, and S.J.D. is supported by the Wellcome Trust.
ABBREVIATIONS
- cCD58
chimeric CD58
- IgSF
Ig superfamily
- sCD2
soluble CD2
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1ccz).
References
- 1.Springer T A, Dustin M L, Kishimoto T K, Marlin SD. Annu Rev Immunol. 1987;5:223–252. doi: 10.1146/annurev.iy.05.040187.001255. [DOI] [PubMed] [Google Scholar]
- 2.Selvaraj P, Plunkett M L, Dustin M, Sanders M E, Shaw S, Springer T A. Nature (London) 1987;326:400–403. doi: 10.1038/326400a0. [DOI] [PubMed] [Google Scholar]
- 3.Sanders M E, Makgoba M W, Sharrow S O, Stephany D, Springer T A, Young H A, Shaw S. J Immunol. 1988;140:1401–1407. [PubMed] [Google Scholar]
- 4.Koyasu S, Lawton T, Novick D, Recny M A, Siliciano R F, Wallner B P, Reinherz E L. Proc Natl Acad Sci USA. 1990;87:2603–2607. doi: 10.1073/pnas.87.7.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teh S-J, Killeen N, Tarakhovsky A, Littman D R, Teh H-S. Blood. 1997;89:1308–1318. [PubMed] [Google Scholar]
- 6.Davis S J, van der Merwe P A. Immunol Today. 1996;17:177–187. doi: 10.1016/0167-5699(96)80617-7. [DOI] [PubMed] [Google Scholar]
- 7.Davis S J, Ikemizu S, Wild M K, van der Merwe P A. Immunol Rev. 1998;163:217–236. doi: 10.1111/j.1600-065x.1998.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 8.Jones E Y, Davis S J, Williams A F, Harlos K, Stuart D I. Nature (London) 1992;360:232–239. doi: 10.1038/360232a0. [DOI] [PubMed] [Google Scholar]
- 9.Bodian D L, Jones E Y, Harlos K, Stuart D I, Davis S J. Structure (London) 1994;2:755–766. doi: 10.1016/s0969-2126(94)00076-x. [DOI] [PubMed] [Google Scholar]
- 10.van der Merwe P A, Barclay A N. Trends Biochem Sci. 1994;19:354–358. doi: 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 11.Davis S J, Davies E A, Tucknott M G, Jones E Y, van der Merwe P A. Proc Natl Acad Sci USA. 1998;95:5490–5494. doi: 10.1073/pnas.95.10.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis S J, Puklavec M J, Ashford D A, Harlos K, Jones E Y, Stuart D I, Williams A F. Protein Eng. 1993;6:229–232. doi: 10.1093/protein/6.2.229. [DOI] [PubMed] [Google Scholar]
- 13.van der Merwe P A, Barclay A N, Mason D W, Davies E A, Morgan B P, Tone M, Krishnam A K, Ianelli C, Davis S J. Biochemistry. 1994;33:10149–10160. doi: 10.1021/bi00199a043. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Navaza Z. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 16.Brünger A T. xplorVersion 3.1: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 17.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski R A, Rullmannn J A, MacArthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 19.Stuart D I, Levine M, Muirhead H, Stammers D K. J Mol Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 20.Esnouf R M. J Mol Graphics. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 21.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 22.Nicholls A, Sharp K, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chothia C, Lesk A M, Tramontano A, Levitt M, Smith-Gill S J, Air G, Sheriff S, Padlan E A, Davies D, Tulip W R, et al. Nature (London) 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 25.Arulanandam A R, Kister A, McGregor M J, Wyss D F, Wagner G, Reinherz E L. J Exp Med. 1994;180:1861–1871. doi: 10.1084/jem.180.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborn L, Day E S, Miller G T, Karpusas M, Tizard R, Meuer S C, Hochman P S. J Exp Med. 1995;181:429–434. doi: 10.1084/jem.181.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leahy D J, Axel R, Hendrickson W A. Cell. 1992;68:1145–1162. doi: 10.1016/0092-8674(92)90085-q. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro L, Doyle J P, Hensley P, Colman D R, Hendrickson W A. Neuron. 1996;17:435–449. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 29.van der Merwe P A, McNamee P N, Davies E A, Barclay A N, Davis S J. Curr Biol. 1995;5:74–84. doi: 10.1016/s0960-9822(95)00019-4. [DOI] [PubMed] [Google Scholar]
- 30.Creighton T E. Proteins: Structures and Molecular Properties. New York: Freeman; 1993. [Google Scholar]
- 31.Schwarz F P, Tello D, Goldbaum F A, Mariuzza R A, Poljak R J. Eur J Biochem. 1995;228:388–394. [PubMed] [Google Scholar]
- 32.Garcia K C, Degano M, Pease L R, Huang M, Peterson P A, Teyton L, Wilson I A. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 33.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]