Abstract
The location of translational initiation factor IF3 bound to the 30S subunit of the Thermus thermophilus ribosome has been determined by cryoelectron microscopy. Both the 30S⋅IF3 complex and control 30S subunit structures were determined to 27-Å resolution. The difference map calculated from the two reconstructions reveals three prominent lobes of positive density. The previously solved crystal structure of IF3 fits very well into two of these lobes, whereas the third lobe probably arises from conformational changes induced in the 30S subunit as a result of IF3 binding. Our placement of IF3 on the 30S subunit allows an understanding in structural terms of the biochemical functions of this initiation factor, namely its ability to dissociate 70S ribosomes into 30S and 50S subunits and the preferential selection of initiator tRNA by IF3 during initiation.
Keywords: cryoelectron microscopy, ribosome
Initiation of translation in prokaryotes is a complex process that requires the 30S subunit to bind the translation initiation region of an mRNA molecule, three translation initiation factors (IF1, IF2, and IF3), initiator tRNA, and GTP (reviewed by Gualerzi and Pon, ref. 1).
Of the three initiation factors, the least is known about IF1. It appears to increase the affinities and activities of the other two factors and is required for cell viability in Escherichia coli (2). It has also been suggested from footprinting data that IF1 may mimic the A-site-bound tRNA and thereby help prevent aminoacyl–tRNA from binding during initiation (3). IF2 is a GTPase that promotes the formation of the ternary complex of fMet–tRNA with the ribosome (1). Both IF1 and IF2 appear to work in concert to promote dissociation of peptidyl–tRNAs charged with short oligopeptides from the P-site of translating ribosomes (4).
The role of IF3 in translational initiation has been studied extensively. It was originally identified as a dissociation factor, because it binds the 30S subunit with high affinity and thereby prevents the formation of 70S couples. This binding results in a shift in the equilibrium toward the dissociation of 70S ribosomes into 50S and 30S subunits (5). The association constant for the 30S⋅IF3 interaction has been estimated at greater than 107 M−1 by steady-state fluorescence polarization techniques (6). IF3 has been shown to select initiator tRNA over other amino-acylated tRNAs on both natural and poly(U) mRNA substrates (7–9). Further experiments showed that this selection arises from a recognition of the anticodon loop and the three unique G:C base pairs of the anticodon stem of the initiator tRNA (10). These studies also suggest that IF3 may discriminate at the third position in the initiation codon on ribosome-bound mRNA. Finally, a more recent study shows that IF3 may be involved in start codon discrimination (11).
Neutron scattering studies showed that IF3 consists of two compact domains separated by approximately 45 Å (12). The structures of both the N- and C-terminal domains of IF3 have been solved by x-ray crystallography (13) and NMR (14, 15). Both domains are largely globular, with an α/β topology. An α-helix that protrudes away from the globular part of the N-terminal domain was postulated to be the linker between the two domains (13). Because the structures of the N- and C-terminal domains were solved separately, the precise orientation of one domain with respect to the other is not known. An NMR study on the E. coli protein showed that the interdomain region was disordered rather than helical in solution, but nevertheless was consistent with the 45-Å separation between the domains (16). However, the conserved residues in the interdomain linker lie along a continuous stripe if this region is assumed to be helical (13), suggesting that they may well adopt a helical conformation on binding the ribosome, even in E. coli. Each domain contains an exposed β-sheet similar to other known RNA-binding proteins (13). Indeed, IF3 is thought to interact primarily with 16S rRNA, and it is the only initiation factor that can be crosslinked to 16S rRNA with any efficiency (1). Further, IF3 has been shown to interfere with the association of naked 16S and 23S rRNA (17).
Given that IF3 consists of two domains separated by a linker and also has at least two known functions, the selection of initiator tRNA and the dissociation of 70S ribosomes, it is tempting to speculate whether each domain of IF3 can be assigned to a distinct function. The N-terminal domain cannot dissociate 70S ribosomes on its own, whereas the C-terminal domain is sufficient for this activity (14). However, it is unclear whether the N- and C-terminal domains function independently, since IF3 with a mutation in the C-terminal globular domain (at a residue that maps near the linker helix) has been shown to be defective in both tRNA selection and 70S dissociation activities (18).
There is a large amount of biochemical data describing regions on 16S rRNA that are affected by IF3 binding. These data include crosslinking to 16S rRNA (19) as well as footprinting studies (3, 20, 21). IF3 has also been crosslinked to ribosomal proteins S7, S11, S12, S18, and S21 by UV irradiation (22). Although there are some disagreements among these studies, most of the existing biochemical data place the binding site for IF3 somewhere on the platform side of the cleft. This region is near sites that are footprinted by P-site tRNA (23, 24). A site-directed mutant at nucleotide 791 showed a 10-fold decrease in affinity for IF3 compared with wild type (25), which is consistent with the central domain of 16S rRNA being important for IF3 binding. Finally, an early immunoelectron microscopy study on IF3 crosslinked to the 30S subunit approximately localized an epitope on IF3 to the cleft region (26).
Over the last few years, the resolution and sensitivity of electron microscopy (EM) has improved to the point that it has been used successfully to map ligand-binding positions on the E. coli ribosome (27–31). In this study, as in our previous work, we refer to surface features based on earlier EM maps of the 70S ribosome (32) and the 30S subunit (33). Here we look directly at the structures of free and IF3-bound 30S subunits by EM, and correlate the differences between them with the crystal structure of IF3 to obtain a definite location and orientation for the factor on the small ribosomal subunit.
MATERIALS AND METHODS
Cloning, Expression, and Purification of IF3.
The coding sequence for IF3 from Thermotoga maritima was introduced into the T7-based expression vector pet-13a and expressed in the E. coli strain BL21 (DE3) by using standard protocols (34, 35). The factor was purified from induced cells by ion exchange, hydroxylapatite, and gel-filtration chromatography in a procedure essentially identical to that described previously for ribosomal proteins (36) and was dialyzed into 10 mM Mg2+/0.1 M NH4Cl/20 mM Tris, pH 7.5. Mass spectrometry showed that the entire gene was expressed. The genes from both Bacillus stearothermophilus and T. maritima correspond to the short form of IF3 from E. coli.
Purification and Characterization of Ribosomal Subunits.
Thermus thermophilus cells were grown in a standard medium for the organism (37) with vigorous shaking at 72°C to an OD600 of 2–4. The cells were harvested by centrifugation and lysed by using a French press. Intact 70S ribosomes were purified by standard procedures (38). Native 30S subunits were purified from 50S subunits by centrifugation through a 10–30% sucrose gradient in 2 mM MgSO4/20 mM Hepes, pH 7.5/50 mM KCl/10 mM NH4Cl/6 mM 2-mercaptoethanol at 24,000 rpm for 17 hr in a Beckman SW28 rotor (Beckman Instruments). The 30S⋅IF3 complexes were purified similarly, except the MgSO4 concentration was 10 mM, and the 70S ribosomes were incubated in an approximately 10-fold molar excess of IF3 for 30 min before being loaded on the sucrose gradient. Both native and IF3-complexed 30S subunits were then dialyzed into 20 mM Hepes, pH 7.5/50 mM KCl/10 mM NH4Cl/10 mM MgSO4/6 mM 2-mercaptoethanol, concentrated by ultrafiltration, and applied to electron microscope grids for flash freezing and cryo-EM.
Cryo-EM.
The reconstructions were computed in the same way except where indicated. The micrographs were recorded as four defocus groups corresponding to 1.05, 1.5, 1.7, 1.95, and 2.7 μm for the control 30S subunit reconstruction and as five defocus groups corresponding to 1.25, 1.4, 1.5, 1.7, and 2.0 μm for the 30S⋅IF3 reconstruction. All micrographs were recorded by using the low-dose protocol (10 e−/Å2) on a Philips EM420 equipped with a low-dose kit and a GATAN model 626 (GATAN, Pleasanton, CA) cryotransfer holder at a magnification of ×52,200 (±2%) as checked by a tobacco mosaic virus standard. Micrographs were checked for astigmatism, drift, and the presence of Thon rings by optical diffraction. Micrographs judged as acceptable were then scanned on a Hi-Scan drum scanner (Eurocore, Saint-Denis, France) and a PSD 1010 microdensitometer (Perkin–Elmer) with a step size of 25 μm, which corresponds to 4.78 Å on the object scale. For the three-dimensional reconstructions, an automated selection procedure (39) was used to select 21,005 particles from 22 micrographs for the 30S control and 16,703 particles from 33 micrographs for the 30S⋅IF3 complex. From these automated selections, real 30S particle candidates were selected from background noise by direct comparison with 83 quasi-evenly spaced projections (40) of an existing structure of the 30S subunit (33). For each defocus group, one step of the three-dimensional projection alignment procedure (40) was applied followed by the computation of a merged contrast transfer function (CTF)-corrected reconstruction (41). The CTF-corrected structure was calculated from 6,963 particles for the free 30S subunit and 7,852 particles for the 30S⋅IF3 complex. Starting with this CTF-corrected reconstruction, the refinement was carried out by using a 2.0° angular step. The effective resolution for both reconstructions was calculated by using the Fourier shell correlation coefficient (42) with a cutoff value of 0.5 (31).
RESULTS
IF3–30S Complex Formation and Preparation.
In this study, we used ribosomes from T. thermophilus, because both 70S ribosomes and small subunits from this species can be produced in a form pure enough to be crystallizable (43, 44), thereby ensuring a homogeneous sample for microscopy. However, because the gene for IF3 from Thermus was still unknown, we used IF3 from another thermophile, T. maritima, whose optimal growth temperature of 80°C is close to the 72°C optimum for T. thermophilus (45). The Thermotoga IF3 is highly conserved when compared with sequences from E. coli and B. stearothermophilus, with 45% of the 176 residues being identical (S.E.G. and V.R., unpublished results). It has been known for over a decade that prokaryotic initiation factors will function in initiation of protein synthesis in heterologous systems (46). Moreover, we show below that IF3 from Thermotoga dissociates 70S Thermus ribosomes and binds tightly and stoichiometrically to 30S subunits, showing that it is biochemically functional in this particular system.
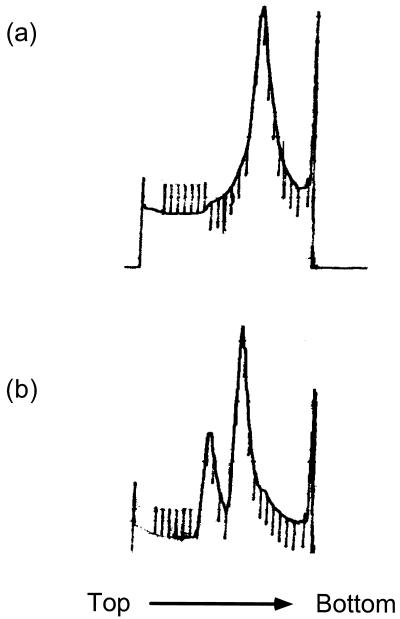
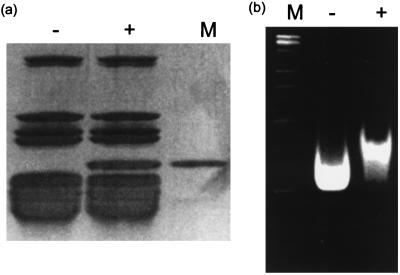
Fig. 1 shows the sucrose gradient sedimentation profile of T. thermophilus 70S ribosomes alone in high (10 mM) Mg2+ and after incubation with IF3 in the same buffer. In buffers containing 10 mM Mg2+, the 70S ribosomes migrate as a single species, while when incubated with an excess of IF3 they are completely dissociated into 30S and 50S subunits. As seen below, the 30S peak actually consists of a stoichiometric 30S⋅IF3 complex. The profile of the dissociated subunits is identical to that obtained when 70S ribosomes were dissociated by incubation and sedimentation in 2 mM Mg2+ in the absence of IF3 (data not shown). Both 30S subunits and 30S⋅IF3 complexes were subjected to SDS/PAGE along with purified IF3 as a marker (Fig. 2a), showing that the 30S⋅IF3 complex isolated on sucrose gradients had a near-stoichiometric amount of IF3 bound to it. The samples were also run on nondenaturing 0.5% agarose–3% acrylamide composite gels (Fig. 2b). The gels show not only that a band-shift occurs on IF3 binding, but also that the IF3⋅30S sample is completely shifted, thereby showing that the binding was stoichiometric and the complex remained intact under centrifugation and electrophoresis. These samples were used for comparison of the structures of free and IF3-bound 30S subunits.
Figure 1.
Sucrose gradient sedimentation profiles of 70S ribosomes in the presence and absence of IF3. (a) Sedimentation profile of 70S ribosomes through a 10–30% sucrose gradient in 10 mM Mg2+. (b) Sedimentation profile of 70S ribosomes incubated with a 10-fold molar excess of IF3 through a 10–30% sucrose gradient in 10 mM Mg2+. The 70S peak is no longer present and the two peaks correspond to the 30S⋅IF3 complex and 50S subunits.
Figure 2.
Characterization of the free and IF3-bound 30S subunits. (a) 10–20% gradient SDS/PAGE: Lane 1, 30S subunits purified by dissociation and sucrose gradient sedimentation of 70S ribosomes in buffers containing low (2 mM) magnesium; Lane 2, 30S⋅IF3 complex purified by dissociation of 70S ribosomes with an excess of IF3 followed by sucrose gradient sedimentation, both in buffers containing high (10 mM) magnesium; Lane 3, purified IF3 as a standard. (b) Nondenaturing agarose–acrylamide composite gel stained with ethidium bromide: Lane 1, molecular weight markers; Lane 2, 30S subunits purified as in lane 1 of (a); Lane 3, 30S⋅IF3 complex purified as in lane 2 of a. The 30S⋅IF3 complex is completely shifted relative to the free 30S subunits, showing that our heterologous IF3 is bound stoichiometrically to the 30S subunit and that the complex is stable under centrifugation and electrophoresis.
EM of Free and IF3-Bound 30S Subunits.
Cryoelectron microscopic maps of free and IF3-bound 30S subunits were calculated as described in Methods. The final resolution, as judged by the point at which the Fourier shell correlation coefficient (42) has a value of 0.5 (31), was about 27 Å for both species. A difference map between the free and IF3-bound 30S subunits was calculated by superimposing the two maps to maximize their correlation coefficient and then scaling the two maps relative to each other. This difference map has contributions both from the presence of IF3 itself and from structural changes induced by the binding of IF3. The latter can have both positive and negative contributions.
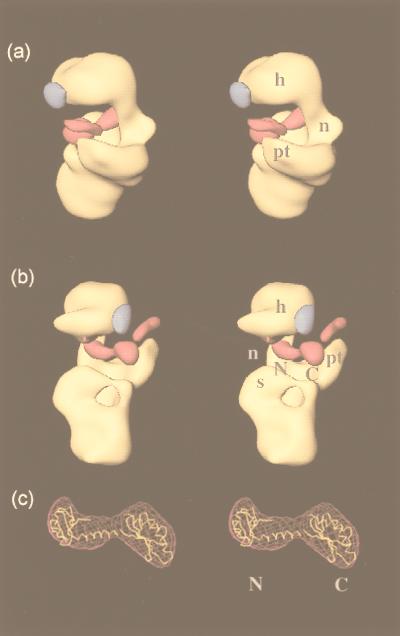
The difference map calculated from the native and IF3-bound 30S subunit reconstructions shows three large lobes of positive density and one lobe of negative density (Fig. 3). When the positive density features were examined, we found that two of these lobes had a shape and separation that was consistent with the bilobed structure of IF3 deduced by x-ray crystallography and NMR (13–15). This still left the question of which domain of IF3 to assign to each of the two lobes. An examination of the shape of the two lobes suggests that the lobe more interior to the cleft has a shape similar to that of the N-terminal domain of IF3 (including the connecting helix between the two domains), while the lobe that is more exterior and closer to the platform has a shape similar to that of the C-terminal domain. Positioning the N- and C-terminal domains of IF3 into these lobes results in an excellent fit (Fig. 3c). The correlation coefficient between the crystal structure and the difference density is 0.81, which is comparable to figures obtained previously for localization of factors and tRNA in the ribosome (28, 31). Reversing the placement leads to a lower correlation coefficient of 0.72. As we discuss below, our placement is also consistent with much of the biochemical data on IF3 binding.
Figure 3.
Stereo views of a 27-Å cryo-EM reconstruction of the 30S subunit bound to IF3. The positive (magenta) and negative (blue) difference densities on IF3 binding are overlaid on the three-dimensional cryo-EM map of the native 30S subunit (yellow). The labels on the 30S subunit indicate the head (h), neck (n), shoulder (s), and platform (pt). The two lobes of positive difference density that we have assigned to the N- and C-terminal domains of IF3 are labeled N and C, respectively. (a) View from the platform side, with the 50S subunit interface on the left; (b) the same structure viewed from the shoulder side, with the 50S subunit interface on the right; (c) the best fit of the x-ray crystal structure of IF3 into the corresponding difference density, with the N- and C-terminal domains indicated by the appropriate letters.
The other region of positive difference density has a narrow sausage-like shape that is incompatible with the shape of IF3. It probably represents a movement of the tip of the platform upwards (i.e., toward the head). A similar movement is seen in the 30S subunit on formation of 70S ribosomes (33, 47). The single region of negative density is distributed on the surface of the head region on the opposite side of the cleft from the platform (Fig. 3) and probably represents a slight movement of the head away from the cleft. This movement is smaller in extent than the movement of the tip of the platform. Taken together, these results suggest that the head and platform move slightly with respect to their positions in the free 30S subunit, with the platform movement being the more significant of the two.
We have ascribed the difference densities near the platform and head to movements of mass in these regions. If these movements occurred strictly as rigid bodies, one would expect to see difference densities of opposite sign elsewhere in the structure. The fact that we do not see such matching densities suggests that the movements are not rigid, but may be the result of subtle conformational changes that are delocalized. Such delocalized difference densities would not be detectable at the current resolution of the structures.
DISCUSSION
Correlation with Biochemical Data.
Our localization places IF3 in a position spanning the region from the 50S subunit side of the platform to the neck of the 30S subunit, adjacent to the cleft. How well does this location explain current biochemical data on IF3?
One problem with comparing three-dimensional location with biochemical data is that the three-dimensional structure of 16S rRNA is not known. A number of three-dimensional models for 16S rRNA have been constructed on the basis of biochemical and biophysical data (48–50). These models allow us to correlate biochemical footprinting data on IF3 with its spatial location on the ribosome. We have used the most recent of these models (50), which is derived by combining biochemical and electron-microscopic data. In doing so, we are aware that neither the model nor all of the biochemical data are likely to be free of errors. Nevertheless, given the resolution of the work here, we think the model is a useful framework to look at the agreement between the footprinting data and the location determined here by EM.
With reference to this model of 16S rRNA (50), it is clear that some of the footprints agree quite well with our placement of IF3, while others cannot readily be explained. In particular, the strong crosslink seen in the 819–859 region of 16S rRNA (19) is not near our location for IF3, while the weaker crosslink seen in that same study is in good agreement with our location. Some biochemical footprinting data (21) also agree well with our placement of IF3. It is quite possible that many of the footprints seen from IF3 (3) that are inconsistent with our location of IF3 were caused not by direct protection but rather by a conformational change in the subunit that reduces or increases reactivity of parts of the 16S rRNA, and our EM shows evidence for such changes on IF3 binding. Such discrepancies could be resolved by mapping IF3 on the 30S subunit with a tethered Fe(II) probe, as was recently done with EF-G (51). Finally, it is also possible that some of the discrepancies between our location of IF3 and the presumed spatial location of various footprints and crosslinks come not from errors in the biochemical data but from the model used for 16S rRNA. In this context, it is worth noting that models for ribosomal RNA continue to improve and be revised based on higher-resolution electron microscopic images (R. Brimacombe, personal communication; A. Malhotra and J.F., unpublished results). Nevertheless, the general thrust of the RNA footprinting and crosslinking data, which suggest that IF3 interacts with the central domain of 16S rRNA, is consistent with our location of IF3.
As mentioned earlier, IF3 has been crosslinked to proteins S7, S11, S12, S18, and S21 (22). The locations of S11, S18, and S21, which lie on the platform side of the cleft of the 30S (52) are close to our location for IF3. Similarly, S7 has recently been modeled into a position on the 30S subunit (53) that would be close to our location for IF3. However, given the spatial location of these five ribosomal proteins (52), it would be difficult for IF3 to be very close to all of them regardless of its location.
We discuss below how our location of IF3 sheds light on two biochemical roles for the factor, inspection of initiator tRNA, and dissociation of 70S subunits.
Inspection of Initiator tRNA.
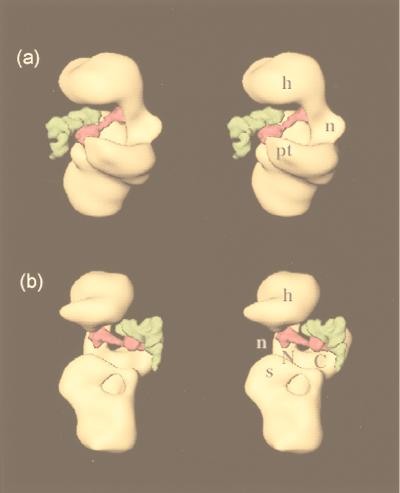
The structure of the E. coli 70S ribosome has previously been visualized in complex with both initiator tRNA and mRNA (31). This structure offers us a way of comparing the relative locations of IF3 and initiator tRNA in the ribosome. Some caution must be exercised in this interpretation, because the 30S subunit undergoes additional changes on binding the 50S subunit (31, 33). However, these changes are unlikely to affect the conclusions from the discussion that follows. We superimposed our reconstruction of the 30S⋅IF3 complex onto the 30S part of the 70S reconstruction with initiator tRNA and mRNA to obtain an approximate relative location of the tRNA with respect to the location of IF3. The results are shown in Fig. 4.
Figure 4.
Stereo views that show the relative positions of IF3 (magenta) and P-site tRNA (green) on the 30S subunit (yellow). The labels on the 30S subunit for the head, neck, shoulder, and platform are the same as in Fig. 3. The molecules of IF3 and tRNA are shown at about 5 Å resolution and represent the best fit of the x-ray crystal structures to the corresponding difference densities of IF3 (Fig. 3c) and P-site tRNA (31). (a) view from the platform side with the 50S subunit interface on the left; (b) view from the shoulder side with the 50S subunit interface on the right.
IF3 is known to recognize the three G:C base pairs of the anticodon stem that are unique to initiator tRNA (10). It can be expected, therefore, that some portion of IF3 should be near the density attributed to the anticodon stem-loop of initiator tRNA in the 70S-initiator tRNA reconstruction. As seen in Fig. 4, the anticodon stem lies directly adjacent to our location for the C-terminal domain of IF3, showing that this domain is probably involved in the recognition of the features of this stem. This observation is consistent with the finding that Lys-110 of E. coli IF3, which maps to the C-terminal domain, is important for tRNA selection and ribosome binding (18). It is also interesting to note that IF3 does not appear to bind the anticodon itself, which is therefore free to bind the start codon on mRNA. The mass attributed to mRNA in the structure of the 70S ribosome bound to initiator tRNA and mRNA (31) seems to come into contact primarily with the linker region of IF3 and the portions of the N and C termini that are near the linker. This result correlates perfectly with the observation that a Tyr-75→Asn mutation (which maps to the linking region between the domains) alters the effect of IF3 on discrimination of the initiation codon (11). Thus our placement of IF3 and its proximity to the anticodon stem-loop of initiator tRNA provide a physical framework for its function in selection of initiator tRNA.
Ribosome Dissociation Activity.
As we have shown, 70S ribosomes in high magnesium concentration migrate as a single stable complex on sucrose gradients, while in the presence of IF3 they are completely dissociated into their subunits. The location of IF3 sheds light on this activity.
In the reconstruction of the 70S ribosome (31), there is a bridge of density connecting the two subunits at a location that corresponds to our placement of the C-terminal domain of IF3. Comparison with the recent 9-Å structure of the 50S subunit from Haloarcula marismortui (54) shows that this density is actually contributed by the 50S subunit. Therefore, there is an intimate docking of the 50S and 30S subunits at exactly the location that we have found for the C-terminal domain. The presence of IF3 at this location would therefore interfere with this docking between the two subunits and prevent the formation of 70S ribosomes. Our placement of the C-terminal domain at the site of an intimate docking between the two subunits agrees with observations that the C-terminal domain by itself is sufficient for the dissociation activity of IF3 (14), while the N-terminal domain lacks this activity.
In contrast to IF3, the observed location of eukaryotic initiation factor 3 (eIF3) on the 40S subunit is not consistent with a model of antiassociation activity by direct physical blockage of the 60S subunit (55). However, the initiation of protein synthesis is quite different in eukaryotes and bacteria (56). Moreover, eIF3 is a 600-kDa complex consisting of eight subunits, none of which have any apparent homology to bacterial IF3. It is not surprising, therefore, that eIF3 and IF3 should act in completely different ways.
This study describes the direct localization of an initiation factor on the bacterial ribosome. Our location for IF3 not only correlates well with the large body of biochemical data, but also sheds light on the structural basis for the roles of the factor in dissociation of 70S ribosomes and inspection of initiator tRNA.
Acknowledgments
We thank Amy B. Heagle for help with illustrations and Irene S. Gabashvili for help with some of the computation. This work was supported by National Institutes of Health grant R01GM 44973 (to V.R. and Stephen W. White) and R37GM 29169 and R01GM 55440 (to J.F.).
ABBREVIATION
- EM
electron microscopy
References
- 1.Gualerzi C O, Pon C L. Biochemistry. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 2.Cummings H S, Hershey J W. J Bacteriol. 1994;176:198–205. doi: 10.1128/jb.176.1.198-205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moazed D, Samaha R R, Gualerzi C, Noller H F. J Mol Biol. 1995;248:207–210. doi: 10.1016/s0022-2836(95)80042-5. [DOI] [PubMed] [Google Scholar]
- 4.Karimi R, Pavlov M Y, Heurgue-Hamard V, Buckingham R H, Ehrenberg M. J Mol Biol. 1998;281:241–252. doi: 10.1006/jmbi.1998.1953. [DOI] [PubMed] [Google Scholar]
- 5.Godefroy-Colburn T, Wolfe A D, Dondon J, Grunberg-Manago M, Dessen P, Pantaloni D. J Mol Biol. 1975;94:461–487. doi: 10.1016/0022-2836(75)90215-6. [DOI] [PubMed] [Google Scholar]
- 6.Weiel J, Hershey J W B. Biochemistry. 1981;20:5859–5865. doi: 10.1021/bi00523a032. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout B, van der Laken C J, van Knippenberg P H. Biochim Biophys Acta. 1986;866:144–153. doi: 10.1016/0167-4781(86)90111-9. [DOI] [PubMed] [Google Scholar]
- 8.Risuleo G, Gualerzi C, Pon C. Eur J Biochem. 1976;67:603–613. doi: 10.1111/j.1432-1033.1976.tb10726.x. [DOI] [PubMed] [Google Scholar]
- 9.Hartz D, McPheeters D S, Gold L. Genes Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- 10.Hartz D, Binkley J, Hollingsworth T, Gold L. Genes Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- 11.Sussman J K, Simons E L, Simons R W. Mol Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 12.Kycia J H, Biou V, Shu F, Gerchman S E, Graziano V, Ramakrishnan V. Biochemistry. 1995;34:6183–6187. doi: 10.1021/bi00018a022. [DOI] [PubMed] [Google Scholar]
- 13.Biou V, Shu F, Ramakrishnan V. EMBO J. 1995;14:4056–4064. doi: 10.1002/j.1460-2075.1995.tb00077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia C, Fortier P L, Blanquet S, Lallemand J Y, Dardel F. J Mol Biol. 1995;254:247–259. doi: 10.1006/jmbi.1995.0615. [DOI] [PubMed] [Google Scholar]
- 15.Garcia C, Fortier P L, Blanquet S, Lallemand J Y, Dardel F. Eur J Biochem. 1995;228:395–402. [PubMed] [Google Scholar]
- 16.Moreau M, de Cock E, Fortier P L, Garcia C, Albaret C, Blanquet S, Lallemand J Y, Dardel F. J Mol Biol. 1997;266:15–22. doi: 10.1006/jmbi.1996.0756. [DOI] [PubMed] [Google Scholar]
- 17.Burma D P, Nag B, Tewari D S. Proc Natl Acad Sci USA. 1983;80:4875–4878. doi: 10.1073/pnas.80.16.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Bellis D, Liveris D, Goss D, Ringquist S, Schwartz I. Biochemistry. 1992;31:11984–11990. doi: 10.1021/bi00163a005. [DOI] [PubMed] [Google Scholar]
- 19.Ehresmann C, Moine H, Mougel M, Dondon J, Grunberg-Manago M, Ebel J P, Ehresmann B. Nucleic Acids Res. 1986;14:4803–4821. doi: 10.1093/nar/14.12.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muralikrishna P, Wickstrom E. Biochemistry. 1989;28:7505–7510. doi: 10.1021/bi00445a002. [DOI] [PubMed] [Google Scholar]
- 21.Laughrea M, Tam J. Biochemistry. 1991;30:11412–11420. doi: 10.1021/bi00112a007. [DOI] [PubMed] [Google Scholar]
- 22.MacKeen L A, Kahan L, Wahba A J, Schwartz I. J Biol Chem. 1980;255:10526–10531. [PubMed] [Google Scholar]
- 23.Moazed D, Noller H F. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 24.Moazed D, Noller H F. J Mol Biol. 1990;211:135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- 25.Tapprich W E, Goss D J, Dahlberg A E. Proc Natl Acad Sci USA. 1989;86:4927–4931. doi: 10.1073/pnas.86.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stöffler G, Bald R, Kastner B, Lührmann R, Stöffler-Meilicke M, Tishendorf G. In: Ribosomes: Structure, Function and Genetics. Chambliss G, Craven G R, Davies J, Davis K, Kahan L, Nomura M, editors. Baltimore, MD: University Park Press; 1980. pp. 171–205. [Google Scholar]
- 27.Agrawal R K, Penczek P, Grassucci R A, Li Y, Leith A, Nierhaus K H, Frank J. Science. 1996;271:1000–1002. doi: 10.1126/science.271.5251.1000. [DOI] [PubMed] [Google Scholar]
- 28.Agrawal R K, Penczek P, Grassucci R A, Frank J. Proc Natl Acad Sci USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stark H, Rodnina M V, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M. Nature (London) 1997;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- 30.Stark H, Orlova E V, Rinke-Appel J, Junke N, Mueller F, Rodnina M, Wintermeyer W, Brimacombe R, van Heel M. Cell. 1997;88:19–28. doi: 10.1016/s0092-8674(00)81854-1. [DOI] [PubMed] [Google Scholar]
- 31.Malhotra A, Penczek P, Agrawal R K, Gabashvili I S, Grassucci R A, Junemann R, Burkhardt N, Nierhaus K H, Frank J. J Mol Biol. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata R K, Agrawal R K. Nature (London) 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 33.Lata K R, Agrawal R K, Penczek P, Grassucci R, Zhu J, Frank J. J Mol Biol. 1996;262:43–52. doi: 10.1006/jmbi.1996.0497. [DOI] [PubMed] [Google Scholar]
- 34.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:61–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 35.Gerchman S E, Graziano V, Ramakrishnan V. Protein Expression Purif. 1994;5:242–251. doi: 10.1006/prep.1994.1037. [DOI] [PubMed] [Google Scholar]
- 36.Wimberly B T, White S W, Ramakrishnan V. Structure (London) 1997;5:1187–1198. doi: 10.1016/s0969-2126(97)00269-4. [DOI] [PubMed] [Google Scholar]
- 37.Williams R A D. In: Thermophilic Bacteria. Kristjansson J K, editor. Boca Raton: CRC; 1992. pp. 51–62. [Google Scholar]
- 38.Spedding G. In: Ribosomes and Protein Synthesis: A Practical Approach. Spedding G, editor. Oxford: Oxford Univ. Press; 1990. pp. 1–27. [Google Scholar]
- 39.Lata K R, Penczek P, Frank J. Ultramicroscopy. 1995;58:381–391. doi: 10.1016/0304-3991(95)00002-i. [DOI] [PubMed] [Google Scholar]
- 40.Penczek P A, Grassucci R A, Frank J. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J, Penczek P A, Schroder R, Frank J. J Struct Biol. 1997;118:197–219. doi: 10.1006/jsbi.1997.3845. [DOI] [PubMed] [Google Scholar]
- 42.van Heel M. Ultramicroscopy. 1987;21:95–100. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 43.Glotz C, Mussig J, Gewitz H S, Makowski I, Arad T, Yonath A, Wittmann H G. Biochem Int. 1987;15:953–960. [PubMed] [Google Scholar]
- 44.Trakhanov S D, Yusupov M M, Agalarov S C, Garber M B, Ryazantsev S N, Tischenko S V, Shirokov V A. FEBS Lett. 1987;220:319–322. [Google Scholar]
- 45.Kristjansson J K, Stetter K O. In: Thermophilic Bacteria. Kristjansson J K, editor. Boca Raton: CRC; 1992. [Google Scholar]
- 46.Brombach M, Gualerzi C O, Nakamura Y, Pon C L. Mol Gen Genet. 1986;205:97–102. doi: 10.1007/BF02428037. [DOI] [PubMed] [Google Scholar]
- 47.Agrawal R K, Lata R K, Frank J. Int J Biochem Cell Biol. 1998;31:243–254. doi: 10.1016/s1357-2725(98)00149-6. [DOI] [PubMed] [Google Scholar]
- 48.Stern S, Weiser B, Noller H F. J Mol Biol. 1988;204:447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- 49.Malhotra A, Harvey S C. J Mol Biol. 1994;240:308–340. doi: 10.1006/jmbi.1994.1448. [DOI] [PubMed] [Google Scholar]
- 50.Müller F, Brimacombe R. J Mol Biol. 1997;271:545–565. doi: 10.1006/jmbi.1997.1211. [DOI] [PubMed] [Google Scholar]
- 51.Wilson K S, Noller H F. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- 52.Capel M S, Engelman D M, Freeborn B R, Kjeldgaard M, Langer J A, Ramakrishnan V, Schindler D G, Schneider D K, Schoenborn B P, Sillers I-Y, et al. Science. 1987;238:1403–1406. doi: 10.1126/science.3317832. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka I, Nakagawa A, Hosaka H, Wakatsuki S, Mueller F, Brimacombe R. RNA. 1998;4:542–550. doi: 10.1017/s1355838298972004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ban N, Freeborn B, Nissen P, Penczek P, Grassucci R A, Sweet R, Frank J, Moore P B, Steitz T A. Cell. 1998;93:1105–1115. doi: 10.1016/s0092-8674(00)81455-5. [DOI] [PubMed] [Google Scholar]
- 55.Srivastava S, Verschoor A, Frank J. J Mol Biol. 1992;226:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- 56.Hershey J W B, Merrick W C. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Press; 1996. pp. 31–69. [Google Scholar]