Abstract
Recently a number of nonclass I genes were discovered in the human MHC class I region. One of these, FAT10, encodes a protein consisting of two domains with homology to ubiquitin. FAT10 mRNA is expressed constitutively in some lymphoblastoid lines and dendritic cells and in certain other cells after γ-interferon induction. FAT10 protein expression is controlled at several levels including transcription, translation, and protein stability. Yeast two-hybrid screening of a human lymphocyte library and immunoprecipitation studies revealed that FAT10 noncovalently associated with MAD2, a protein implicated in a cell-cycle checkpoint for spindle assembly during anaphase. Thus, FAT10 may modulate cell growth during B cell or dendritic cell development and activation.
The human MHC Class I region was recently extensively studied and was shown to contain a number of nonclass I genes belonging to a variety of structural families (1, 2). One of these, FAT10, encodes a protein that is homologous to diubiquitin but is substantially different from other members of the ubiquitin family (2–4).
Ubiquitin is best known for its role in protein degradation in all eucaryotic cells (5). Regulated proteolysis of cell-cycle regulatory proteins by the ubiquitin degradation system occurs at critical steps in cell-cycle progression (6). The ubiquitin family now includes many ubiquitin-like (UBL) proteins with diverse functions in cell-growth regulation (7–9). One group of UBL proteins contains one or more N-terminal UBL domains in a larger protein. Examples are elongin B (10), RAD23 (11), Dsk2 (12), and Bag1 (13). These UBL domains were found to be important for transcription elongation (elongin B) and for excision repair of UV-damaged DNA (RAD23). A T cell-derived nonspecific monoclonal suppressor factor was reported to be a fusion protein of a UBL and a ribosomal protein (14).
A second group of UBL proteins contains only domains with homology to a monomer or dimer of ubiquitin. This group includes SUMO/PIC/Sentrin/UBL1/SMT3 (SUMO) (7), NEDD8/RUB1 (NEDD8) (9), and ubiquitin cross-reactive protein (UCRP/ISG15) (15). SUMO is involved in many cellular functions, including nucleo–cytoplasmic transport (16, 17), cell-cycle regulation (18, 19), DNA repair (20), prevention of IκBα degradation (21), and apoptosis (22). UCRP can be induced by interferon and may be partly secreted with an effect on natural killer cell activity (14, 23). The second group of UBL proteins can be processed to have a free C-terminal glycine doublet for conjugation to other proteins. SUMO and NEDD8 use the heterodimeric complexes Aos1/Uba2 and APP-Bb1/Uba3, respectively, for E1 activity instead of a monomeric protein (24–26). The E2-conjugating enzymes for SUMO and NEDD8 are also different from the ubiquitin-specific E2.
Many of these UBL proteins are associated with cell cycle-related events. SMT3 is a suppressor of mutations in MIF2, a centromere protein gene required for mitotic spindle integrity during anaphase (27). The human counterpart of SMT3, SUMO, is associated with the oncogene PML (18, 19) and the death domain in Fas/APO-1 and tumor necrosis factor (TNF) receptors (22). Elongin B is in a complex with Elongin C that is homologous to yeast Skp1 in the Skp1-cullin-F-box protein (SCF)Cdc4 complex (28). The elongin B/C complex is associated with the von Hippel–Lindau tumor suppressor protein. Modification of Cdc53p (another component of the SCFCdc4 complex) by NEDD8 also affects the function of the SCFCdc4 complex (25). DSK2 is involved in duplication of the spindle pole body (12).
In the present study, we have characterized the structure and expression of the FAT10 gene and its encoded protein. FAT10 protein associated with the human spindle assembly checkpoint protein, MAD2. Thus, FAT10 may modulate cell cycling during B cell or dendritic cell development and activation.
MATERIALS AND METHODS
Yeast Artificial Chromosome (YAC) Clones and Cell Lines.
Human MHC YAC 903B9 has been described (2, 29). The mouse MHC C09 and E8.2 YAC (30) were from Ruma Chowdhury (The University of Cambridge, U.K.). Human cell lines JY, X50–7, 1123, B958, Lou (B lymphoblastoid), Jurkat (T cell), MC116 (undifferentiated lymphoma), ST486 (Burkitt’s lymphoma), Ramos (RA1) (Burkitt’s lymphoma), K562 (erythroleukemia), Reh (acute lymphocytic leukemia), BjaB (lymphoma), and HL60 (Promyelocytic leukemia) were grown as described (3).
Reagents and the cDNA Library.
TNF-α and γ-IFN were from R & D Systems. The proteasome inhibitor, acetyl-leucyl-leucinal-norleucinal (ALLN) was from Calbiochem. Human spleen marathon cDNA (CLONTECH) was used as the template for rapid amplification of cDNA ends (RACE) for the 5′ end of the FAT10 gene. The human spleen cDNA library was used to screen for FAT10 cDNA as described (3, 29).
Anti-FAT10 Antibody.
Initially recombinant FAT10 protein, missing the N-terminal 19 amino acids, was produced from pQE8, a His-tagged vector (Qiagen, Chatsworth, CA), in Escherichia coli. Rabbit antibody was raised against recombinant His-FAT10 protein. A peptide, amino acids 28 (Asp) to 43 (Val), was chemically synthesized and used to raise antipeptide antibody in rabbits at Research Genetics (Huntsville, AL). Both antibodies were further immune affinity purified. A 500-fold dilution of both purified antibodies was generally used for Western blot analysis.
Expression of FAT10 Protein.
Various primers were used in PCR reactions to amplify the FAT10 gene (see Results for more details). The resulting PCR FAT10 coding sequences were cloned into a pCDNA3.1 vector. The coupled transcription and translation (rabbit reticulocyte lysate) system (Promega) was used to produce FAT10 protein in vitro. Transient transfection assays were performed with COS-7 and HeLa cells for 48 to 72 hr by using Lipofectin (BRL).
In Situ Hybridization.
Both the mouse FAT10 coding and the 3′ untranslated regions (UTR) were used to make sense and antisense digoxigenin (DIG)-labeled FAT10 RNA probes. Hybridizations were at 65°C overnight in a humid chamber (31). The slides were washed at 60°C for 15 min twice in 2× SSC, 1× SSC, and 0.2× SSC. The DIG-hybrid was detected immunologically with the anti-DIG antibody (1:500), as suggested by Boehringer Mannheim.
Yeast Two-Hybrid Screening.
The FAT10 coding sequence, missing the first 19 amino acids, was cloned into pGBT9 (CLONTECH) and tested with the human B lymphocyte Matchmaker cDNA library (CLONTECH) in yeast Y190 cells. Selection was made on the plates with minimal synthetic dropout medium lacking leucine, tryptophan, and histidine (SD/-Leu/-Trp/-His). Colonies positive for β-galactosidase activity were further streaked onto plates for a second colony filter assay. Recovered plasmids were further tested for interaction with FAT10-pGBT9 and with pLam5–1, a nonspecific bait containing human lamin C, in another round of transformation of Y190 and H7fc strains and colony filter assay.
RESULTS
FAT10 Gene.
From the human spleen cDNA library screening and 5′ rapid amplification of cDNA (5′ RACE) ends extension, the full length of FAT10 cDNA was obtained and is shown in Fig. 1A. Primer extension experiments were carried out to determine the extreme 5′ upstream sequence. The FAT10 genomic sequence, from the MHC YAC930B minilibrary, was used to correlate the cDNA and the intron sequences.
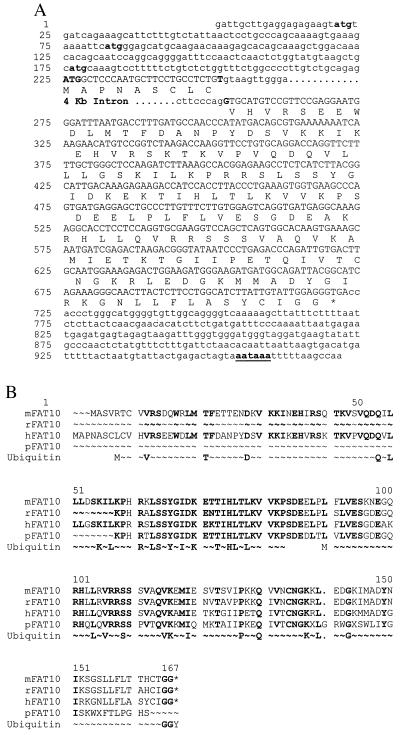
Figure 1.
The sequence of human FAT10 gene. (A) The human FAT10 gene contains 971 base pairs spanning two exons. There are three short uORFs (starting at nucleotides 21, 82, and 176) preceding the long FAT10 ORF (225 to 722). Potential translation initiation codons for the short uORF are indicated by lower-case bold letters and that for FAT10 by upper-case bold letters. The FAT10 coding sequence represented by capital letters encodes a 166-aa polypeptide. The four-kilobase intron is located between codons 9 and 10, represented by bold letters. The poly(A) signal of AATAAA is underlined at the 3′ end of FAT10 gene. The access number for this sequence is AF123050. (B) Comparison of human, murine, partial rat, and porcine FAT10 proteins with classical ubiquitin amino acids sequences. The ubiquitin sequence is shown twice to compare with the two FAT10 UBL domains, and only the conserved amino acids are presented in the diagram.
The 5′ upstream UTR includes three relatively short ORFs starting at positions 21, 82, and 176. The last two short ORFs overlap the initial codons of the FAT10 ORF that begins at residue 225 in a consensus Kozak sequence. The encoded FAT10 protein is composed of two UBL domains (amino acids 8–83 and 91–166). The amino acid identities (similarity) to ubiquitin are 32% (47%) and 40% (52%), respectively. Both UBL domains contain conserved lysine residues including one equivalent to ubiquitin Lys-48, the site for polyubiquitination. The free C-terminal glycine doublet in the FAT10 protein is a potential site for conjugation to other proteins.
The expressed sequence tag (EST) database contains murine, rat, and porcine sequence homologues of FAT10. We used the murine EST sequence as a probe to obtain a near full-length cDNA sequence from murine spleen RNA and murine genomic sequences from the DNA of murine MHC YAC. The extreme 5′ sequence was not further pursued. There is extensive amino acid conservation (69% identity) between murine and human FAT10 proteins (Fig. 1B). The predicted amino acid sequence of the murine analogue retained similarly placed lysine residues and the free glycine doublet at the C terminus. The extended murine FAT10 cDNA sequence also contains an upstream AUG in the putative 5′ UTR, while the murine FAT10 genomic DNA shows more potential AUGs further upstream. The amino acid sequences of these upstream ORFs (uORFs) were not conserved between human and murine FAT10 mRNA, but parts of the nucleotide sequence were.
Expression of FAT10 Gene.
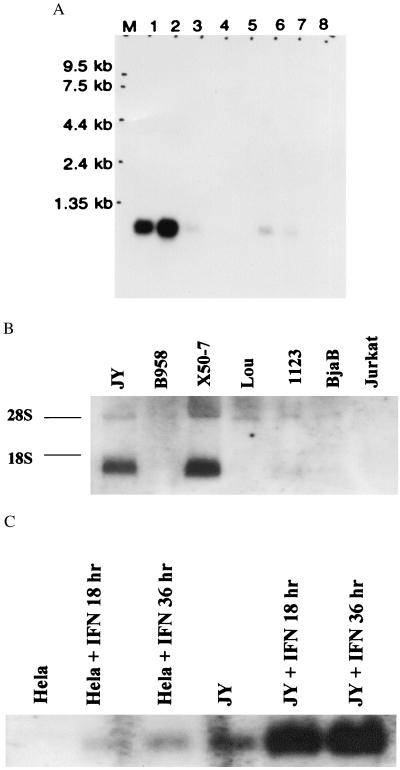
Northern blots showed that FAT10 mRNA was most strongly expressed in thymus and spleen, with only a trace level of expression in other tissues (Fig. 2A). We studied a wider variety of lymphoblastoid cell lines and lymphomas (see Materials and Methods) and found the highest FAT10 mRNA level in X50–7 cells and a slightly lower level in JY cells (Fig. 2B).
Figure 2.
Northern blot analysis of FAT10 mRNA. (A) The multiple human normal tissues Northern blot (CLONTECH). Lane 1, spleen; 2, thymus; 3, prostate; 4, testis; 5, ovary; 6, small intestine; 7, colon mucosas lining, and 8, peripheral blood leukocytes. Lane M is the RNA marker as labeled. (B) The analysis of total RNA blots from various cell lines. The RNA preparation and hybridization were performed as described (29). The origins of cell lines are indicated at the top and the two ribosomal RNA bands are indicated at the side as 28S and 18S. (C) γ-IFN treatment (3,000 u/ml) (0, 18, and 36 hr) induces progressive increases in FAT10 mRNA expression in JY and HeLa cells.
γ-IFN treatment increased the levels of FAT10 mRNA in JY cells with a progressive rise in mRNA level on longer treatment, up to 36 and 48 hr (Fig. 2C). The upstream probe and the coding region probe gave identical Northern patterns with control- and γ-IFN-treated JY cells, suggesting there is no alternative splicing and no alternative promoter usage on γ-IFN treatment. Similar treatment induced a low level of mRNA in HeLa cells and a higher level in endothelial cells, but did not induce mRNA in Jurkat T or Lou cells (not shown). Neither α-IFN nor TNF-α increased FAT10 mRNA level in JY and X50–7 cells (not shown).
The Expression of FAT10 Protein in Transfected COS and HeLa Cells.
Both affinity-purified anti-FAT10 and antipeptide antibodies reacted strongly with bacterially produced recombinant His-FAT10, while preimmune sera were entirely unreactive. However, Western blots with these antibodies detected no specific protein band of any size that was present in JY and X50–7 cell lysate but was absent from Jurkat and HeLa cell lysates (data not shown).
To test translational regulation, we prepared the following four constructs (see Materials and Methods for more details): (i) The intact FAT10 coding region with the original C and N termini (FAT10); (ii) A 6× his-tag attached to the first Met of FAT10 (N-His-FAT10); (iii) A 6× his-tag at the C terminus of the FAT10 deleting the original C-terminal glycine doublet (C-His-FAT10); and (iv) a longer FAT10 cDNA bearing the short uORF (starting from nucleotide 82) at the 5′ UTR followed by the FAT10 coding region (uORF-FAT10). We cloned these PCR products separately into the eukaryotic expression vector, pCDNA 3.1, to express FAT10 protein in transient transfection assays in COS and HeLa cells.
The DNA constructs FAT10, N-His-Fat10, and C-His-FAT10 produced the same FAT10 monomer. Both anti-FAT10 and anti-6× His-tag monoclonal antibodies (Qiagen, 1:3,000; data not shown) detected the same single protein bands for C-His-FAT10 (Fig. 3A) and N-His-tag FAT10 (not shown). We did not observe significant conjugates of FAT10 as larger-sized bands that were present in FAT10 and N-His-FAT10 but absent in C-His-FAT10 transfected cells, although in the latter cells C-His-FAT10 was unavailable for conjugation because of the deletion of the terminal glycine doublet.
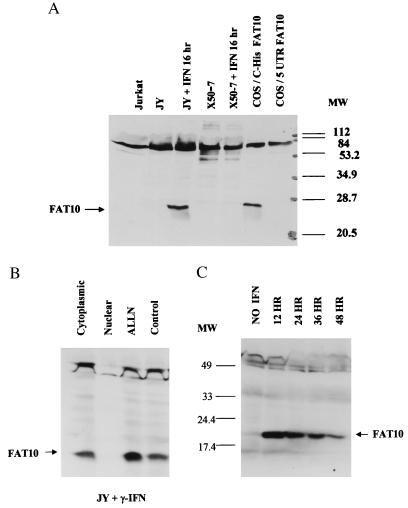
Figure 3.
Western analysis of FAT10 proteins in various cell lines. (A) The Western analysis of JY and X50–7 cells in 20% SDS/PAGE shows that γ-IFN (3,000 u/ml) can induce FAT10 protein in JY cells, but not in Jurkat or γ-IFN-treated X50–7 cells. The C-His-FAT10 construct produces a single his-tagged FAT10 protein in transfected COS cells, while the uORF-FAT10 produces no detectable FAT10 protein. (B) Western analysis of FAT10 protein in 12% SDS/PAGE shows that the majority of FAT10 protein induced by γ-IFN is in the cytoplasmic fraction. Short treatment (1.5 hr) of JY cells with ALLN (260 μM) after overnight γ-IFN induction also increases the FAT10 protein level. (C) Induction of FAT10 protein by γ-IFN with JY cells peaks between 12 and 24 hr induction and then gradually declines with time.
The uORF-FAT10 construct showed little detectable FAT10 protein in the same transfections, even though abundant uORF-FAT10 mRNA was made (not shown). In in vitro transcription and translation assays (data not shown), a large amount of 35S-labeled FAT10 protein was made by using the DNA constructs FAT10, N-His-FAT10, and C-His-FAT10 but a very low level of 35S-labeled FAT10 protein was made by using the uORF-FAT10 construct.
Overexpression of C-His-FAT10 protein in HeLa cells led to massive cell death during G418 selection and the very few colonies of surviving cells no longer expressed any detectable FAT10 proteins (not shown).
Cytokine Effect on FAT10 Protein Level.
After overnight treatment with γ-IFN, a prominent protein of 22 kDa became evident in JY cells (Fig. 3A). Subcellular fractionation showed that FAT10 protein was present mainly in the cytoplasm (Fig. 3B). FAT10 protein became prominent after 12-hr γ-IFN treatment and decreased progressively after 24 hr or 48 hr of treatment (Fig. 3C). The FAT10 protein level did not correlate well with the FAT10 mRNA level (Fig. 2C). In contrast to JY cells, the same γ-IFN treatment did not induce any clear FAT10 protein in X50–7 cells (Fig. 3A), even though it increased FAT10 mRNA level. However, TNF-α induced high levels of FAT10 protein in both JY and X50–7 cells after 6 hr of treatment (not shown), after which the levels rapidly decreased to a very low level. This TNF-α induction of FAT10 protein occurs without substantial changes in FAT10 mRNA level in JY and X50–7 cells (not shown).
To investigate the stability of the FAT10 protein, we treated JY and X50–7 with a proteasome specific inhibitor (ALLN) and other serine protease inhibitors (not shown). After γ-IFN induction, ALLN treatment enhanced the level of FAT10 protein (Fig. 3B). Applying inhibitors alone did not induce FAT10 protein expression in JY cells or increase the amount of FAT10 present in the lysate of transfected COS and HeLa cells (data not shown).
Under no conditions was there any convincing evidence of the presence of any proteins larger than FAT10 monomer that reacted with antibody and was not present in cells lacking FAT10 mRNA.
In Situ Hybridization.
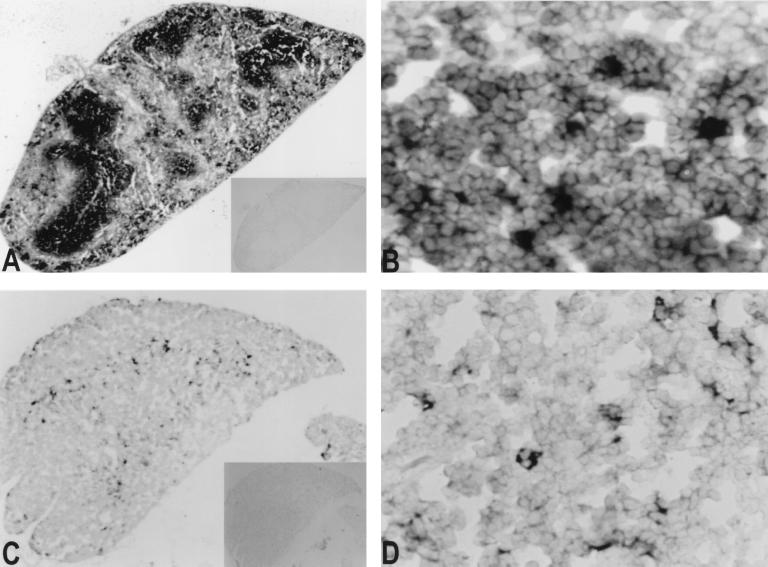
FAT10 mRNA can be readily detected in mouse spleen and thymus tissue (Fig. 4). In spleen, FAT10 mRNA expression is mainly in the white pulp, with heavily stained cells scattered inside the white pulp. In the spleen sections from mouse injected with γ-IFN, the overall FAT10 mRNA expression in the white pulp is much more prominent (not shown). In thymus, most of the heavily stained cells are scattered in the cortico-medullary junction. The apparent morphology of the expanded cytoplasm in heavily stained cells in thymus suggests that much of the mFAT10 mRNA may be in interdigitating dendritic cells (Fig. 4D).
Figure 4.
In situ hybridization on mouse tissue sections. Mouse thin sections (5 μ) were hybridized with FAT10 antisense (A and B, spleen; C and D, thymus) and sense probes (insets in A and C). Expression of murine FAT10 mRNA was seen only with antisense probe. The heavily stained cells are scattered in the white pulp in the spleen (A and B). In mouse thymus, the positive cells were mainly in cortico-medullary junction (C). At higher resolution (D), most FAT10 mRNA was in cells with expanded cytoplasm.
Interaction of FAT10 and MAD2 Proteins.
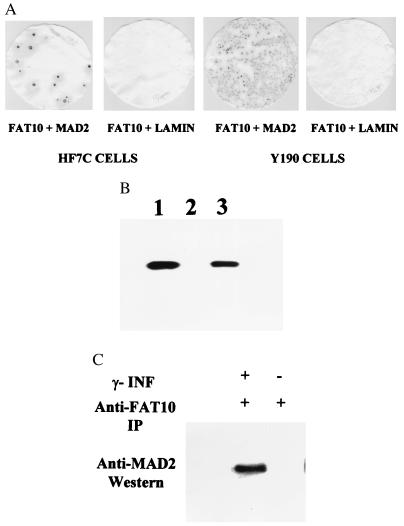
To investigate possible functions of FAT10, we used the yeast two-hybrid system to screen a B cell Matchmaker cDNA library. Because the FAT10 DNA sequence was present in this library by PCR analysis (not shown), its protein partners were likely to be present as well. Two strong positive isolates were different cDNA clones encoding the human MAD2 gene. In yeast HF7c and Y190 cells, FAT10 and MAD2 were shown to interact specifically (Fig. 5A). In vitro studies with 35S-labeled MAD2 and bacterially produced glutathione S-transferase (GST)-FAT10 fusion protein confirmed that MAD2 can specifically bind to GST-FAT10 fusion protein on the GST column, but not to the GST protein itself on the GST column (Fig. 5B). Furthermore, immunoprecipitation of induced JY cell lysate with anti-FAT10 antibody also coprecipitated the MAD2 protein, as determined by subsequent anti-MAD2 Western analysis (Fig. 5C), indicating that the interaction FAT10 and MAD2 occurred in vivo.
Figure 5.
The specific interaction of FAT10 with MAD2 in a yeast two-hybrid screen (A), in vitro on a GST column (B), and in vivo in immunoprecipitates (C). (A) The β-galactosidase assays show positive (blue) for specific interaction between FAT10 and MAD2 but negative (white) for FAT10 and human lamin C. (B) The FAT10-GST fusion proteins were produced with the GST expression vector, pGEX-4T-1 (Pharmacia). The 35S-labeled MAD2 was added to the GST column in RPMI 1640 medium. The 35S-labeled MAD2 retained on GST columns was washed, eluted, and analyzed in 10% SDS/PAGE. (1) 35S- labeled MAD2 control, (2) 35S MAD2 eluted from GST protein bound GST column, (3) 35S MAD2 from FAT10-GST bound GST column. (C) Anti-FAT10 polyclonal antibody was used to immunoprecipitate FAT10 in the JY cell lysates in 50 mM Tris (pH 8)/150 mM NaCl/1% Triton X-100/100 μg/ml PMSF and a protease inhibitor mixture (Boehringer Mannheim). The antibody–FAT10 complex was pulled down with protein A Sepharose 6B (Pharmacia) resin. After extensive wash, the final immunoprecipitate was analyzed by Western blot with anti-MAD2 antibody (Santa Cruz Biotechnology). The MAD2 clearly coprecipitates with FAT10 in the γ-IFN induced JY cell lysate but not in the uninduced JY cell lysate.
DISCUSSION
The FAT10 protein sequence clearly is as homologous to classical ubiquitin and other members of the UBL protein family as most of these proteins are to one another. It has a glycine doublet at the C terminus, a site that is essential for ubiquitin to covalently conjugate to its substrates. FAT10 is unique in the ubiquitin and UBL gene family, as it encodes a free C-terminal glycine doublet before posttranslational processing. The internal lysine, which is equivalent to the ubiquitin Lys-48, a site for polyubiquitination, is also preserved in both UBL domains of FAT10. These two important conjugation sites, C-terminal glycine doublet and the internal lysine, are well preserved among human, murine and rat FAT10 genes. All these data suggest that FAT10 is capable of conjugation to other protein molecules.
The increases in FAT10 protein after treatment with various protease inhibitors indicate that FAT10 protein is rapidly degraded in JY cells. The short window of FAT10 protein appearance and its rapid disappearance also indicate that it is labile. This transient nature of FAT10 protein is in contrast to the relative stability of ubiquitin and UBL proteins, and it makes detection of any FAT10 conjugate with other proteins difficult.
γ-IFN and TNF-α regulate FAT10 mRNA and protein expression in JY, X50–7, and endothelial cells. One major effect of these cytokines is translational regulation, which is consistent with the presence of uORFs at the 5′ UTR of FAT10 mRNA. This configuration is characteristic for many genes involved in regulation of growth and differentiation (32, 33). It has been shown to hinder the translation from the downstream authentic AUG start codon (34). The tight regulations of FAT10 gene at the level of transcription and translation, the transient nature of FAT10 protein, the association of FAT10 with MAD2, and the difficulty in obtaining stable transfection cells suggest that FAT10 might be a strong regulator of cell growth.
MAD2 binds to unattached kinetochores in mitotic cells and is part of the apparatus that causes a delay of anaphase until the kinetochores properly attach to the spindle (35, 36). Fang et al. (37) propose that MAD2 binds to anaphase promoting complex via CDC20 and regulates its activity in mitosis. In Schizosaccharomyces pombe, overexpression of MAD2 can arrest cells at metaphase (38). Microinjection of anti-MAD2 antibody into mammalian cells in mitosis can induce premature anaphase (39).
MAD2 functions as a tetramer, and this oligomerization is independent of MAD2 monomer concentration in vitro (37). Therefore, binding of FAT10 to MAD2 even at substoichiometric levels might modulate MAD2 functions. There is other evidence suggesting that these spindle assembly checkpoint proteins may have some regulatory role in cell growth. Human T cell leukemia virus oncoprotein Tax targets MAD1, which is in a complex with MAD2 (40). The C terminus of the inactive insulin receptor can also bind to MAD2 (41). Some tumor cell lines may be defective in MAD2 expression (35).
Our in situ analysis showed some heavily stained cells scattered in the white pulp in mouse spleen and some interdigitating dendritic cells in mouse thymus. FAT10 mRNA was dramatically increased in the mouse spleen stimulated with γ-IFN treatment. Bates et al. (4) have also recently found FAT10 mRNA mainly in germinal center-derived mature B cell lines, dendritic cells derived in vitro from CD34+ cells or monocytes, and epithelial cells scattered in germinal centers in inflamed tonsils. The association of FAT10 with MAD2 and with this in vivo expression pattern may suggest a functional role for FAT10 in lymphocyte development and in immune responses. Dendritic cells are terminal nondividing cells, and their cytoskeletal structure might interfere with successful transition through anaphase, creating a role for cell-cycle arrest before anaphase. Elucidation of the overall physiologic role for FAT10 will require more biochemical and in vivo study. Nevertheless, the action of the FAT10 gene product represents another potential role for the MHC in the immune response (1).
Acknowledgments
We are grateful to Drs. D. Brash and M. Solomon for critical reading of this manuscript. We would also like to thank members in Dr. Weissman’s laboratory for technical help and insightful discussions and, in particular, Dr. A. Raghunathan for providing the tissues from mice treated with γ-interferon. This work was supported by National Institutes of Health grants (CA42556-10 to S.M.W. and HL43331 to J.R.B.) and by an Anna Fuller Fellowship (to Y.-C.L.).
ABBREVIATIONS
- UBL
ubiquitin-like
- TNF
tumor necrosis factor
- YAC
yeast artificial chromosome
- ALLN
acetyl-leucinyl-leucinal-norleucinal
- UTR
untranslated region
- GST
glutathione S-transferase
- MAD
mitotic arrest deficiency
- uORF
upstream ORF
References
- 1.Gruen J R, Weissman S M. Blood. 1997;90:4252–4265. [PubMed] [Google Scholar]
- 2.Gruen J R, Nalabolu S R, Chu T W, Bowlus C, Fan W, Goei V L, Wei H, Sivakamasundari R, Liu Y-C, Xu H, et al. Genomics. 1996;36:70–85. doi: 10.1006/geno.1996.0427. [DOI] [PubMed] [Google Scholar]
- 3.Fan W, Cai W, Parimoo S, Lennon G G, Weissman S M. Immunogenetics. 1996;44:97–103. doi: 10.1007/BF02660056. [DOI] [PubMed] [Google Scholar]
- 4.Bates E E, Ravel O, Dieu M C, Ho S, Guret C, Bridon J M, Ait-Yahia S, Briere F, Caux C, Banchereau J, et al. Eur J Immunol. 1977;10:2471–2477. doi: 10.1002/eji.1830271002. [DOI] [PubMed] [Google Scholar]
- 5.Hochstrasser M. Annu Rev Genet. 1997;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 6.King R W, Deshaies R J, Peters J M, Kirschner M W. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 7.Saitoh H, Pu R T, Dasso M. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 8.Johnson P R, Hochstrasser M. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser M. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 10.Garrett K P, Aso T, Bradsher J H, Foundling S I, Lane W S, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watkins J F, Sung P, Prakash L, Prakash S. Mol Cell Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biggins S, Ivanovska I, Rose M D. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan J A, Reed J C. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura M, Xavier R M, Tsunematsu T, Tanigawa Y. Proc Natl Acad Sci USA. 1995;92:3463–3467. doi: 10.1073/pnas.92.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas A L, Ahrens P, Bright P M, Ankel H. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 16.Matunis M J, Coutavas W, Blobel G. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 18.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 19.Muller S, Matunis M J, Dejean A. EMBO J. 1997;147:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen Z, Pardington-Purtymun P E, Comeauz J C, Moyzis R K, Chen D J. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 21.Desterro J M, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 22.Okura T, Gong L, Kamituni T, Wada T, Okura I, Wei C-F, Chang H-M, Yeh T H. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- 23.D’Cunha J, Knight E, Jr, Haas A L, Truitt R L, Borden E C. Proc Natl Acad Sci USA. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson E S, Schwienhorst I, Dohmen R J, Blobel G. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lammer D, Mathias N, Laplaza J M, Jiang W, Liu Y, Callis J, Goeble M, Estele M. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meluh P B, Koshland D. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaelin W G, Iliopoulos O, Lonergan K M, Ohh M. J Intern Med. 1998;243:535–539. doi: 10.1046/j.1365-2796.1998.00335.x. [DOI] [PubMed] [Google Scholar]
- 29.Fan W, Liu Y-C, Parimoo S, Weissman S M. Genomics. 1995;27:119–123. doi: 10.1006/geno.1995.1013. [DOI] [PubMed] [Google Scholar]
- 30.Jones E P, Xiao H, Schutlz R A, Flaherty L, Trachtulec Z, Vincek V, Larin Z, Lehrach H, Lindahl K F. Genomics. 1995;27:40–51. doi: 10.1006/geno.1995.1006. [DOI] [PubMed] [Google Scholar]
- 31.Schaeren-Wieners N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 32.Morris D R. Prog Nucleic Acid Res Mol Biol. 1995;51:339–363. doi: 10.1016/s0079-6603(08)60883-1. [DOI] [PubMed] [Google Scholar]
- 33.Geballe A P. In: Translational Control. Hershley J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 173–197. [Google Scholar]
- 34.Marth J D, Overell R W, Meier C E, Drebs E G, Perlmutter R M. Nature (London) 1988;332:171–173. doi: 10.1038/332171a0. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Benezra R. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 36.Chen R-H, Waters J C, Salmon E D, Murray A W. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 37.Fang G, Yu H, Kirschner M W. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X, Patterson T E, Sazer S. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorbsky G J, Chen R H, Murray A W. J Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin D-Y, Spencer F, Jeang K-T. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 41.O’Neill T J, Zhu Y, Gustafson T A. J Biol Chem. 1997;272:10035–10040. doi: 10.1074/jbc.272.15.10035. [DOI] [PubMed] [Google Scholar]