Abstract
Retroviral entry into cells depends on envelope glycoproteins, whereby receptor binding to the surface-exposed subunit triggers membrane fusion by the transmembrane protein (TM) subunit. We determined the crystal structure at 2.5-Å resolution of the ectodomain of gp21, the TM from human T cell leukemia virus type 1. The gp21 fragment was crystallized as a maltose-binding protein chimera, and the maltose-binding protein domain was used to solve the initial phases by the method of molecular replacement. The structure of gp21 comprises an N-terminal trimeric coiled coil, an adjacent disulfide-bonded loop that stabilizes a chain reversal, and a C-terminal sequence structurally distinct from HIV type 1/simian immunodeficiency virus gp41 that packs against the coil in an extended antiparallel fashion. Comparison of the gp21 structure with the structures of other retroviral TMs contrasts the conserved nature of the coiled coil-forming region and adjacent disulfide-bonded loop with the variable nature of the C-terminal ectodomain segment. The structure points to these features having evolved to enable the dual roles of retroviral TMs: conserved fusion function and an ability to anchor diverse surface-exposed subunit structures to the virion envelope and infected cell surface. The structure of gp21 implies that the N-terminal fusion peptide is in close proximity to the C-terminal transmembrane domain and likely represents a postfusion conformation.
Human T cell leukemia virus type 1 (HTLV-1) is a retrovirus with a wide geographic distribution associated with adult T cell leukemia (1) and tropical spastic paraparesis/HTLV-1-associated myelopathy (2). Retroviral envelope glycoproteins (Env) mediate the attachment and subsequent membrane fusion of virions and infected cells with target cells. Cell-free HTLV-1 virions have low infectivity, and viral spread appears to occur mainly between infected and uninfected cells (3, 4). The native HTLV-1 Env consists of a receptor-binding surface-exposed subunit (SU), gp46, and a noncovalently associated transmembrane protein (TM), gp21, formed by proteolytic cleavage of a polyprotein precursor, gp62 (5). Receptor binding triggers conformational changes in retroviral SU-TM complexes that result in TM-mediated fusion (6). Although the cellular receptors for HIV-1, CD4, and the chemokine receptors have been extensively studied, the HTLV-1 gp46 receptor(s) are unknown.
Despite considerable sequence diversity, retroviral TMs share similar gross structural features. The extraviral domain (ectodomain) comprises an N-terminal hydrophobic fusion peptide, an adjacent coiled coil-forming sequence, a short disulfide-bonded loop, and a C-terminal segment containing α-helical elements. The transmembrane domain and cytoplasmic tail are C terminally located (7). The available structures of Moloney murine leukemia virus (MoMLV) (8), HIV type 1 (HIV-1) (9–11), and simian immunodeficiency virus (SIV) (12, 13) TM ectodomain fragments lack either 37 C-terminal ectodomain residues (MoMLV), or an intact disulfide-bonded region (HIV and SIV). The disulfide-bonded region is critical for retroviral infectivity (14), overlaps with an immunosuppressive sequence (15), and in the case of HIV-1, contains the principal immunodominant antibody epitope (16). The knob-like structure formed by the disulfide-bonded loop has been proposed to fill a cavity in SU to stabilize the SU-TM complex (17).
We report the crystal structure of an HTLV-1 gp21 ectodomain segment (Met-338–Thr-425, gp62 numbering) that places the coiled coil-forming sequence, the disulfide-bonded loop, and the C-terminal ectodomain segment in a more complete structural context than has previously been described for other retroviral TMs. We used a strategy for crystallization and structure determination whereby the gp21 segment was linked to maltose-binding protein (MBP) as a crystallization tag of known three-dimensional structure (18). The structure of gp21 shows the expected N-terminal trimeric coiled coil, the adjacent disulfide-bonded loop similar to MoMLV (8) that stabilizes a chain reversal, and a C-terminal sequence structurally distinct from HIV-1/SIV gp41 (9–13) that packs against the coil in an extended antiparallel fashion. The structure of gp21 likely represents the fusion-activated or postfusion conformation. The observed structural divergence of the C-terminal regions of retroviral TMs mirrors the diversity of retroviral SU receptors on target cells.
MATERIALS AND METHODS
Expression, Purification, Crystallization, and Diffraction Data Collection.
MBP/gp21 comprising MBP, the three-alanine linker, and gp62 residues 338–425 was expressed, purified, and crystallized as described (18). Diffraction data were collected from a single crystal [soaked for 10 sec in 20% ethylene glycol/22% PEG 4,000/0.2 M ammonium sulfate/0.1 M sodium acetate, pH 4.7 before flash-freezing at 100 K in a liquid nitrogen stream (Oxford Cryosystems, Oxford, U.K.)], by using MAR-Research image plate detector and CuKα radiation from a Rigaku (Tokyo) RU-200 rotating anode generator. Data were auto-indexed and processed with the hkl suite (19). The crystals have the symmetry of the rhombohedral space group R3 with cell dimensions a = b = 102.9 Å, c = 118.0 Å and angles α = β = 90°, γ = 120°. A total of 150,336 diffraction measurements were collected to yield 19,248 unique reflections, 91% completeness, and Rmerge = 0.068 between infinity and 2.29-Å resolution (65% completeness, and Rmerge = 0.364 between 2.37- and 2.29-Å resolution). Rmerge = Σhkl(Σi(|I hkl, i − <I hkl >|))/Σhkl, i <I hkl>, where Ihkl, i is the intensity of an individual measurement of the reflection with Miller indices h, k, and l, and <Ihkl> is the mean intensity of that reflection; calculated for I > −3σ(I).
Structure Determination.
The crystal structure of MBP/gp21 was determined by molecular replacement (resolution range 10–3.5 Å) by using the structure of MBP (20) as the search model (program x-plor version 3.851). The top solution in the direct rotation function followed by Patterson correlation refinement (21) (correlation coefficient 0.155, highest false peak 0.067) resulted in an unambiguous translation function solution (Rcryst = 44%) with good crystal packing and revealed electron density corresponding to the gp21 segment (Fig. 1A). The missing portion of the model was built by using the program o (22) and improved as judged by the free R factor (23) by rounds of crystallographic refinement using the program x-plor (version 3.851) with bulk solvent correction and an anisotropic overall B factor, and manual rebuilding. Solvent molecules were added with the program arp (24). The current model contains MBP residues 6–368, gp21 residues 338–421 (3,483 protein atoms), one maltose molecule, and 39 solvent molecules (two putatively assigned as chloride ions based on the similarity to the coordination with the chloride in MoMLV TM; ref. 8); residues 1–5 of MBP and residue 422–425 of gp21 have no interpretable electron density and presumably are disordered. Refinement statistics were: resolution, 40–2.5 Å (reflections beyond 2.5 Å were not used in refinement because of weakness and poor completeness); F>0, number of reflections, 15,268; Rcryst = 22.3%; Rfree = 30.5% (test set size 7%); F>3σ(F), Rcryst =19.7%, Rfree = 28.3%; rms deviations from ideal values, bond lengths 0.006 Å, bond angles 1.1°. No residues are in disallowed regions, and 83% residues are in most favored regions of the Ramachandran plot (25). The average B factors are 65.1 and 67.5 Å2 for the MBP and gp21 moieties, respectively. The rms deviation of 366 Cα atoms of MBP between MBP/gp21 and Protein Data Bank entry 1ANF (20) is 0.61 Å. Rcryst = Σhkl(||Fobshkl| − |Fcalchkl||)/|Fobshkl|, where |Fobshkl| and |Fcalchkl| are the observed and calculated structure factor amplitudes; Rfree is equivalent to Rcryst but calculated with reflections omitted from the refinement process.
Figure 1.
(A) Structure determination. Electron density map (22) (magenta) calculated with coefficients |Fobs| − |Fcalc| (40–2.5 Å resolution) and phases based on the MBP positioned by molecular replacement, contoured at 1.5 SDs. Superimposed is the Cα trace of the refined model of MBP/gp21 trimer (MBP green, gp21 yellow). The path of the gp21 chain is clearly visible in this map. (B) Structure of MBP/gp21 chimera. Ribbon diagram of a trimer [drawn with programs molscript (48) and raster3d (49)]. MBP is green, the three-alanine linker is yellow, and gp21 is blue. The view is down the crystallographic 3-fold axis (rotated 90° around the horizontal axis relative to the view in A).
RESULTS AND DISCUSSION
Structure of MBP/gp21 Chimera.
A gp21 ectodomain segment lacking the fusion peptide and 20 C-terminal residues was expressed and crystallized as a chimera with Escherichia coli MBP; the two moieties were linked via a tri-alanine linker (18). The structure of MBP (20) was used to obtain the initial phases by molecular replacement (Fig. 1A), and the model was refined at 2.5-Å resolution. A trimeric structure is conferred by the gp21 sequences, consistent with the quaternary structure of the HTLV-1 envelope protein precursor produced in mammalian cells (18).
The MBP/gp21 chimera trimer has a mushroom shape, with the gp21 sequences forming the stalk (Fig. 2 a and b), and the MBP moieties comprising the head (Fig. 1 A and B). A stereo diagram of a Ca trace of a gp21 trimer with each monomer in a different color and selected residues indicated by balls and numbers is published as supplemental data on the PNAS web site, www.pnas.org. The core of gp21 consists of a ≈70 Å-long, parallel trimeric coiled coil, positioning three monomers around a crystallographic symmetry axis. The N-terminal α-helix of gp21 is oriented roughly perpendicular to the C-terminal helix of MBP via the linker sequence N368AAAMS339 in an extended conformation (gp62 numbering is used throughout; to convert to gp21 numbering subtract 312; to convert to MBP/gp21 chimera numbering add 34). The linker sequence runs over the nearest gp21 helix, connecting each MBP moiety to a distal and not the most proximal gp21 sequence in the structure (Fig. 1B).
Figure 2.
Structure of gp21. (a) Ribbon diagram of the gp21 monomer with the coiled coil-forming helix magenta, the disulfide-bonded domain CX6CC green, the C-terminal helix cyan, and the rest gray. The side chains of Met-338, Gly-343, Leu-346, and Val-350 with unusual packing (yellow), the a and d residues in the coiled coil (magenta), the hydrophobic residues in the base of the coiled coil (gray), and the cysteine residues (green) are shown. (b) As in a but showing trimer. (c) Ribbon diagram of MoMLV TM trimer (8) with the coiled coil-forming helix magenta and the disulfide-bonded domain CX6CC green. The side chains of the a and d residues in the coiled coil (magenta), the hydrophobic residues in the base of the coiled coil (gray), and the cysteine residues (green) are shown. (d) Ribbon diagram of HIV-1 gp41 trimer (10) with the coiled coil-forming helix magenta and the C-terminal helix cyan. The side chains of Gln-41 in the glutamine-rich layer (yellow), and the a and d residues in the coiled coil (magenta) are shown. GCN4 region is not shown. (e) Ribbon diagram of the TBHA2 trimer (26) with the coiled coil-forming helix magenta, the C-terminal helix cyan, the HA1 residues blue, and the rest gray. The side chains of Thr-59 with x-type packing in a region where loop-to-helix transition occurs at low pH (yellow), and the a and d residues in the coiled coil (magenta) are shown. (f) Molecular surface (probe radius 1.4 Å) of the trimeric coiled coil of gp21 (residues 338–387) color-coded according to surface complementarity (50) with the C-terminal segment (residues 390–421). Red, Sc (shape correlation statistic) > 0.76; yellow, 0.76 > Sc > 0.3; green, 0.3 > Sc > −0.3; light blue, Sc < −0.3. The overall Sc equals 0.65. The C-terminal segment is shown in magenta, with the interacting residues (contacts < 4 Å) in white. Drawn by grasp (51).
Structure of gp21.
The hydrophobic core of the coiled coil consists of 14 layers of homotrimeric interactions. The majority contains regular knobs-into-holes packing of residues in a and d positions (I354LINLIANLL385) (Fig. 2 a and b). This type of packing is altered in the N-terminal region where Val-350 shows x-type packing (26) and Leu-346 packs in a d-type fashion (Leu-347 otherwise would occupy an a position). The cavity formed by Gly-343 in a d position is partially filled by the side chain of Met-338. The glycine is substituted by serine in HTLV-2 and simian T lymphotropic virus type 2 (STLV-2) TMs, and the accompanying Met-338–Leu substitution suggests that the Met-338 side chain requires a cavity at position 343.
At the base of the coiled coil (Trp-387), the chain turns perpendicular to the axis of the coil by forming a short 310-helix followed by a short α-helix, and then again turns 90° to continue antiparallel to the coil. The region of the chain reversal is stabilized by both a disulfide bridge (residues 393–400; Cys-401 is reduced) and a salt bridge between Glu-398 within the disulfide-bonded loop and Arg-380 close to the base of the coiled coil. This region contains several solvent-exposed hydrophobic residues (e.g., Leu-384, Phe-386, and Trp-387) at the base of the coil. The C-terminal sequence running antiparallel to the coiled coil is extended between residues 397 and 407 [the Asn-404 side chain, which is glycosylated in the native protein (5), points into solvent]. Residues 408–415 form a short helix that packs against the segment 356–365 in the N-terminal helix. The helix is terminated by two prolines (417–418) whereas the rest of the chain is extended and poorly ordered. In some HTLV-1 isolates, Ser-411 is substituted by Pro (27), suggesting a shorter helix may be tolerated in these cases. A large complementary surface area (2,700 Å2) is buried between the C-terminal sequence (residues 409–421 of a monomer) and the coiled coil (residues 338–387 of the trimer) (Fig. 2f). A table summarizing the contacts between the C-terminal sequence and the coiled coil is published as supplemental data on the PNAS web site, www.pnas.org.
The gp21 structure provides a framework for interpreting earlier mutational studies of native Env function (28). Nonconservative mutation of residues in a and d positions of the coiled coil (Leu-368–Arg and Ala-375–Asp) or of residues that mediate contacts between the coiled coil and the C-terminal segment (Asp-351–His, Arg-379–Gly, Asp-383–Tyr, Glu-398–Val, and Leu-419–Arg) result in decreased gp62 precursor processing, indicating that these residues are important in both the prefusogenic and fusion-activated Env structures. Mutation of other residues within either of these regions (Ala-360–Glu, Ile-361–Arg, Ile-361–Leu, Ala-375–Val, Gln-377–Leu, and Asn-407–Tyr) causes reduced Env fusogenicity and viral infectivity but allows normal Env maturation, suggesting roles in fusion. Gln-377 and Asn-407 appear to be key residues in the interface between the coiled coil and C-terminal segment and illustrate the functional importance of polar interactions between the glutamine side chain and the C-terminal segment. The effects of most other mutations are likely caused by the introduction of steric hindrance through mutation to a larger side chain.
Structural Diversity of Retroviral TMs.
A comparison of HTLV-1, MoMLV, and HIV-1 (Fig. 2 b–d) and SIV TM (12, 13) ectodomain structures reveals that the N-terminal coiled coil-forming sequences have a conserved oligomerization function. The disulfide-bonded regions are very similar in HTLV-1 and MoMLV, although the chains exit the coiled coil in slightly different directions (rms deviation 0.86 Å for 53 Cα atoms). The C-terminal ectodomain sequences beyond the chain reversal are less conserved structurally between HTLV-1 and HIV-1/SIV. The extended chain-α-helix-extended chain arrangement in gp21 is substituted in HIV-1 and SIV gp41 by a longer α-helix. The segment of approximately 20 residues of gp41 that links the disulfide-bonded loop to the C-terminal α-helix contains multiple glycosylation sites (Fig. 3) and may form a protease-sensitive substructure on the outside of the gp41 core (29, 30).
Figure 3.
Structural diversity of retroviral TMs. Comparison of known (cylinders; gp21, red; MoMLV TM, light green; gp41, dark green) and predicted (boxed) (31) α-helices present in the TM ectodomains of the retroviridae. Extended/random coil is shown as a green line and 310-helix as a blue cylinder. Residues in the a and d positions of the heptad repeat are highlighted in magenta and residues involved in irregular packing at the N terminus of the coiled coil are highlighted in yellow. Acidic residues are red, basic residues blue, aromatic residues cyan, and cysteines highlighted in olive. Known and potential N-linked glycosylation sites are italic and underlined. The retrovirus type species used are HTLV-13–19-3 (HTLV), STLV-L (STLV, GenBank accession no. Y07616), BLV (K02120), MoMLV (J02255), SMRV, (M23385); MPMV (AF033815), RSV (AF033808); HIV-1HXB2R (HIV, K03455), SIVAGMtan (SIV, U58991); and mouse mammary tumor virus (MMTV) (D16249). *RSV has an internal fusion peptide: IFASILAPGVAAA.
Comparison of known and predicted (31) secondary structural elements within the TMs of diverse retroviruses (Fig. 3) suggests that gp21 is most closely related structurally to the TMs of STLV-L and bovine leukemia virus (BLV, HTLV/BLV genus), the mammalian C-type retroviruses [e.g., MoMLV and squirrel monkey sarcoma virus (SMRV)], and D-type retroviruses [e.g., Mason-Pfizer monkey virus (MPMV)], all of which contain a conserved disulfide-bonded CX4EXCCF motif adjacent to their N-terminal coiled coil-forming sequences. The HTLV-1 and MoMLV TM structures indicate that the salt bridge formed between Glu-398 and Arg-380 may be a conserved mechanism for stabilizing the chain reversal in these viruses (8). By analogy with gp21, the basic residue within the disulfide-bonded loop of divergent primate lentiviruses (e.g., HIV-1HXB2R and SIVAGMtan), CX2K/RX2C, could form a salt bridge with the conserved glutamate corresponding to Arg-379 of gp21. The solution structure of a SIV gp41 with Cys-86 and Cys-92 mutated to alanine determined by NMR (13) showed that the sequence linking N- and C-terminal α-helices forms a hairpin at the tip of the helical core that differs remarkably from the structures of corresponding sequences in HTLV-1 and MoMLV TMs. However, it should be noted that: (i) the alanines substituting for the cysteines that would normally form the disulfide bond are >9 Å apart in the structure; (ii) the corresponding region is disordered in the crystal structure of an identical construct†; and (iii) the substitution of the corresponding cysteines of HIV-1 gp41 abolished Env maturation and viral infectivity (14). Therefore, this region may be in a non-native conformation in the described SIV gp41 solution structure (13).
Comparisons of known and predicted secondary structural elements suggest the greatest diversity in the C-terminal ectodomain sequences. The eight-residue α-helix in gp21 is substituted by predicted 13–17-residue, highly charged α-helices in MoMLV, SMRV, MPMV, and Rous sarcoma virus (RSV), whereas the B-type mouse mammary tumor virus is expected to contain short helices interspersed with random coils. The conserved nature of the coiled coil-forming regions and adjacent disulfide-bonded loops contrasts with the variable nature of C-terminal ectodomain segments. These features reflect the dual roles of retroviral TMs: conserved fusion function, and the ability to anchor SUs with diverse receptor binding specificities to virions.
Mechanism of Membrane Fusion.
There are striking similarities between gp21 and the fusion-activated structure (TBHA2) of influenza virus hemagglutinin (HA), the archetype TM subunit (26) (Fig. 2 b and e). These similarities include the presence of an N-terminal trimeric coiled coil, a region of chain reversal, and a C-terminal extended chain-α-helix-extended chain arrangement (in this respect, gp21 resembles TBHA2 better than HIV and SIV gp41). The similarities between the high-resolution structures of HA, gp21, and other retroviral TMs, and Ebola virus GP2 (32) and the TM of paramyxovirus (33), as deduced from biochemical studies, suggest a common membrane fusion mechanism. Both prefusogenic (34) and fusion-activated (26) HA structures are assembled into trimers via a triple-stranded coiled coil. Exposure of HA to endosomal pH (pH 5–6) induces fusogenic activity. This fusion activation involves a helical extension of the central coiled-coil N terminus to relocate the fusion peptide from the HA core to an envelope-distal location for insertion into the target cellular membrane. The envelope-proximal portion of the central coiled coil is transformed into a random coil-α-helix arrangement that mediates the chain reversal for the antiparallel packing of the C-terminal ectodomain sequence on the outside of the coil. The ability of HA to undergo this conformational change is most likely caused by the prefusogenic form folding into a metastable conformation that is destabilized by low pH, resulting in the adoption of the most stable structure (35, 36).
Receptor binding by SU is considered to trigger the membrane fusion activity of retroviral TMs (6). The coiled coil likely represents the core of retroviral Envs throughout the fusion process (9–11, 37). In influenza HA, irregularities in the 4–3 periodicity are observed near the regions where loop-to-helix transition is triggered by fusion activation (26, 34) (Fig. 2e); in gp21, fusion peptide exposure for insertion into the target membrane therefore may occur via helical extension at the N terminus of the coiled coil between residues Leu-340 and Val-350 where irregular packing is also found (Fig. 2b). The structures of gp21, MoMLV TM, and Ebola virus GP2 highlight the pivotal role of the conserved disulfide-bonded loop in stabilizing the chain reversal, and by analogy with HA, may be a key structural feature of the transition from the prefusogenic to fusion-activated TM conformation. During the structural transition, C-terminal ectodomain sequences presumably refold for antiparallel packing on the outside of the coiled coil. This refolding will relocate the adjacent membrane-anchoring sequences to induce a highly curved, destabilized region of the viral membrane in close proximity to the target membrane facilitating fusion, pore formation, and viral entry (38–41). Highly curved membrane structures have indeed been observed at sites of influenza virus-liposome fusion (38). This refolding process may be inhibited by the binding of peptides derived from HTLV-1 and HIV-1 C-terminal TM sequences to the coiled coil in fusogenic TM intermediates (refs. 42–44 and references therein; ref. 52). Because the structure of gp21 places the fusion peptide and the membrane-anchoring sequence in close proximity, it likely represents the postfusion conformation, i.e., following the fusion of viral and target membranes, with fusion peptide and transmembrane sequences within the composite membrane. This postfusion conformation is also probable for the structures of gp41, MoMLV TM, and TBHA2. Although it is in principle possible that the missing C-terminal 25 residues of gp21 could reverse the chain direction and place the transmembrane regions on the other side of the rod, it is unlikely in the context of all the available structures.
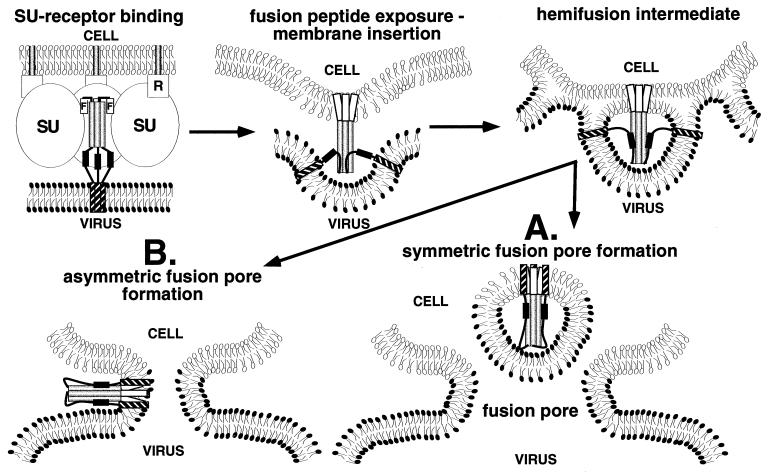
A model for retroviral membrane fusion recently was proposed by Weissenhorn et al. (10). Assembly of the C-terminal segment antiparallel to the coiled coil forces into close apposition the N-terminal fusion peptides and the C-terminal transmembrane anchors that reside in the target and viral/infected cell membranes, respectively. The flexible links between these membrane-embedded sequences and the central rod may allow nonsymmetrical bending at the end of the rod while the two membranes are drawn into close proximity for the initiation of fusion. However, it is also possible that the trimeric symmetry of the retroviral protein is maintained throughout the fusion process (Fig. 4). This scenario would require embedding of the TM in the membrane, consistent with the intramembrane particles observed by Kanaseki et al. (38) for influenza virus fusion. The exposed hydrophobic residues found at the lower tip of the rod of gp21, MoMLV TM (8), TBHA2 (26), and Ebola virus GP2 (32) (Fig. 2 b and c) may play a role in this step. Symmetrical relocation of transmembrane sequences may induce the high positive curvature in the inner membrane leaflet of the virus or infected cell that was shown to be required for the transition from hemifusion induced by glycosylphosphatidylinositol-anchored HA to full fusion (40, 41). At least two scenarios seem possible to complete the fusion process. If the entire area surrounding the TM resulted in pore formation, a membrane vesicle containing the TM will be produced (Fig. 4A); vesicles sometimes are observed during influenza virus fusion (38). However, the observed pore flickering stage of the fusion process (45) suggests that initial pore formation is reversible, and only part of the area of membrane destabilization eventually may form a stable pore. This alternative (Fig. 4B) is more obviously consistent with electron microscopic analysis of influenza HA in the fusion pH conformation (46), as well as the observed cooperativity of at least three HA trimers at a late stage of influenza virus fusion (47); the perturbation of the membrane between several TMs could increase the probability of an initial pore to advance and form a stable one.
Figure 4.
Model of retroviral membrane fusion in which the trimeric symmetry of the TM is maintained during the fusion process. R, cellular receptor. In the TM subunit, fusion peptides are indicated by white cylinders marked F, the N-terminal coiled coil by light gray cylinders, the C-terminal segment is black and the transmembrane domain is hatched. The cytoplasmic tails are not shown. Two scenarios (A and B) are possible for fusion pore formation (see text). The model is based on evidence accumulated for membrane fusion in retroviruses and influenza virus; experimental evidence for conformational changes in gp21 induced by receptor binding is not yet available.
The gp21 structure reveals suitable conserved target sites for inhibitors both in the interface between the N-terminal coiled coil and the C-terminal sequences, as previously proposed for HIV-1/SIV gp41 (9–12), and in the region of chain reversal, such as the pocket containing the Glu-398–Arg-380 salt bridge (Fig. 2f). Such inhibitors may be useful as therapeutic agents and as a means to help us understand the fusion process.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council, National Institutes of Health, and the Wellcome Trust. B.K. is a Wellcome Senior Research Fellow in Medical Science in Australia. R.J.C. is a Commonwealth AIDS Research Grant scholar, and B.E.K. is a National Health and Medical Research Council Research Fellow.
ABBREVIATIONS
- SU
surface-exposed subunit
- TM
transmembrane protein
- HTLV-1
human T cell leukemia virus type 1
- MBP
maltose-binding protein
- Env
envelope glycoprotein
- MoMLV
Moloney murine leukemia virus
- HA
influenza virus hemagglutinin
- HIV-1
HIV type 1
- SIV
simian immunodeficiency virus
- STLV-2
simian T lymphotropic virus type 2
- BLV
bovine leukemia virus
- SMRV
squirrel monkey sarcoma virus
- MPMV
Mason-Pfizer monkey virus
- RSV
Rous sarcoma virus
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The coordinates reported in this paper have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1MG1).
Yang, Z.-N., Kaufman, J., Stahl, S. J., Wingfield, P. T. & Hyde, C. C., American Crystallographic Association Annual Meeting, July 18–23, 1998, Arlington, VA, p. 153 (abstr.).
References
- 1.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osame M, Usuku K, Izumo S, Ijishi N, Amitani H, Igata A, Matsumoto M, Tara M. Lancet. 1986;1:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 3.Fan N, Gavalchin J, Paul B, Wells K, Lane M J, Poiesz B. J Clin Microbiol. 1992;30:905–910. doi: 10.1128/jcm.30.4.905-910.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donegan E, Lee H, Operskalski E A, Shaw G M, Kleinman S H, Busch M P, Stevens C E, Schiff E R, Nowicki M J, Hollingsworth C G, et al. Transfusion. 1994;34:478–483. doi: 10.1046/j.1537-2995.1994.34694295061.x. [DOI] [PubMed] [Google Scholar]
- 5.Pique C, Pham D, Tursz T, Dokhélar M-C. J Virol. 1992;66:906–913. doi: 10.1128/jvi.66.2.906-913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez L D, Peters R J, Delos S E, Young J A T, Agard D A, White J M. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 8.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 9.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 10.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 11.Tan K, Liu J-H, Wang J-H, Shen S, Lu M. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malashkevich V N, Chan D C, Chutkowski C T, Kim P S. Proc Natl Acad Sci USA. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caffrey M, Cai M, Kaufman J, Stahl S J, Wingfield P T, Covell D G, Gronenborn A M, Clore G M. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syu W-J, Lee W-R, Du B, Yu Q-C, Essex M, Lee T-H. J Virol. 1991;65:6349–6352. doi: 10.1128/jvi.65.11.6349-6352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klasse P J, Pipkorn R, Blomberg J. Proc Natl Acad Sci USA. 1988;85:5225–5229. doi: 10.1073/pnas.85.14.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J J, Steel S, Wisniewolski R, Wang C Y. Proc Natl Acad Sci USA. 1986;83:6159–6163. doi: 10.1073/pnas.83.16.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz T F, Jameson B A, Lopalco L, Siccardi A G, Weiss R A, Moore J P. AIDS Res Hum Retroviruses. 1992;8:1571–1580. doi: 10.1089/aid.1992.8.1571. [DOI] [PubMed] [Google Scholar]
- 18.Center R J, Kobe B, Wilson K A, Teh T, Howlett G J, Kemp B E, Poumbourios P. Protein Sci. 1998;7:1612–1619. doi: 10.1002/pro.5560070715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Daresbury Laboratory, Warrington, U.K.: Science and Engineering Research Council; 1993. pp. 56–62. [Google Scholar]
- 20.Quiocho F A, Spurlino J C, Rodseth L E. Structure (London) 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 21.DeLano W L, Brünger A T. Acta Crystallogr D. 1995;51:740–748. doi: 10.1107/S0907444995001284. [DOI] [PubMed] [Google Scholar]
- 22.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Brünger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 24.Lamzin V S, Wilson K S. Acta Crystallogr D. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 26.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 27.Mahieux R, Ibrahim F, Mauclere P, Herve V, Michel P, Tekaia F, Chappey C, Garin B, Van Der Ryst E, Guillemaine B, et al. J Virol. 1997;71:1317–1333. doi: 10.1128/jvi.71.2.1317-1333.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg A R, Delamarre L, Pique C, Pham D, Dokhélar M-C. J Virol. 1997;71:7180–7186. doi: 10.1128/jvi.71.10.7180-7186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M, Blacklow S C, Kim P S. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 30.Blacklow S C, Lu M, Kim P S. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 31.Rost B. Methods Enzymol. 1996;266:525–559. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 32.Weissenhorn W, Carfi A, Lee K H, Skehel J J, Wiley D C. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 33.Joshi S B, Dutch R E, Lamb R A. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 34.Wilson I A, Skehel J J, Wiley D C. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Wharton S A, Weissenhorn W, Calder L J, Hughson F M, Skehel J J, Wiley D C. Proc Natl Acad Sci USA. 1995;92:12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr C M, Chaudhry C, Kim P S. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poumbourios P, Wilson K A, Center R J, El Ahmar W, Kemp B E. J Virol. 1997;71:2041–2049. doi: 10.1128/jvi.71.3.2041-2049.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanaseki T, Kawasaki K, Murata M, Ikeuchi Y, Ohnishi S. J Cell Biol. 1997;137:1041–1056. doi: 10.1083/jcb.137.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chernomordik L, Kozlov M M, Zimmerberg J. J Membr Biol. 1995;146:1–14. doi: 10.1007/BF00232676. [DOI] [PubMed] [Google Scholar]
- 40.Kemble G W, Danielli T, White J M. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 41.Melikyan G B, Brener S A, Ok D C, Cohen F S. J Cell Biol. 1997;136:995–1005. doi: 10.1083/jcb.136.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagara Y, Inoue Y, Shiraki H, Jinno A, Hoshino H, Maeda Y. J Virol. 1996;70:1564–1569. doi: 10.1128/jvi.70.3.1564-1569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furuta R A, Wild C T, Weng Y, Weiss C D. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 44.Rimsky L T, Shugars D C, Matthews T J. J Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spruce A E, Iwata A, White J M, Almers W. Nature (London) 1989;342:555–558. doi: 10.1038/342555a0. [DOI] [PubMed] [Google Scholar]
- 46.Wharton S A, Calder L J, Ruigrok R W H, Skehel J J, Steinhauer D A, Wiley D C. EMBO J. 1995;14:240–246. doi: 10.1002/j.1460-2075.1995.tb06997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danieli T, Pelletier S L, Henis Y I, White J M. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 49.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–525. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 50.Lawrence M C, Colman P M. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 51.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 52.Kemp, B. E., McPhee, D. A. & Doherty, R. R. (1988) Eur. Patent Appl. 88,312,216.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.