Abstract
The Escherichia coli BglG protein antiterminates transcription at two terminator sites within the bgl operon in response to the presence of β-glucosides in the growth medium. BglG was previously shown to be an RNA-binding protein that recognizes a specific sequence located just upstream of each of the terminators and partially overlapping with them. We show here that BglG also binds to the E. coli RNA polymerase, both in vivo and in vitro. By using several techniques, we identified the β′ subunit of RNA polymerase as the target for BglG binding. The region that contains the binding site for BglG was mapped to the N-terminal region of β′. The β′ subunit, produced in excess, prevented BglG activity as a transcriptional antiterminator. Possible roles of the interaction between BglG and the polymerase β′ subunit are discussed.
The bgl operon in Escherichia coli, induced by β-glucosides, is regulated by two of its gene products, BglG, a transcriptional regulator, and BglF, a membrane-bound sensor (1). Transcription from the bgl promoter initiates constitutively, but in the absence of inducer, most transcripts terminate prematurely at one of two ρ-independent terminators within the operon; in the presence of inducer, BglG prevents termination at these sites (2, 3). The mechanism by which BglG antiterminates transcription differs from the antitermination mechanisms which operate in λ and the mechanisms of attenuation at amino acid biosynthetic operons, as well as at the pyrB1 and ampC operons (4). BglG is an RNA-binding protein that recognizes and binds to a specific sequence on the bgl transcript, which partially overlaps with each of the bgl terminators (5). BglF, the β-glucosides phosphotransferase, regulates the activity of BglG by reversible phosphorylation (6–9), which modulates BglG dimeric state (10). The phosphorylation and dimerization sites on BglG were recently mapped (11–13).
Systems that resemble the bgl system were identified in different organisms (14). The most studied are the two sac systems in Bacillus subtilis, which regulate transcription of sacB and sacPA according to sucrose availability. Expression of these loci is regulated at the level of transcription antitermination by the two BglG homologues, SacY and SacT, respectively (15–17). Similarly to BglG, SacY is reversibly phosphorylated in vivo (18). The RNA sequences recognized by SacY and SacT closely resemble the target site for BglG in the bgl transcript (19, 16) and also have the potential to fold into a stem–loop structure that partially overlaps with the respective terminators (5, 20).† It was suggested that the antiterminators of the BglG family block the formation of the terminator structures by stabilizing an alternative RNA conformation. The question then arises as to whether interaction with the nascent RNA chain is sufficient for implementing antitermination or additional interactions with the transcription machinery are required. In this paper we provide both in vivo and in vitro evidence for the interaction of BglG with the β′ subunit of E. coli RNA polymerase (RNAP). The interaction site is contained within the first 517 aa of β′. Overexpression of β′ titrated BglG in vivo and therefore prevented it from functioning as a transcriptional antiterminator.
MATERIALS AND METHODS
Plasmids.
Some plasmids used in this work are listed in Table 1. pT7FH-G carries the bglG gene (6). pGP1–2 carries the T7 RNAP gene under control of λCI857 repressor (BRL). pMBP–BglG, obtained from A. Wright, carries a fusion between malE and the entire bglG gene cloned downstream of Ptac promoter. Plasmids pLAX185 (21), pMKSe2 (22), and pT7β′ (23) encode the α, β, and β′ subunits of E. coli RNAP, respectively. pMKA201 (24) codes for β′ with a C-terminal hexahistidine tag; the rpoC gene is cloned downstream from tandem Plac and PT7 (Φ10P) promoters. A pMKA201 derivative, which expresses β′ first 517 residues fused to its last 142 residues [β′-(1–517 and 1266–1407)], was obtained from K. Severinov. The plasmids that express either β′ first 876 residues or β′ last 863 residues [β′-(1–876) and β′-(545–1407), respectively] were derived from pMKA201 (25). The plasmid that encodes β′ last 547 residues [β′-(877–1407)] was obtained by treating pT7β′ with HindIII (cuts once downstream of the rpoC gene) and SalI (cuts once at rpoC codon 877), flushing the ends with Klenow DNA polymerase, and religating.
Table 1.
Plasmids
| Plasmids | Relevant characteristics | Source or ref. |
|---|---|---|
| pT7FH-G | ApR; ori pBR322; Φ10P-bglG | 6 |
| pMBP-BglG | ApR; ori pBR322; tacP-malE-bglG | A. Wright |
| pLAX185 | ApR; ori pBR322; lppP-lacP-rpoA | 22 |
| pMKSe2 | ApR; ori pBR322; lacP-rpoB | 23 |
| pT7β′ | ApR; ori pBR322; Φ10P-rpoC | 24 |
| pMKA201 | ApR; ori pBR322; lacP-Φ10P-rpoC | 25 |
In plasmid pMKA201, only the Plac promoter was used for the expression of rpoC because of the lack of T7 polymerase in the host strain. ApR, ampicillin resistance.
Media.
Enriched and minimal media were prepared as described (26). The minimal medium used for 35S-labeling (27) contained 0.4% succinate as carbon source. Ampicillin (200 μg/ml), kanamycin (30 μg/ml), or chloramphenicol (30 μg/ml) were added as appropriate.
Preparation of Cell Extracts Enriched for 35S-Labeled BglG.
A culture of E. coli strain K38 (HfrC trpR thiλ+, from C. Richardson) containing pT7FH-G and pGP1–2 was induced and labeled with [35S]methionine in the presence of rifampicin (27). Cells were harvested by centrifugation, washed with 10 mM Tris⋅HCl (pH 7.9), and resuspended in 20% sucrose (wt/vol) and 0.03 M Tris⋅HCl followed by incubation with 0.1 mg lysozyme in 0.1 M EDTA for 30 min on ice. Extracts were frozen and thawed four times and then centrifuged at 9,000 × g for 5 min. The supernatant was stored at −70°C or used directly in the coimmunoprecipitation assays.
Purification of Maltose-Binding Protein (MBP)–BglG.
Expression and purification of MBP–BglG were carried out in E. coli strain MC1061 [hsdR mcrB araD 139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi] as described in Chen et al. (9).
Purification of β and β′ Subunits.
RNAP β subunit was overproduced in E. coli strains HB101 [supE44 hsdS20(rB−mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1] containing pMKSe2 and pLacIQ, and BL21(DE3) [hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene1) (Stratagene)] containing pT7β′ and pLacIQ (28). Proteins were recovered from inclusion bodies (29) and dialyzed for 16 h at 4°C against reconstitution buffer [50 mM Tris⋅HCl, pH 7.9/0.2 M KCl/20% glycerol (vol/vol)/10 mM MgCl2/10 μM ZnCl2/1 mM EDTA/1 mM DTT].
Preparation of Cell Extracts Enriched for Truncated β′ Proteins.
E. coli XL1-Blue cells [supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac− F′ (proAB+ lacIq lacZΔM15 Tn10), (Stratagene)] containing pMKA201 and its derivatives, which encode the truncated β′ proteins, β′-(1–517 and 1266–1407), β′-(1–876) and β′-(545–1407), and E. coli BL21(DE3) cells containing a plasmid that encodes the truncated β′ protein β′-(877–1407) were grown at 30°C to an OD600 of 0.5 in LB containing ampicillin. Isoprophyl β-d-thiogalactoside (IPTG) was added to a final concentration of 0.5 mM. Cells were grown for an additional 2.5 h (the OD600 was ≈1) and harvested by centrifugation.
Electrophoresis and Immunoblotting.
Proteins were fractionated on an SDS/8% polyacrylamide gel (30) and blotted onto a nitrocellulose filter (31). After blocking with 1% fat milk for 1 h at room temperature, the filter was incubated with either anti-MBP antiserum (New England Biolabs) or anti-RNAP holoenzyme antiserum (from A. Ishihama), diluted 1:10,000 in 1% fat milk, or with monoclonal antibodies against the individual RNAP subunits (from A. Goldfarb), diluted 1:5,000, for 16 h at 4°C with gentle mixing. After three washes of 5 min in PBS containing 0.01% Tween 20, the filter was incubated for 30 min at room temperature with horseradish peroxidase-conjugated goat anti-rabbit IgGs or IgMs (Jackon ImmunoResearch), which were diluted 1:20,000 in PBS containing 0.1% Tween 20. The filter was then washed three times in PBS for 5 min. Binding of the antibodies to the filter was probed by using the enhanced chemiluminescence (ECL) light-based detection procedure (Amersham Pharmacia) and visualized by autoradiography.
Coimmunoprecipitation.
RNAP holoenzyme (Boehringer Mannheim) or individual RNAP subunits, α, β, or β′, were mixed with either MBP–BglG or MBP in equimolar amounts in binding buffer (25 mM Tris⋅HCl, pH 7.5/150 mM NaCl/0.25% Triton X-100/5 mM EDTA/0.1% BSA/0.2 mM PMSF). The protein mixtures (200 μl) were rotated gently at 4°C for 16 h. In parallel, Protein A-Sepharose (Amersham Pharmacia; 25 μl) was incubated with either anti-MBP antiserum or anti-RNAP holoenzyme antiserum or mAb against the individual RNAP subunits for 16 h, at 4°C in PBS. The Ab-coated Protein A was added to each of the above protein mixtures, and incubation was continued at room temperature for 2 h with slow mixing.
Immunoprecipitation of [35S]BglG with RNAP from cell extracts, using anti-RNAP holoenzyme antiserum, was carried out essentially as described (6).
The Protein A-Sepharose-bound proteins, obtained by centrifugation, were washed three times with binding buffer and resuspended in electrophoresis sample buffer (30). After boiling and removal of insoluble material by centrifugation, the supernatants were analyzed by using SDS/PAGE. When using [35S]BglG, the gels were dried and exposed to Kodak XAR-5-ray film. Otherwise, antibodies against the appropriate proteins were used to probe for precipitation as described above.
Far-Western Analysis.
SDS/polyacrylamide gel-fractionated proteins (approximately 5 μg of protein per lane) were transferred to a nitrocellulose filter. Denaturation, renaturation, and blocking with BSA were done as described (32). The filter was incubated with 5 μg of the appropriate secondary protein (indicated in the text) for 16 h at 4°C, followed by three washes of 5 min in PBS containing 0.1% Tween 20. To detect binding of the secondary protein to the blotted protein, we used antibodies directed against the secondary protein followed by the ECL light-based detection procedure.
Affinity Chromatography.
MBP–BglG (3 μg) in 200 μl of column buffer (20 mM Tris⋅HCl, pH 7.5/200 mM NaCl/1 mM EDTA/20 mM PMSF) were mixed gently with 20 μl of amylose resin matrix (New England Biolabs) for 2 h at 4°C. The resin was then centrifuged for 30 sec at 5,000 × g, washed five times with 1 ml of column buffer, and incubated with RNAP holoenzyme (2 μg) or its individual subunits for 16 h at 4°C. After washing the resin five times with 1 ml of column buffer, bound proteins were eluted by gently mixing with column buffer containing 10 mM maltose for 10 min. After centrifugation, the supernatants were analyzed by using Western blot analysis.
Measurements of β-Galactosidase Activity.
Plasmids expressing the different RNAP subunits were introduced into the E. coli MA200–1 strain, which carries a bgl′-lacZ fusion on its chromosome and a defective bglF gene [F− ΔlacX74 thi srl∷Tn10 recA56 bglR11 (bglR∷IS1) (Bgl+) bglF201(λ bglR7 bglG′ lacZ+ lacY+ Φ(bgl-lac) (33)]. Fresh colonies were inoculated into LB, which was supplemented with the appropriate antibiotics, and grown at 37°C for 2 h. Assays for β-galactosidase activity were carried out as described by Miller (26).
RESULTS
BglG Associates with RNAP in the Cell.
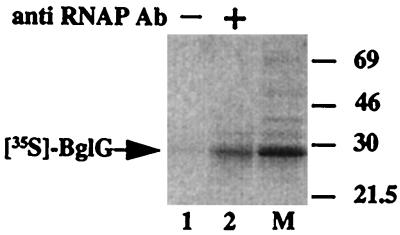
To determine whether BglG is found in a complex with RNAP in the cell, we tested whether BglG coprecipitates with RNAP when extracts of cells overproducing BglG are incubated with anti-RNAP antibodies. To this end, the bglG gene was overexpressed and labeled with [35S]methionine. BglG was almost exclusively labeled under the conditions employed (Fig. 1, lane M). The cellular proteins were extracted by a procedure that does not involve denaturation. Incubation of this extract with antibodies against RNAP led to precipitation of [35S]BglG (Fig. 1, lane 2). No [35S]BglG was detected when anti-RNAP antibodies were not included in the incubation (Fig. 1, lane 1).
Figure 1.
BglG and RNAP can be coimmunoprecipitated from cellular extracts. Extracts of cells enriched for [35S]BglG were prepared as described in Material and Methods. The major labeled product was BglG (lane M). The extracts were incubated either without (lane 1) or with (lane 2) antibodies against RNAP. Samples were analyzed by using SDS/PAGE followed by autoradiography. Molecular masses of protein standards are given in kDa.
BglG Binds Directly to RNAP.
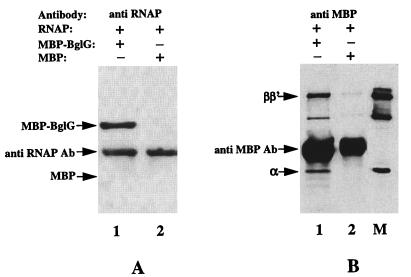
To examine whether BglG can interact directly with RNAP or their association requires accessory proteins, we carried out immunoprecipitation experiments with purified BglG fused to maltose binding protein (MBP–BglG) and purified RNAP holoenzyme. These proteins were mixed and incubated with antibodies against MBP or against RNAP holoenzyme. Precipitation of MBP–BglG or RNAP was detected by probing the blotted proteins with anti-MBP or anti-RNAP antibodies, respectively. MBP–BglG precipitated after incubation with RNAP and anti-RNAP antibodies (Fig. 2A, lane 1). In the reciprocal experiment, RNAP precipitated after incubation with MBP–BglG and antibodies against MBP (Fig. 2B, lane 1). To rule out the possibility that precipitation occurred because of interaction of the MBP moiety of MBP–BglG with RNAP, we repeated these experiments with purified MBP protein instead of MBP–BglG. Precipitation of MBP with RNAP and anti-RNAP antibodies was not detected (Fig. 2A, lane 2), nor was precipitation of RNAP with MBP and antibodies against MBP (Fig. 2B, lane 2). These results imply that the interaction of BglG with RNAP is direct and does not require auxiliary proteins.
Figure 2.
BglG directly interacts with RNAP. Purified MBP–BglG (lanes 1) or MBP (lanes 2) was incubated with purified RNAP and antibodies against RNAP holoenzyme (A) or against MBP (B), both raised in rabbit. Immunoprecipitated proteins were fractionated by using SDS/PAGE, blotted onto nitrocellulose filters, and probed with antibodies against MBP (A) or against RNAP holoenzyme (B). The filter was reacted with a secondary antibody, goat-anti rabbit, which was detected as described in Material and Methods. The first antibody, which is included in each incubation, also fluoresces up because of the interaction with the secondary antibody. Lane M contains RNAP as a marker.
BglG Interacts with the β′ Subunit of RNAP.
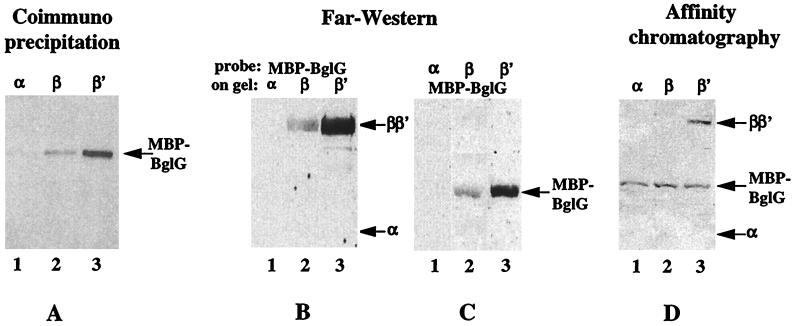
To identify the RNAP subunit that interacts with BglG, we used three techniques: immunoprecipitation, Far Western analysis, and affinity chromatography. The purpose of pursuing the three techniques, in parallel, was to circumvent the possibility that binding of one of the components to antibodies (during immunoprecipitation) or to a matrix (of an affinity column) or denaturation of a component (before blotting for Far Western), will sequester or destroy the binding domain(s), at least partially, or give artifacts. In the experiments described below we used purified MBP–BglG as well as purified RNAP subunits.
We first asked which RNAP subunit can lead to precipitation of MBP–BglG when incubated with antibodies against the respective subunit. The blotted immunoprecipitated proteins were probed with antibodies against MBP. The results are presented in Fig. 3A. MBP–BglG did not coprecipitate with the α subunit (Fig. 3A, lane 1), some precipitation of MBP–BglG was detected with the β subunit (Fig. 3A, lane 2), and a strong signal was observed after incubation with the β′ subunit (Fig. 3A, lane 3). In the reciprocal experiment, MBP–BglG was incubated with each of the subunits in the presence of anti-MBP antibodies, and the blotted immunoprecipitated proteins were probed with antibodies against the respective subunit. The results were essentially the same, i.e., no signal with α, a weak signal with β, and a strong signal with β′ (data not shown). When MBP, instead of MBP–BglG, was incubated with the purified RNAP subunits, no precipitation of MBP was detected with any of the subunits (data not shown). The results presented here are in agreement with our finding that after the incubation of heat-denatured RNAP with MBP–BglG and anti-MBP antibodies, a signal was detected only with the comigrating β and β′ subunits (data not shown).
Figure 3.
BglG interacts with the β′ subunit of RNAP. (A) Purified MBP–BglG was incubated with equimolar amounts of purified α (lane 1), β (lane 2), or β′ (lane 3) subunits of RNAP, in the presence of mAbs against the respective subunit. The immunoprecipitated MBP–BglG was detected after SDS/PAGE and Western blot analysis by probing the blot with antibodies against MBP. (B) The individual purified RNAP subunits were fractionated by using SDS/PAGE, blotted onto a nitrocellulose filter, and probed with MBP–BglG, and then with anti-MBP antibodies. (C) Purified MBP–BglG was blotted onto a nitrocellulose filter after SDS/PAGE and probed with the individual RNAP subunits and then with antibodies against the RNAP holoenzyme. (D) Purified α (lane 1), β (lane 2), or β′ (lane 3) were added to MBP–BglG immobilized on amylose resin. After washes, the proteins that eluted with maltose were fractionated by using SDS/PAGE, blotted onto a nitrocellulose filter, and probed with anti-RNAP and anti-MBP antibodies.
We next tested the interaction between BglG and the RNAP subunits by using the Far Western technique. The blotted individual subunits were probed with MBP–BglG and then with antibodies against MBP. Again, no signal was detected with the α subunit (Fig. 3B, lane 1), a weak signal was detected with β (Fig. 3B, lane 2) and a strong signal was observed with β′ (Fig. 3B, lane 3). The reciprocal experiment, in which a filter-immobilized MBP–BglG was probed with the individual subunits and then with antibodies against RNAP holoenzyme, gave the same results (Fig. 3C). When MBP, instead of MBP–BglG, was used to probe the different RNAP subunits or was probed by them, no binding was observed (data not shown). These results are in agreement with results of earlier experiments: when RNAP was fractionated, blotted, and probed with MBP–BglG and antibodies against MBP, only the comigrating β and β′ subunits were detected; when strips of filter with MBP–BglG were incubated with heat-denatured RNAP and antibodies against the individual subunits, only binding to β′, but not to α and β, could be detected (not shown).
Last, we tested which RNAP subunit can bind to MBP–BglG, which was immobilized on amylose columns, and elute with the MBP–BglG on the addition of maltose (which replaces the amylose in binding the MBP). When the blotted eluted proteins were probed with antibodies against the different RNAP subunits, only β′ was detected (Fig. 3D).
The results, obtained with the three different techniques, demonstrate unequivocally that BglG binds to the β′ subunit of RNAP.
BglG Interacts with the N-Terminal Region of β′ Subunit.
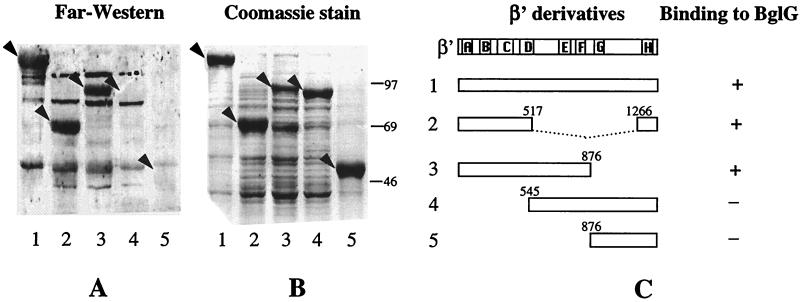
To identify the region on β′ that contains the site to which BglG binds, we examined the interaction of BglG with β′ proteins, which were truncated from either the N or the C termini, by using the Far Western technique. Cellular extracts, enriched for β′ or for one of the four truncated β′ proteins, β′-(1–517 and 1266–1407), β′-(1–876), β′-(545–1407), and β′-(877–1407), were fractionated by using SDS/PAGE, blotted onto a nitrocellulose filter, and probed with MBP–BglG and then with antibodies against MBP. The results are presented in Fig. 4. Strong signals were detected with wild-type β′ and with two truncated proteins that contain the N-terminal region of β′ (Fig. 4A, lanes 1–3). However, no binding of BglG to truncated β′ proteins that lack the N-terminal region was observed (Fig. 4A, lanes 4 and 5). The failure to detect binding with these proteins was not due to their expression at a lower level (Fig. 4B, compare the levels of the different overproduced proteins). Therefore, we conclude that BglG interacts with a site contained within the first 517 residues of β′.
Figure 4.
BglG interacts with the N-terminal region of β′ subunit. (A) Extracts of cells overproducing β′ (1) or one of the truncated β′ proteins, β′-(1–517 and 1266–1407) (2), β′-(1–876) (3), β′-(545–1407) (4), and β′-(877–1407) (5), were fractionated by using SDS/PAGE, blotted onto a nitrocellulose filter, and probed with MBP–BglG and then with antibodies against MBP (A) or stained with Coomassie blue (B). Arrowheads indicate the positions of β′ and its derivatives. (C) Schematic representation of β′ and its derivatives. Numbers of amino acids are indicated. Boxed areas A to H indicate evolutionarily conserved regions (56). + indicates binding of BglG to the β′ derivative; − indicates lack of binding.
The β′ Subunit of RNAP Competes for BglG Binding in Vivo.
The results described thus far demonstrate that BglG interacts with RNAP and that the target for BglG interaction, at least in vitro, is the β′ subunit. To establish which polymerase subunit binds BglG in vivo, we tested whether overproduction of β′, or any other polymerase subunit, can compete for binding of the cellular BglG and therefore reduce the level of BglG available for transcriptional antitermination (in vivo titration). To this end, we made use of strain MA200–1, which carries a bgl′-lacZ fusion (a fusion of the bgl promoter and transcription terminator to the lacZ gene) (33). Because of the constitutive expression of BglG in this strain, lacZ is constitutively expressed. Therefore, MA200–1 forms red colonies on MacConkey–lactose plates and gives high production of β-galactosidase units (Table 2). Plasmids expressing the individual RNAP subunits were introduced into MA200–1 containing pLacIQ. Expression of the RNAP subunits from the different plasmids was from the IPTG-inducible Plac promoter. The ability of BglG to act as a transcriptional antiterminator in the transformed strains was tested by growing them on MacConkey–lactose plates and by measuring β-galactosidase activity. The results are presented in Table 2. Overexpression of the α and β subunits had no effect on lacZ expression, indicating that these subunits cannot titrate BglG. Overproduction of β′ inhibited bgl′-lacZ expression, as demonstrated by the growth of white colonies on MacConkey–lactose plates and by the dramatic reduction in the level of β-galactosidase (8 units vs. >100 units in all of the other cases). Based on these results, we concluded that β′, produced in excess, was acting as an efficient competitor for BglG binding in vivo, whereas the other polymerase subunits were not. Overexpression of β′ apparently also had an effect on the level of BglG in the cell, because removal of BglG by titration precludes not only transcription of bgl-lacZ, but also of the bgl operon. β′ is therefore the target for BglG binding not only in vitro, but also in vivo.
Table 2.
The β′ subunit of RNAP titrates BglG in vivo
| Plasmid | Plasmid-encoded RNAP subunit | Phenotype on MacConkey–lactose plates* | β-gal activity†, units
|
|
|---|---|---|---|---|
| −IPTG | +IPTG | |||
| pLAX185 | α | red | 156 | 105 |
| pMKSe2 | β | red | 153 | 102 |
| pMKA201 | β′ | white | 102 | 8 |
| pBR322 | — | red | 138 | ND |
The experiment was carried out in MA200-1 which carries a bgl-lacZ transcriptional fusion and expresses bglG constitutively because of a mutation in bglF (33).
IPTG was added to a final concentration of 0.5 mM to plates and 1 mM to liquid medium.
The values represent the average of four independent measurements. IPTG, isopropyl β-d-thiogalactoside. ND, not determined.
DISCUSSION
The various proteins known to regulate transcription termination in prokaryotic systems, like termination factor Rho of E. coli and antitermination factors N and Q of bacteriophage λ, were shown to associate, either directly or through auxiliary proteins, with RNAP (4). At the same time, these factors recognize and bind to specific sequences in nucleic acids; whereas Rho and N bind to mRNA (34–36), Q binds to DNA (37). The RNA-binding proteins of the BglG family antiterminate transcription by a mechanism that differs from the ones suggested for N- and Q-mediated antitermination. The three-dimensional structure of the RNA-binding domain of a BglG homologue from B. subtilis, SacY, was determined and it does not resemble known RNA-binding motifs (20, 38). Antiterminators of this family were shown to bind to specific target sites on the mRNA chains that partially overlap with the terminators and that have the potential to fold into an alternative secondary structure (5, 20).† It was therefore suggested that binding of the BglG-like proteins to their target sites on the mRNA physically blocks formation of the terminator structures. This model does not necessitate interaction of these proteins with RNAP as part of the mechanism that leads to antitermination. It is reminiscent of the models suggested for the trp and other amino acid biosynthetic operons, as well as for the pyrB1 and ampC operons, in which the ribosomes function as transcriptional antiterminators, which prevent the formation of the RNA terminator structures by stabilizing alternative RNA conformations (4). However, it was also shown that BglG can recognize and bind to its target site on the mRNA only if transcription of the terminator sequence has not been completed, implying that BglG and RNAP are in very close proximity, probably physically touching each other (5). In fact, the partial overlap of the BglG-binding sequence with the terminator sequence, which makes their presence mutually exclusive, demands that the BglG recognition sequence is bound by BglG immediately as it emerges from the RNAP. Thus, unlike the case of attenuation at amino acid biosynthetic operons and pyrB1, when the close proximity of ribosomes is determined by an RNAP pause, in the case of the bgl system, BglG must be recruited to the polymerase. The results presented in this paper demonstrate that BglG interacts with the transcription machinery and that this interaction is specific and occurs with a site that is contained within the 517 N-terminal residues of β′ subunit of RNAP.
The reason that BglG needs to dimerize to act as a transcriptional antiterminator is not understood. Many DNA-binding proteins that regulate transcription act as dimers. However, these proteins bind to DNA sequences that exhibit dyad symmetry, i.e., they contain two binding sites, whereas the RNA sequence to which BglG binds contains only one binding site. Yet, much less is known about the mode of action of RNA-binding proteins in comparison to DNA-binding proteins. The need for BglG dimerization to bring about antitermination may reflect a concomitant interaction of BglG with RNA and with RNAP, each mediated by one of the dimer subunits. Alternatively, dimerization of BglG may be needed for the formation of the RNA-binding site. In the latter case, one of the RNA-bound dimer subunits, or both, may concomitantly interact with the RNAP. In the case of class I promoters, although both of the identical subunits of the E. coli catabolite gene activator protein (CAP) dimer bind to DNA, only one of them interacts with the α subunit of the RNAP (39).
According to our results, the binding of BglG to RNAP, at least in vitro, can occur in the absence of RNA. Nevertheless, it is possible that in vivo the binding of BglG to the RNA transcript is required for, or facilitates, the BglG–RNAP recognition or binding. Because in the uninduced conditions, there are only few BglG molecules in the cell, there should be a mechanism that will direct these molecules to the small subset of RNAP molecules engaged in transcribing the bgl operon. So far, the only known property of BglG that might facilitate polymerase choice is its ability to bind the to the bgl mRNA. Thus, it is possible that at physiological concentrations of BglG, there is little, if any, binding to RNAP, but under inducing conditions binding is facilitated by BglG–mRNA interaction, which juxtaposes BglG with polymerase molecules engaged in bgl transcription. The low concentration of BglG in the cell makes the opposite model, i.e., that BglG is recruited to the transcript because of its affinity to RNAP, less likely. The fact that overexpression of BglG affects cell growth does not help in discerning between these models, because this phenomenon can result from perturbation of general transcription because of binding to RNAP or from nonspecific binding to RNA or to other, as-yet-unknown, BglG-binding proteins. Another option is that BglG binding to RNAP may be independent of the RNA-binding property of BglG. We have preliminary evidence that demonstrates that truncated BglG proteins lacking the RNA-binding site did not lose the ability to bind to the RNAP in vitro (unpublished results). It was shown by L. Rothman-Denes and coworkers (40) that the single-stranded binding activity of the N4SSB protein from bacteriophage N4 is not required for the ability of this protein to bind to the β′ subunit of RNAP and to activate transcription. One of the explanations suggested by the investigators for their results is that N4SSB does not activate transcription by “tethering” the RNAP to the promoter but rather by facilitating subsequent steps in transcription. One might speculate that the role of BglG binding to the polymerase may be to facilitate subsequent transcriptional steps rather than in antitermination per se. For example, interaction of BglG with RNAP may activate the elongation complex (post-antitermination), thus guaranteeing full transcription of the operon. If BglG has a role in post-antitermination transcriptional steps, it could imply that BglG continues to interact with the polymerase and travels with it beyond the antitermination site. An in vitro system with purified components, not available yet, that faithfully imitates the in vivo events during antitermination in the bgl operon, will hopefully enable us to address these questions in the future and whether proteins other than BglG participate in the BglG-promoted transcription antitermination and/or elongation.
Accumulating evidence indicates that the β′ subunit of RNAP is involved in transcription elongation and termination. First, β′ was shown to interact both with DNA and with RNA, specifically with the RNA 3′ terminus (41–45). Many of these interactions are with sequences that are known to influence pausing and termination (46). Second, substitutions in homologous segments in β′ and in the eukaryotic RNA polymerase II largest subunit, which is a homologue of β′, were shown to confer resistance to antibiotics that block transcription elongation [streptolydigin and α-amanitin, respectively (47, 48)]. Third, a mutation in β′ was shown to suppress the rho201, allele which produces a defective Rho termination factor (49, 50). Fourth, mutations in rpoC, which block Nun-dependent termination at λ nutR have been isolated (51). Fifth, discrete regions in β′ that are involved in transcript elongation and termination were identified by the characterization of termination-altering amino acid substitutions in this subunit (52). Most of these regions are conserved in the eukaryotic homologues of β′. Finally, substitutions in the putative zinc-finger domain in the N-terminal portion of β′ were shown to block factor-independent antitermination and increase termination in coliphage HK022 DNA (53, 54). The involvement of different regions of β′ in transcription elongation and termination/antitermination seem to reflect the different interactions of β′ with the different components of the transcription complex, i.e., DNA, RNA, or transcription factors. The fact that the BglG-binding site is contained within the N-terminal portion of β′, to which the mutations that block factor-independent antitermination of phage HK022 transcription and suppress the rho201 allele, were also localized, may be significant. The localization of the BglG-binding site to the N-terminal region of β′ may explain the weak binding observed with some techniques between BglG and β, because the first 330 residues in β′ and β show weak sequence similarity (55).
Acknowledgments
We thank A. Goldfarb and K. Severinov for the gift of plasmids pMKSe2, pT7β′, pMKA201, and plasmids encoding truncated β′ proteins and for the gifts of purified RNAP α subunit and mAbs against the individual RNAP subunits. We also thank them for their advice on the purification of the β and β′ subunits of RNAP. We thank A. Ishihama for the gifts of plasmid pLAX185 and the antiserum against E. coli RNAP holoenzyme and A. Wright for the gift of plasmid pMBP–BglG. This research was supported by Grant 91-00125 from the United States-Israel Binational Science Foundation (BSF) and by a grant from the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities–Charles H. Revson Research Foundation. A.N.-S. was supported by a postdoctoral fellowship from the Israel Ministry of Science.
Footnotes
Abbrevations: RNAP, RNA polymerase; MBP, maltose binding protein.
Singh, J., Bailey, M., Diaz-Torres, M. & Wright, A., Meeting on Molecular Genetics of Bacteria and Phage, August 2–7, 1994, Madison, WI, abstr. 70.
References
- 1.Amster-Choder O, Wright A. J Cell Biochem. 1993;51:83–90. doi: 10.1002/jcb.240510115. [DOI] [PubMed] [Google Scholar]
- 2.Mahadevan S, Wright A. Cell. 1987;50:485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- 3.Schnetz K, Rak B. EMBO J. 1988;7:3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson J P. Crit Rev Biochem Mol Biol. 1993;28:1–30. doi: 10.3109/10409239309082571. [DOI] [PubMed] [Google Scholar]
- 5.Houman F, Diaz-Torres M R, Wright A. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 6.Amster-Choder O, Houman F, Wright A. Cell. 1989;58:847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- 7.Amster-Choder O, Wright A. Science. 1990;249:540–542. doi: 10.1126/science.2200123. [DOI] [PubMed] [Google Scholar]
- 8.Schnetz K, Rak B. Proc Natl Acad Sci USA. 1990;87:5074–5078. doi: 10.1073/pnas.87.13.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Arents J C, Bader R, Postma P, Amster-Choder O. EMBO J. 1997;16:4617–4627. doi: 10.1093/emboj/16.15.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amster-Choder O, Wright A. Science. 1992;257:1395–1398. doi: 10.1126/science.1382312. [DOI] [PubMed] [Google Scholar]
- 11.Amster-Choder O, Wright A. J Bacteriol. 1997;179:5621–5624. doi: 10.1128/jb.179.17.5621-5624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, Engelberg-Kulka H, Amster-Choder O. J Biol Chem. 1997;272:17263–17268. doi: 10.1074/jbc.272.28.17263. [DOI] [PubMed] [Google Scholar]
- 13.Boss, A. Nussbaum-Shochat, A. & Amster-Choder, O. (1999) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 14.Stulke J, Arnaud M, Rapoport G, Martin-Vestraete A. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 15.Crutz A M, Steinmetz M, Aymerich S, Richter R, Le-Coq D. J Bacteriol. 1990;172:1043–1050. doi: 10.1128/jb.172.2.1043-1050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debarbouille M, Arnaud M, Fouet A, Klier A, Rapoport G. J Bacteriol. 1990;172:3966–3973. doi: 10.1128/jb.172.7.3966-3973.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnaud M, Debarbouille M, Rapoport G, Saier M H J, Reizer J. J Biol Chem. 1996;271:18966–18972. doi: 10.1074/jbc.271.31.18966. [DOI] [PubMed] [Google Scholar]
- 18.Idelson M, Amster-Choder O. J Bacteriol. 1998;180:660–666. doi: 10.1128/jb.180.3.660-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aymerich S, Steinmetz M. Proc Natl Acad Sci USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manival X, Yang Y, Strub M P, Kochoyan M, Steinmetz M, Aymerich S. EMBO J. 1997;16:5019–5029. doi: 10.1093/emboj/16.16.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayward R S, Igarashi K, Ishihama A. J Mol Biol. 1991;221:23–29. doi: 10.1016/0022-2836(91)80197-3. [DOI] [PubMed] [Google Scholar]
- 22.Severinov K, Soushko M, Goldfarb A, Nikiforov V. J Biol Chem. 1993;268:14820–14825. [PubMed] [Google Scholar]
- 23.Zalenskaya K, Lee J, Gujuluva C N, Shin Y K, Slutsky M, Goldfarb A. Gene. 1990;89:7–12. doi: 10.1016/0378-1119(90)90199-2. [DOI] [PubMed] [Google Scholar]
- 24.Kashlev M, Martin E, Polyakov A, Severinov K, Nikiforov V, Goldfarb A. Gene. 1993;130:9–14. doi: 10.1016/0378-1119(93)90340-9. [DOI] [PubMed] [Google Scholar]
- 25.Severinov K, Mustaev A, Kukarin A, Muzzin O, Bass I, Darst S A, Goldfarb A. J Biol Chem. 1996;271:27969–27974. doi: 10.1074/jbc.271.44.27969. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 27.Tabor R, Richardson C C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKinney J D, Lee J, O’Neill R E, Goldfarb A. Gene. 1987;58:13–18. doi: 10.1016/0378-1119(87)90024-2. [DOI] [PubMed] [Google Scholar]
- 29.Borukhov S, Goldfarb A. Protein Expression Purif. 1993;4:503–511. doi: 10.1006/prep.1993.1066. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciufo D M, Mullen M-A, Hayward G S. J Virol. 1994;68:3267–3282. doi: 10.1128/jvi.68.5.3267-3282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahadevan S, Reynolds A E, Wright A. J Bacteriol. 1987;169:2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowery C, Richardson J P. J Biol Chem. 1977;252:1381–1385. [PubMed] [Google Scholar]
- 35.Roberts J W. Cell. 1988;52:5–6. doi: 10.1016/0092-8674(88)90523-5. [DOI] [PubMed] [Google Scholar]
- 36.Lazinski D, Grzadzielska E, Das A. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- 37.Yarnell W S, Roberts J W. Cell. 1992;69:1181–1189. doi: 10.1016/0092-8674(92)90639-t. [DOI] [PubMed] [Google Scholar]
- 38.van Tilbeurgh H, Manival X, Aymerich S, Lhoste J-M, Dumas C, Kochoyan M. EMBO J. 1997;16:5030–5036. doi: 10.1093/emboj/16.16.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Busby S, Ebright R H. Cell. 1993;73:375–379. doi: 10.1016/0092-8674(93)90236-j. [DOI] [PubMed] [Google Scholar]
- 40.Miller A, Wood D, Ebright R H, Rothman-Denes L B. Science. 1997;275:1655–1657. doi: 10.1126/science.275.5306.1655. [DOI] [PubMed] [Google Scholar]
- 41.Okada M, Vergne J, Brahms J. Nucleic Acids Res. 1978;5:1845–1862. doi: 10.1093/nar/5.6.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chenchik A, Bibiashvili R S, Mirzabekov A D, Shik V. Mol Biol (Moscow) 1982;16:35–46. [PubMed] [Google Scholar]
- 43.Hanna M M, Meares C F. Proc Natl Acad Sci USA. 1983;80:4238–4242. doi: 10.1073/pnas.80.14.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dissinger S, Hannah M M. J Biol Chem. 1991;219:11–25. doi: 10.1016/0022-2836(91)90853-x. [DOI] [PubMed] [Google Scholar]
- 45.Borukhov S, Lee J, Goldfarb A. J Biol Chem. 1991;266:23932–23935. [PubMed] [Google Scholar]
- 46.Mooney R A, Artismovitch I, Landick R. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst S E, Goldfarb A. J Biol Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 48.Sentenac A. CRC Crit Rev Biochem. 1985;18:31–91. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- 49.Jin D J, Gross C A. Mol Gen Genet. 1989;216:269–275. doi: 10.1007/BF00334365. [DOI] [PubMed] [Google Scholar]
- 50.Heisler L M, Feng G, Jin D J, Gross C A, Landick R. J Biol Chem. 1996;271:14572–14583. doi: 10.1074/jbc.271.24.14572. [DOI] [PubMed] [Google Scholar]
- 51.Robledo R, Atkinson B L, Gottesman M E. J Mol Biol. 1991;220:613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- 52.Weilbaecher R, Hebron C, Feng G, Landick R. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 53.Clerget M, Jin D J, Weisberg R A. J Mol Biol. 1995;248:768–780. doi: 10.1006/jmbi.1995.0259. [DOI] [PubMed] [Google Scholar]
- 54.King R A, Banik-Maiti S, Jin D J, Weisberg R A. Cell. 1996;87:893–903. doi: 10.1016/s0092-8674(00)81996-0. [DOI] [PubMed] [Google Scholar]
- 55.Ohnishi K. Nucleic Acids Symp Ser. 1985;22:253–256. [PubMed] [Google Scholar]
- 56.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]