Abstract
Binding of initiator methionyl-tRNA to ribosomes is catalyzed in prokaryotes by initiation factor (IF) IF2 and in eukaryotes by eIF2. The discovery of both IF2 and eIF2 homologs in yeast and archaea suggested that these microbes possess an evolutionarily intermediate protein synthesis apparatus. We describe the identification of a human IF2 homolog, and we demonstrate by using in vivo and in vitro assays that human IF2 functions as a translation factor. In addition, we show that archaea IF2 can substitute for its yeast homolog both in vivo and in vitro. We propose a universally conserved function for IF2 in facilitating the proper binding of initiator methionyl-tRNA to the ribosomal P site.
Keywords: eukaryotic translation initiation factor, FUN12, initiator tRNA
In eukaryotes, successful translation initiation requires the efficient and accurate delivery of the initiator methionyl-tRNA (Met-tRNAiMet) to the 40S ribosomal subunit. The heterotrimeric eukaryotic initiation factor eIF2 forms a ternary complex with GTP and Met-tRNAiMet and facilitates the binding of the Met-tRNAiMet to the ribosome to form a 43S preinitiation complex (1). This 43S complex then binds to an mRNA and scans to the AUG start codon. Hydrolysis of the GTP of the ternary complex results in release of eIF2 in the binary complex eIF2⋅GDP, and the guanine nucleotide exchange factor eIF2B is required to catalyze the exchange of GTP for GDP on eIF2 to regenerate the eIF2⋅GTP complex (1, 2). This nucleotide exchange reaction is an important control point of translation and phosphorylation of the α subunit of eIF2 on Ser-51 inhibits translation by converting eIF2 from a substrate to a competitive inhibitor of eIF2B (2). In prokaryotes, the single polypeptide factor IF2 mediates the binding of Met-tRNAiMet to the ribosome in a reaction facilitated by the factor IF1. IF2 has a conserved GTP-binding domain near the center of the protein, and GTP hydrolysis is required for IF2 release after subunit joining (3, 4). In contrast to eIF2, which forms a stable ternary complex with GTP and Met-tRNAiMet, IF2 does not form a stable complex with either GTP or Met-tRNAiMet in the absence of the ribosome and mRNA (3, 4). It has been shown that the N-terminal region of Escherichia coli IF2 is dispensable for cell viability (5), demonstrating that the essential functions of IF2 in translation initiation are present in the GTP-binding domain and C-terminal region of the protein.
Based on the distinct structures and activities of IF2 and eIF2, it has been thought that prokaryotes and eukaryotes have evolved different mechanisms for binding Met-tRNAiMet to the ribosome during translation initiation. However, the genome sequences of Methanococcus jannaschii (6) and Saccharomyces cerevisiae (7) revealed the presence of IF2 homologs in addition to all three eIF2 subunits in this archaeon and eukaryote, respectively. The identification of both IF2 and eIF2 homologs in archaea was surprising and contributed to speculation that archaea may possess a translation apparatus intermediate between prokaryotes and eukaryotes (8). The presence of the IF2 homolog in yeast suggests instead that IF2 has been conserved throughout the evolution of microbes from bacteria to the lower eukaryotes. Recently, we have found that the yeast IF2 homolog, yIF2, plays an important role in cellular translation initiation (9). Polyribosome profiles from yeast strains lacking the FUN12 gene, which encodes yIF2, showed reduced amounts of polyribosomes and increased levels of 80S monosomes consistent with defects in translation initiation. Extracts from fun12Δ strains were defective for translating a reporter mRNA, and this defect could be rescued by adding back purified yIF2. These results indicated that yIF2 is a general translation factor in yeast. The function of yIF2 was localized to the Met-tRNAiMet binding step of translation initiation, as yIF2 could stimulate a model assay of first peptide bond synthesis. Finally, fun12Δ strains showed defects in the translational regulation of the GCN4 mRNA, which critically depends on the rate of Met-tRNAiMet binding to 40S ribosomes. These results all are consistent with yIF2 functioning as a general translation IF in yeast cells by promoting the recruitment of Met-tRNAiMet to the ribosome.
In this study we have examined the universal conservation of IF2. We report the molecular cloning and the genetic and biochemical characterization of hIF2, a human IF2 homolog. The expression of hIF2 complemented the slow-growth phenotype of a fun12Δ yeast strain and purified hIF2 substituted for yIF2 to stimulate the translational activity of extracts from a fun12Δ strain. In addition, in vivo and in vitro analyses of hIF2 mutants containing point mutations in the GTP-binding domain, the most conserved region among the IF2 homologs, indicated that GTP binding and hydrolysis are necessary for full activity of hIF2. We also demonstrate that an archaeal IF2 homolog (aIF2) will substitute for yIF2. These results reveal a functional equivalence between archaeal and eukaryotic IF2 proteins, and they demonstrate that the IF2 protein and activity have been universally conserved throughout evolution.
MATERIALS AND METHODS
hIF2 Cloning and Plasmid Construction.
The clones 627534, 469971, and 593665, containing portions of cDNAs for hIF2, were obtained from the I.M.A.G.E. Consortium (Lawrence Livermore National Laboratory, Berkeley, CA). Primers were designed to PCR-amplify and incorporate restriction sites to ligate the clones. The three PCR products were digested with the appropriate enzymes and then ligated to the vector pGEX-4T-2 (Pharmacia) digested with BamHI and SalI to generate the glutathione S-transferase (GST)-hIF2 expression vector pC592. The same PCR products also were ligated to the vector pMAL-c2 (New England Biolabs) to generate the maltose-binding protein (MBP)-hIF2 expression vector pC597. The inserts in both plasmids were sequenced to insure no mutations were introduced by PCR. In addition, an internal SacI–SalI fragment isolated from a human testis hIF2 cDNA clone, which was isolated as described below, was inserted in pC592 and pC597 in place of the corresponding fragments to generate pC598 and pC604, respectively. Finally, the BamHI–SalI fragment from pC598 was subcloned to the yeast expression vector pEMBLyex4, creating pC602. The three expression vectors described above (pC598, pC604, and pC602) will express only the C-terminal half of hIF2, residues 587-1220 (see Fig. 1A).
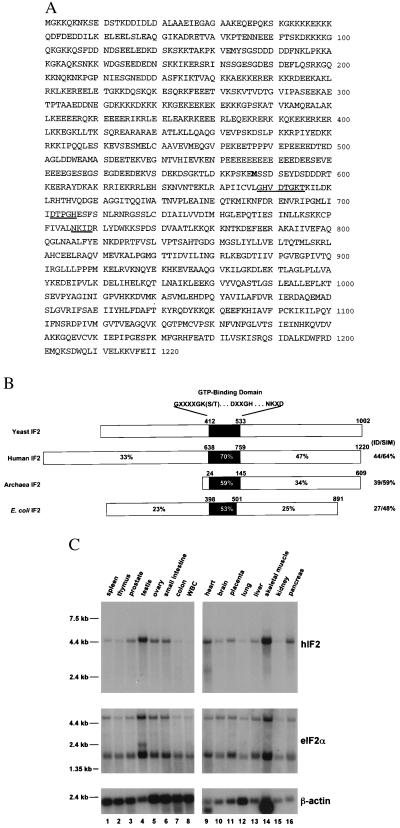
Figure 1.
Predicted amino acid sequence of hIF2 and homology of human, yeast, archaeal, and prokaryotic IF2 proteins. (A) The deduced amino acid sequence of hIF2 based on the nucleotide sequence of cDNA clones from human testis and skeletal muscle is shown. The three consensus motifs of the GTP-binding domain (12) are underlined and indicated in bold is Met-587, the start site of the N-terminal-truncated hIF2 proteins. (B) Structural conservation among the eukaryotic, archaeal, and prokaryotic IF2 proteins. Indicated on the cartoons of hIF2, aIF2 (M. jannaschii) and E. coli IF2 proteins are the percentages of amino acid sequence identities, as determined by the program bestfit (Genetics Computer Group, Madison, WI), to yIF2 in the N-terminal region, the GTP-binding domain, and the C-terminal region of the proteins. Also indicated (at the right) are the amino acid sequence identity (ID) and similarity (SIM) of the full-length proteins compared with yIF2. The black box in the cartoons identifies the location of the GTP-binding domain with the indicated consensus sequence motifs, and the numbers above the cartoons refer to the amino acid residues in each protein. (C) Northern blot analysis of RNA from human tissues probed with a portion of an hIF2 cDNA. A multiple tissue Northern blot (CLONTECH) containing 2 μg of poly(A)+ RNA isolated from the indicated human tissues was probed with a 0.7-kb fragment from a testis hIF2 cDNA encoding amino acids 587–825 (Top), with a 535-bp fragment from a human eIF2α cDNA (Middle), or with a 2-kb fragment of a human β-actin cDNA (Bottom), according to the vendor’s instructions. The positions of size standards are shown on the left.
To screen for longer hIF2 cDNA clones, a 238-bp fragment from clone 627534 was amplified by PCR, labeled by using the random priming procedure, and then used to screen 1 × 106 plaques of a human testis cDNA library in λgt10 (CLONTECH). Four positive clones were identified, and the inserts were subcloned into pGEX-4T-2 and sequenced on both strands. Because the longest clones did not encode the AUG translation initiation site, 5′-rapid amplification of cDNA ends (RACE; Life Technologies, Gaithersburg, MD) was performed by using 1 μg of human skeletal muscle or human testis poly(A)+ RNA (CLONTECH) as the template. First-strand cDNA was synthesized by using SuperScript II reverse transcriptase and a gene-specific primer. The 2.0-kb cDNA then was amplified by using the vendor’s abridged anchor primer and an antisense gene-specific primer. Four independent PCR products from both the human skeletal muscle and testis RACE cDNAs were subcloned in pGEM-T (Promega) and sequenced.
Point mutations were introduced in the hIF2 GTP-binding domain by using fusion PCR with primers containing the appropriate mutations. All new constructs were confirmed by DNA sequencing. Val-640 and His-706 were mutated to Gly and Glu by changing the GTG Val codon to GGG and the CAT His codon to GAA, respectively. His-706 and Asp-759 were mutated to Gln and Asn by changing the CAT His codon to CAA and the GAT Asp codon to AAT, respectively. SacI–StuI fragments encoding the mutations were isolated from the PCR fusion products and subcloned into SacI–StuI digested pC598, creating the plasmids pC610 (GST-hIF2-V640G), pC611 (GST-hIF2-H706E), pC899 (GST-hIF2-D759N), and pC904 (GST-hIF2-H706Q). SacI–SalI fragments isolated from pC610 and pC611 were inserted between the same sites of pC602 to create the plasmids pC612 and pC613, respectively.
aIF2 Plasmids.
In M. jannaschii the aIF2 protein (MJ0262) is expressed as a precursor containing an intein after amino acid residue Lys-30 (6). To produce aIF2 in yeast and bacteria, a DNA fragment encoding aIF2 lacking the intein was generated by PCR. A 0.35-kb PCR product encoding the aIF2 residues Met-1 to Lys-30 was obtained by using The Institute for Genomic Research (TIGR) clone AMJFK55 as a template and then ligated to the vector pProEX-HTb (Life Technologies) between the EheI and XbaI restriction sites generating the plasmid pJH169. A 1.8-kb PCR product encoding the aIF2 residues His-28 to the C terminus was obtained by using the TIGR clone AMJBT06 as a template and then ligated to pJH169 between the NcoI and XbaI sites to generate the plasmid pC688. The aIF2 insert in pC688 was isolated as an EheI–XhoI fragment and inserted between the SmaI and SalI sites of pEMBLyex4, creating the plasmid pC690, and inserted between the SmaI and XhoI sites of pGEX-4T-2, creating the plasmid pC689. Finally, the BamHI–XhoI fragment was isolated from pC689 and inserted between the same sites of pGEX-6P-1, creating the plasmid pC687. GST-aIF2 fusion protein was purified from E. coli containing the plasmid pC687, the fusion protein was cleaved with PreScission protease (Amersham Pharmacia), and the aIF2 protein was purified as will be described elsewhere.
Preparation of hIF2-Specific Antibodies.
MBP-hIF2 and GST-hIF2 expression was induced in E. coli, and the fusion proteins were purified following the vendors’ protocols. The hIF2-specific peptide SLEAQGIKADRETVAVKPT (residues 66–84) was synthesized by BioSynthesis (Lewisville, TX). Two New Zealand White rabbits were injected with 0.4 mg of MBP-hIF2 or hIF2-specific peptide and boosted at week 3 and, subsequently, at 2-week intervals by Covance Laboratories (Denver, PA). Antibodies raised against the MBP-hIF2 fusion protein were affinity purified by using the GST-hIF2 fusion protein according to the protocol of Harlow and Lane (10) with slight modifications.
In Vitro Translation Assay.
Translation reactions were performed essentially as described (9) by using in vitro translation extracts (11) prepared from the fun12Δ strain J133 (9) carrying the low copy-number FUN12 plasmid pC479 (9) or the vector pRS316 only.
RESULTS
Cloning of cDNA for a hIF2 Homolog.
Comparison of the yIF2 amino acid sequence with expressed sequence tag (EST) databases revealed significant similarities with several human EST DNAs, EST627534, EST593665, and EST469971. These three clones were obtained from the I.M.A.G.E. Consortium, and by using a set of specific nested primers for EST627534 a PCR product was generated and used to probe a multiple tissue Northern blot. As described later, the strongest expression of hIF2 mRNA was found in skeletal muscle and testis. In an attempt to obtain a full-length cDNA clone, a testis cDNA library was probed with a 700-bp fragment from the 5′ end of the most 5′ EST clone (EST627534). Of 1 × 106 plaque-forming units probed, four positive plaques remained after three rounds of screening. Based on DNA sequence analysis, the 5′ end of the longest clone appeared to lack the N terminus of hIF2. Therefore, we performed a 5′-RACE reaction using total testis mRNA as a template and obtained a 2-kb DNA fragment. Three independent 5′-RACE products were sequenced and found to have identical nucleotide sequences. Analysis of the DNA sequence of the 5′-RACE product in combination with the hIF2 cDNA clone revealed that the hIF2 mRNA extends over 4,000 nt and contains a 5′ untranslated region (UTR) of at least 118 nt, a continuous ORF of 3,660 nt, and a 3′ UTR of at least 222 nt. An in-frame stop codon was found 9 nt before the predicted start codon at nucleotide 118. In addition, the sequences surrounding this AUG codon exactly matched EST sequences from human skeletal muscle and mouse. The complete ORF of hIF2 encoded a protein of 1,220 aa with a calculated molecular mass of 139 kDa, considerably larger than the 100-kDa yIF2 (Fig. 1A). As expected, the hIF2 sequence contains three motifs characteristic for GTP-binding proteins (12) (Fig. 1 A and B). The deduced amino acid sequence of hIF2 shows 44% identity to the S. cerevisiae yIF2 and 39% identity to the aIF2 from M. jannaschii (6) (Fig. 1B). The similarity is even greater in the GTP-binding domains where hIF2 shows 70% identity to yIF2 and 54% identity to aIF2. The similarity is also strong in the C-terminal regions of the proteins. The N-terminal regions of hIF2 and yIF2 showed little sequence similarity; however, both proteins contain a large number of charged residues. In fact, of the first 637 aa in hIF2, 347 are charged residues with the majority being either Glu or Lys (Fig. 1A). Long stretches of poly-Glu and poly-Lys can be found in the N-terminal regions of both hIF2 and yIF2.
To verify that the sequences of the 5′-RACE product and the hIF2 cDNA were present on the same mRNA, we probed a Northern blot containing poly(A)+ RNA from various human tissues with probes derived from the 5′-RACE product (encoding hIF2 residues 8–187) and the hIF2 cDNA (encoding amino acids 587–825). Both probes detected a 4.5-kb transcript in the human tissues as shown in Fig. 1C (Top) for the probe derived from the hIF2 cDNA. The 4.5-kb length of the RNA is consistent with the lengths of the cDNA and 5′-RACE product. In addition, the expression pattern detected by the two probes was identical with highest expression in testis and skeletal muscle. A strikingly similar expression pattern was observed when the same blot was probed with a fragment of a human eIF2α cDNA (Fig. 1C, Middle). The expression pattern observed here for eIF2α is consistent with the pattern reported previously for human and mouse eIF2α (13), and the expression patterns for hIF2 and eIF2α are also similar to those previously reported for the translation IFs eIF4GI and eIF4GII (14), consistent with the idea that hIF2 is a human translation IF.
To examine hIF2 protein expression, antibodies were generated against a MBP-hIF2 fusion protein that contains the C-terminal half of hIF2. The antibodies were affinity-purified and then tested in an immunoblot analysis of a HeLa cell extract. A single crossreactive polypeptide of 175 kDa was detected (Fig. 2A, lane 1), whereas the predicted size of the protein was 139 kDa. This size discrepancy may be the result of posttranslational modifications of hIF2 or, perhaps more likely, of unusual behavior in SDS/PAGE caused by the highly charged N-terminal region of hIF2. We have previously observed that the highly charged eIF2β and eIF1A translation factors migrate in SDS/PAGE with a slower mobility than expected based on the amino acid sequence of the proteins (T.E.D. and W. C. Merrick, unpublished observations). To further confirm that the 5′-RACE product and the hIF2 cDNA encode the same protein, we raised antibodies against an N-terminal polypeptide from hIF2 (residues 66–84). This antipeptide antiserum detected a 175-kDa polypeptide in HeLa cell extracts that perfectly comigrated with the protein detected by using the anti-hIF2 antibodies (Fig. 2A, compare lane 2 to lane 1). These results confirm the authenticity of the hIF2 clone and demonstrate that the 175-kDa polypeptide is hIF2. Similar 175-kDa polypeptides also were detected in immunoblot analyses of extracts from mouse NIH 3T3 cells and from rabbit reticulocyte lysates (data not shown).
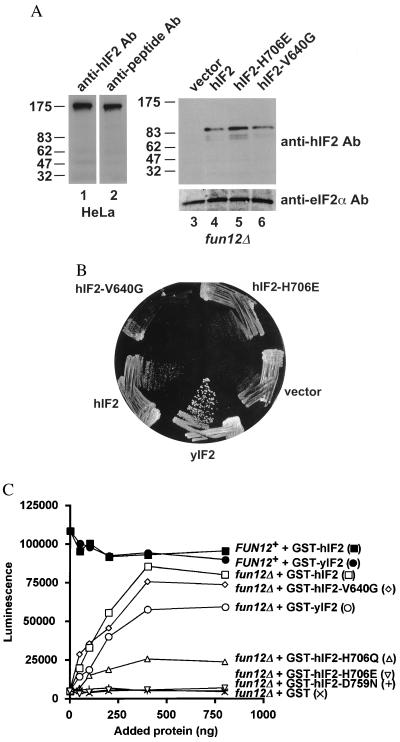
Figure 2.
hIF2 substitutes for yIF2 to stimulate translation. (A) Protein immunoblot analysis of hIF2. (Left) Twenty micrograms of crude HeLa cell extract was subjected to SDS/PAGE and then blotted to a nitrocellulose membrane. The blot was probed with affinity-purified anti-hIF2 antibodies that were raised against a MBP-hIF2 fusion protein containing the C-terminal half of hIF2 (lane 1), then the blot was stripped and probed with antiserum raised against an N-terminal hIF2 peptide (lane 2). (Right, lanes 3–6) 20 μg of crude yeast cell extracts from a fun12Δ strain expressing the indicated hIF2 proteins was subjected to SDS/PAGE and then blotted to a nitrocellulose membrane. The blot was probed with the affinity-purified anti-hIF2 antibodies, and then stripped and probed with anti-eIF2α antiserum, as indicated. Positions of protein molecular mass markers (kDa) are indicated on the left. (B) Expression of hIF2 in yeast complements the slow-growth phenotype of a fun12Δ strain. The fun12Δ strain J133 [Mataura3–52 leu2–3 leu2–112 fun12Δ] was transformed with the vector pEMBLyex4 (vector), the pEMBLyex4-hIF2 expression plasmids pC602 (hIF2), pC612 (hIF2-V640G), or pC613 (hIF2-H706E), or the pEGKT-GST-yIF2 plasmid pC485 (yIF2) (9). The indicated strains were streaked on synthetic minimal medium containing 10% galactose plus the required nutrient supplements, and the plates were incubated at 30°C for 5 days. (C) Restoration of translational activity in extracts from fun12Δ strains by addition of purified hIF2. In vitro translation extracts were prepared from the fun12Δ strain J133 carrying the low-copy-number plasmid pC479 containing wild-type FUN12 (FUN12+) or the vector pRS316 only (fun12Δ). Extracts were incubated with 200 ng of luciferase mRNA and the indicated amounts of recombinant GST, GST-yIF2, or the indicated GST-hIF2 wild-type or mutant fusion proteins. Translational activity was determined by measuring luminescence after 15-min incubation at 26°C. The results presented are representative of at least three independent experiments.
hIF2 Functionally Substitutes for yIF2 Both in Vivo and in Vitro.
Based on the high level of sequence similarity between hIF2 and yIF2, we proposed that hIF2 is the human homolog of yIF2. To test this hypothesis we asked whether hIF2 could substitute for yIF2 in yeast cells. Strains lacking the FUN12 gene encoding yIF2 show a severe slow-growth phenotype. Expression of an N-terminally truncated form of yIF2 under the control of a galactose-inducible GAL promoter fully complemented the slow-growth phenotype of a fun12Δ strain (Fig. 2B, compare sectors labeled vector and yIF2, and data not shown). As shown in Fig. 2B, expression of an N-terminally truncated version of hIF2 under the control of the same GAL promoter partially complemented the slow-growth phenotype of the fun12Δ stain on galactose medium. Thus, hIF2 is a functional homolog of yIF2. These results demonstrate that IF2 function has been conserved between yeast and mammalian cells, and that IF2 is not simply a microbe-specific translation factor.
To determine whether the in vivo complementation by hIF2 in the fun12Δ strain resulted from a restoration of translational activity, we next examined the ability of purified hIF2 to rescue the translation defect in extracts from a fun12Δ strain. A luciferase reporter mRNA was added to in vitro translation extracts prepared from isogenic wild-type and fun12Δ strains. Translational activity was determined by monitoring luminescence. As reported previously, and as shown in Fig. 2C, an extract from a wild-type strain had roughly 20-fold greater translational activity than an extract from a fun12Δ strain. This translational defect was rescued in a dose-dependent manner by the addition of purified recombinant GST-yIF2 fusion protein or by addition of purified recombinant GST-hIF2 (Fig. 2C). In contrast to the in vivo complementation studies, GST-hIF2 appeared more active than GST-yIF2 in the luciferase assay. However, this finding may reflect a difference in the sensitivities of the in vivo and in vitro assays; while the in vivo complementation requires the efficient synthesis of many proteins, the in vitro assay examines the translation of only a single mRNA under less competitive conditions. Based on the ability of GST-hIF2 to stimulate protein synthesis in this in vitro assay we conclude that hIF2 is a translation IF and that the function of IF2 in protein synthesis has been conserved between yeast and human.
Mutations in the GTP-Binding Domain of hIF2 Affect its Ability to Promote Translation.
All IF2 proteins from bacteria to humans contain the three consensus sequence motifs of a GTP-binding domain (12). To test whether the GTP-binding domain is critical for translational activity of hIF2, we introduced point mutations in the hIF2 motifs I and II, which are primarily responsible for interactions with the phosphates of GDP or GTP, and in motif III, which makes interactions with the guanine base (12). Val-640 of hIF2 in motif I corresponds to Val-20 of the translation elongation factor EF-Tu from E. coli and to Gly-12 of the oncoprotein ras. Mutations at Gly-12 in ras lead to a transformed phenotype and mutation of Val-20 in EF-Tu to Gly significantly impairs the GTPase activity and appears to lock EF-Tu in an active conformation. An EF-Tu-V20G mutant protein retains some activity in model translation assays (15, 16). Expression of an hIF2-V640G mutant protein not only failed to complement the slow-growth phenotype of a fun12Δ strain, but also appeared to exacerbate the slow-growth phenotype in this strain (Fig. 2B). Perhaps the mutant protein is poisoning translation by binding to the ribosome and interfering with the inefficient mechanism of translation initiation that occurs in the absence of any IF2 protein. When this same mutation was introduced into a GST-hIF2 fusion protein and then tested for its effect on the restoration of translational activity in extracts from a fun12Δ strain, it was found to retain the full activity observed with the wild-type hIF2 fusion protein (Fig. 2C). This in vitro activity of the hIF2-V640G mutant protein is consistent with the partial activity in model translation assays of the corresponding EF-Tu-V20G mutant protein and may indicate that the in vitro luciferase translation assay is a single-round assay and that GTP hydrolysis by hIF2 is required in vivo for reutilization of the factor in additional rounds of translation initiation. His-706 of hIF2 in GTP-binding domain motif II corresponds to His-85 of EF-Tu. Mutation of His-85 in EF-Tu to Gln reduced the activity of the protein in model translation assays, whereas mutation of His-85 to Leu inactivated EF-Tu (17). Similar to the EF-Tu-H85L mutation, an hIF2-H706E mutant was completely inactive both in vivo (Fig. 2B) and in vitro (Fig. 2C). Mutation of His-706 in hIF2 to Gln similarly abolished hIF2 function in vivo (data not shown); however, the hIF2-H706Q protein retained partial activity in vitro (Fig. 2C), as was the case for the EF-Tu-H85Q mutation (17). Finally, mutation of hIF2 Asp-759 in motif III to Asn resulted in the complete inactivation of the factor (Fig. 2C), consistent with the loss of activity in a polyphenylalanine synthesis assay observed for the corresponding Asp-138 to Asn mutation in EF-Tu (18). The inability of the various hIF2 mutant proteins to complement the slow-growth phenotype of the fun12Δ strain was not caused by poor protein expression. The hIF2 mutant proteins were expressed at levels comparable to the wild-type protein (Fig. 2A, lanes 4–6, and data not shown). The failure of these mutant hIF2 proteins to function in yeast is consistent with the lethal phenotype observed when the corresponding bacterial IF2 mutant proteins were examined in E. coli (19). The analysis of these various hIF2 mutants indicates that GTP binding and hydrolysis are essential for the translational stimulatory activity of hIF2, and that the corresponding mutations in hIF2 and EF-Tu have similar effects on the factors’ activities.
An aIF2 Homolog Functionally Substitutes for yIF2.
The M. jannaschii genome sequence revealed an ORF (MJ0262) showing significant similarity to bacterial IF2 proteins and even greater similarity to yIF2 (Fig. 1B). The aIF2 from M. jannaschii contains an intein that interrupts the GTP-binding domain of the protein. To test whether aIF2 could substitute for yIF2 we expressed M. jannaschii aIF2, lacking the intein, in a fun12Δ yeast strain. The aIF2 was expressed under the control of a galactose-inducible GAL promoter, and as a control we also expressed N-terminally truncated forms of yIF2 and hIF2 by using the same galactose-inducible promoter. As shown in Fig. 3A, introduction of a low-copy-number plasmid carrying the wild-type FUN12 gene, encoding full-length yIF2, fully complemented the growth defect in the fun12Δ strain (sector labeled WT). Expression of the truncated yIF2 also fully complemented the slow-growth phenotype of the fun12Δ stain (Fig. 3A, sector labeled yIF2). When aIF2 expression was induced on galactose medium, it was found to partially complement the slow-growth phenotype of the fun12Δ strain. The level of complementation observed with aIF2 is similar to what we previously observed with hIF2 (see Fig. 3A, sectors aIF2 and hIF2). When tested in the in vitro translation assay, bacterially expressed aIF2 was able to partially rescue the translation defect in extracts from a fun12Δ strain (Fig. 3B). This partial rescue is consistent with the results of the in vivo complementation assays, and together these results indicate that aIF2 is a functional homolog of yIF2 and that the function of IF2 has been conserved between archaea and eukaryotes.
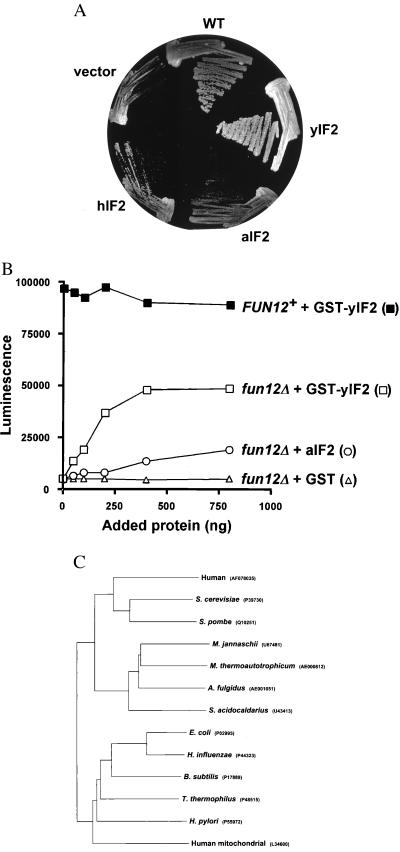
Figure 3.
aIF2 substitutes for yIF2. (A) Expression of aIF2 in yeast complements the slow-growth phenotype of a fun12Δ strain. The fun12Δ strain J133 was transformed with the vector pEMBLyex4 (vector), the pEMBLyex4-hIF2 expression plasmid pC602 (hIF2), the pEMBLyex4-aIF2 expression plasmid pC690 (aIF2), the pEGKT-GST-yIF2 plasmid pC485 (yIF2) (9), or the wild-type yeast FUN12 gene on low-copy URA3 plasmid (pC479, WT) (9). The indicated strains were streaked on synthetic minimal medium containing 10% galactose plus the required nutrient supplements, and the plates were incubated at 30°C for 5 days. (B) Addition of aIF2 partially restores translational activity in extracts from a fun12Δ strain. In vitro translation extracts were prepared from the fun12Δ strain J133 carrying the low-copy-number plasmid pC479 containing wild-type FUN12 (FUN12+) or the vector pRS316 only (fun12Δ). Extracts were incubated with 200 ng of luciferase mRNA and the indicated amounts of recombinant GST, GST-yIF2, and aIF2 proteins. Translational activity was determined by measuring luminescence after 15-min incubation at 26°C. The results presented are representative of at least three independent experiments. (C) Phylogenetic tree of eukaryotic, archaeal, prokaryotic, and mitochondrial IF2 proteins. The full-length sequences of IF2 proteins from the indicated organisms were aligned and a neighbor-joining (NJ) tree was calculated by using the program clustal w (version 1.7) (27). The tree was plotted by using the program njplot (Manolo Gouy, University of Lyon, France). Following the name of each organism is an accession number for the sequence.
DISCUSSION
In this study we isolated and characterized a human homolog of yIF2, and we demonstrated that hIF2 as well as an aIF2 homolog could functionally replace yIF2 in vivo and in vitro, demonstrating that the IF2 function has been conserved throughout evolution. Database searches have revealed the presence of bacterial, archaeal or eukaryotic IF2 homologs in a variety of organisms. Full-length IF2 protein sequences have been obtained from a large number of prokaryotes, several archaeal species, and three eukaryotes. Based on alignments using these full-length IF2 sequences, a phylogenetic tree can be constructed. As shown in Fig. 3C, the prokaryotic and mitochondrial IF2 proteins form one branch of the tree, while the archaeal and eukaryotic IF2 proteins are on a separate branch. This clustering of the archaeal and eukaryotic IF2 proteins is in accord with previous reports showing that the archaea and eukaryotes are sister groups (20). Despite significant sequence similarity between the bacterial and eukaryotic IF2 proteins, expression of E. coli IF2 failed to complement the slow-growth phenotype of a fun12Δ strain (data not shown). Likewise, the yeast, archaea, and human IF2 proteins were unable to support the growth of an E. coli strain lacking IF2 (data not shown). These results provide functional data to support the clustering of the archaeal and eukaryotic IF2 proteins on a branch distinct from the prokaryotic IF2 homologs on the phylogenetic tree in Fig. 3C. In addition to identifying IF2 homologs in humans and the yeasts S. cerevisiae and Schizosaccharomyces pombe, partial cDNA sequences encoding IF2 homologs can be found from rabbit, mouse, zebrafish, and maize. Thus it appears that IF2 is a universally conserved translation IF from prokaryotes to archaea and to eukaryotes, including both animals and plants. Additional, albeit indirect, support for our assertions regarding IF2 functional homology among prokaryotes, archaea, and eukaryotes comes from the work of Sander and Schneider (21), who demonstrated that two proteins with 25% (or greater) sequence identity will have the same three-dimensional structure. As shown in Fig. 1B, the various IF2 proteins show at least 27% amino acid sequence identity to yIF2, indicating that these proteins will have the same structure. In addition, within the GTP-binding domain the minimum identity is 53%, which indicates that the chemical properties of this domain (in terms of GTP binding), are likely to be extremely similar.
The discovery of IF2 homologs in archaea and yeast raised the possibility that these organisms possessed a translation apparatus intermediate between prokaryotes and the higher eukaryotes. However, the identification of an IF2 homolog in humans as reported here, as well as the identification of IF2 homologs, distinct from mitochondrial IF2, in other higher eukaryotes reveals that IF2 is a universally conserved translation factor. Current models for translation initiation have proposed that the heterotrimeric eIF2 complex in eukaryotes serves the same function as IF2 in prokaryotes. However, the identification of IF2 homologs in all organisms, combined with the fact that efficient translation of a normal mRNA in yeast requires both eIF2 and yIF2 (9), necessitates rethinking of the model. We propose that the IF2 homologs in archaea and eukaryotes function to direct binding of Met-tRNAiMet to the ribosomal P site, perhaps by binding to the ribosomal A site using a molecular mimicry domain as hypothesized for translation elongation factor EF-G (22) and as recently proposed for bacterial IF2 (23). The crystal structure of EF-G shows striking similarity to the structure of the EF-Tu⋅GTP⋅Phe-tRNA ternary complex with EF-G domains IV and V occupying similar positions as the central and anticodon domains of the tRNA in the EF-Tu ternary complex (22). This finding has led to the proposal that EF-G binds to the ribosomal A site using domains IV and V that structurally resemble a tRNA. Recently, Brock et al. (23) extended this molecular mimicry hypothesis to the initiation phase of protein synthesis when they found that together bacterial IF2 and IF1 share sequence similarity with domains IV and V of EF-G. They proposed that IF2 and IF1 would together bind to the ribosomal A site and direct the fMet-tRNAiMet to the ribosomal P site during translation initiation. We compared the sequences of the archaeal and eukaryotic IF2 homologs to EF-G, and its eukaryotic counterpart EF-2, and found that like bacterial IF2, aIF2, yIF2, and hIF2 possess an interesting sequence similarity to EF-G (EF-2) domains IV and V (data not shown). In addition, Kyrpides and Woese (24) recently have noted sequence similarity between bacterial IF1 and the archaeal and eukaryotic translation IF eIF1A. Based on these homologies we propose that the molecular mimicry hypothesis can be extended to describe the function of the universally conserved IF2 proteins. According to this model, the IF2 homologs in archaea and eukaryotes in combination with eIF1A would structurally mimic the anticodon arm of a tRNA and bind to the ribosomal A site and direct the binding of the eIF2⋅GTP⋅Met-tRNAiMet ternary complex to the ribosomal P site. Moreover, the IF2 homologs may stabilize the binding of Met-tRNAiMet to the small ribosomal subunit as shown for bacterial IF2 (25), or aid in the correct positioning of the Met-tRNAiMet in the P site to facilitate AUG codon recognition and subunit joining. These models are consistent with recent biochemical data showing the eIF1A is required for stable binding of eIF2⋅GTP⋅Met-tRNAiMet ternary complexes to the small ribosomal subunit (26). Further analysis of the various IF2 proteins and characterization of their role in translation initiation should prove very interesting and help us better understand the mechanism and evolution of the translation process.
Acknowledgments
We thank Alan Hinnebusch for advice and comments on the manuscript; Bill Merrick, Tatyana Pestova, Christopher Hellen, Terri Kinzy, and members of the Dever and Hinnebusch laboratories for helpful discussions; Alan Sachs for advice on the in vitro translation assays; Charles Moehle and Simon Green (Ribogene) for the luciferase plasmid; Matt Marton for his input on the Northern blot; and Sang Won Kang for HeLa cells. A.R.-M. was supported by a National Science Foundation Graduate Fellowship.
ABBREVIATIONS
- IF
initiation factor
- eIF2
eukaryotic translation IF 2
- IF2
prokaryotic translation IF 2
- aIF2
archaeal IF2 homolog
- yIF2
yeast IF2 homolog
- hIF2
human IF2 homolog
- GST
glutathione S-transferase
- MBP
maltose-binding protein
- RACE
rapid amplification of cDNA ends
- EST
expressed sequence tag
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF078035).
References
- 1.Merrick W C. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trachsel H. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 113–138. [Google Scholar]
- 3.Maitra U, Stringer E A, Chaudhuri A. Annu Rev Biochem. 1982;51:869–900. doi: 10.1146/annurev.bi.51.070182.004253. [DOI] [PubMed] [Google Scholar]
- 4.Hershey J W B. In: Translation in Eukaryotes. Trachsel H, editor. Boca Raton, FL: CRC; 1991. pp. 353–374. [Google Scholar]
- 5.Laalami S, Putzer H, Plumbridge J A, Grunberg-Manago M. J Mol Biol. 1991;220:335–349. doi: 10.1016/0022-2836(91)90017-z. [DOI] [PubMed] [Google Scholar]
- 6.White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Bussey H, Kaback D B, Zhong W W, Vo D T, Clark M W, Fortin N, Hall J, Ouellette B F F, Keng T, Barton A B, et al. Proc Natl Acad Sci USA. 1995;92:3809–3813. doi: 10.1073/pnas.92.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis P P. Cell. 1997;89:1007–1010. doi: 10.1016/s0092-8674(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 9.Choi S K, Lee J H, Zoll W L, Merrick W C, Dever T E. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 11.Tarun S Z, Sachs A B. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 12.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto S, Chiorini J A, Urcelay E, Safer B. Biochem J. 1996;315:791–798. doi: 10.1042/bj3150791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquet E, Parmeggiani A. EMBO J. 1988;7:2861–2867. doi: 10.1002/j.1460-2075.1988.tb03142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacquet E, Parmeggiani A. Eur J Biochem. 1989;185:341–346. doi: 10.1111/j.1432-1033.1989.tb15121.x. [DOI] [PubMed] [Google Scholar]
- 17.Zeidler W, Egle C, Ribeiro S, Wagner A, Katunin V, Kreutzer R, Rodnina M, Wintermeyer W, Sprinzl M. Eur J Biochem. 1995;229:596–604. doi: 10.1111/j.1432-1033.1995.tb20503.x. [DOI] [PubMed] [Google Scholar]
- 18.Weijland A, Parmeggiani A. Science. 1993;259:1311–1314. doi: 10.1126/science.8446899. [DOI] [PubMed] [Google Scholar]
- 19.Laalami S, Timofeev Andrei V, Putzer H, Leautey J, Grunberg-Manago M. Mol Microbiol. 1994;11:293–302. doi: 10.1111/j.1365-2958.1994.tb00309.x. [DOI] [PubMed] [Google Scholar]
- 20.Keeling P J, Baldauf S L, Doolittle W F, Zillig W, Klenk H-P. Syst Appl Microbiol. 1996;19:312–321. [Google Scholar]
- 21.Sander C, Schneider R. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 22.Nyborg J, Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Clark B F C. Trends Biochem Sci. 1996;21:81–82. [PubMed] [Google Scholar]
- 23.Brock S, Szkaradkiewicz K, Sprinzl M. Mol Microbiol. 1998;29:409–417. doi: 10.1046/j.1365-2958.1998.00893.x. [DOI] [PubMed] [Google Scholar]
- 24.Kyrpides N C, Woese C R. Proc Natl Acad Sci USA. 1998;95:224–228. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gualerzi C O, Severini M, Spurio R, la Teana A, Pon C L. J Biol Chem. 1991;266:16356–16362. [PubMed] [Google Scholar]
- 26.Chaudhuri J, Si K, Maitra U. J Biol Chem. 1997;272:7883–7891. doi: 10.1074/jbc.272.12.7883. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]