Abstract
The MWFE polypeptide of mammalian complex I (the proton-translocating NADH-quinone oxidoreductase) is 70 amino acids long, and it is predicted to be a membrane protein. The NDUFA1 gene encoding the MWFE polypeptide is located on the X chromosome. This polypeptide is 1 of approximately 28 “accessory proteins” identified in complex I, which is composed of 42 unlike subunits. It was considered accessory, because it is not one of the 14 polypeptides making up the core complex I; a homologous set of 14 polypeptides can make a fully functional proton-translocating NADH-quinone oxidoreductase in prokaryotes. One MWFE mutant has been identified and isolated from a collection of respiration-deficient Chinese hamster cell mutants. The CCL16-B2 mutant has suffered a deletion that would produce a truncated and abnormal MWFE protein. In these mutant cells, complex I activity is reduced severely (<10%). Complementation with hamster NDUFA1 cDNA restored the rotenone-sensitive complex I activity of these mutant cells to ≈100% of the parent cell activity. Thus, it is established that the MWFE polypeptide is absolutely essential for an active complex I in mammals.
Multisubunit NADH-quinone oxidoreductases have been described in vertebrate and plant mitochondria as well as in prokaryotes (1–6) but curiously not in all fungi. Multisubunit NADH-quinone oxidoreductases constitute complex I of the mitochondrial electron transport chain. Complex I is notably absent in the yeast Saccharomyces cerevisiae. The bacterial complex has a total of 14 subunits (7, 8) for which homologous peptides can be identified in mammals and plants. These core peptides therefore are considered to be essential for the oxidation of NADH, electron transport, proton translocation across the membrane, and reduction of the quinone (3). The mammalian complex I has long been known to have an exceptionally large number of subunits, and the most recent data suggest the total to be ≈42, 7 of which (ND1, ND2, ND3, ND4, ND4L, ND5, and ND6) are encoded by the mitochondrial genome; they are made in the mitochondrial matrix and assembled with the other 35 peptides in a complex made up of several major subdomains (9–11). One such subdomain contains all of the integral membrane proteins, including the mitochondrial peptides. Another domain, which may be fractionated further by mild dissociation conditions, extends into the mitochondrial matrix. This domain includes the active site for the substrate NADH, a flavin mononucleotide as the hydride acceptor, and four of the five electron paramagnetic resonance-detectable iron-sulfur clusters (12). The function of the remainder of the peptides in the complex of vertebrates and plants is still quite obscure (13). The peptides may assist in the assembly of the complex, contribute to its stability, have some role in regulation of activity, and even carry out biochemical functions like the acyl carrier protein (14). An official nomenclature for many of these peptides remains to be established, but following a suggestion made by the Walker laboratory (14), they can be referred to conveniently by listing the first four amino acids from the N terminus.
A collection of respiration-deficient (res−) Chinese hamster mutant cell lines has been described by our laboratory. They fall into at least seven complementation groups, as determined by pairwise somatic cell hybridizations (15). A biochemical analysis of these mutants has identified a mutant cell line defective in mitochondrial protein synthesis (16, 17), a mutant cell line defective in complex II (succinate dehydrogenase; refs. 18 and 19), and at least two complementation groups (I and II) of cells with defects in complex I (NADH-quinone oxidoreductase; refs. 15 and 20). All of the mutations that we identified were in nuclear genes and were recessive in intraspecies hybrids. The large number of potential genes made it difficult to identify a priori the corresponding defective peptide in the complex I mutants. A genetic analysis of intraspecies and interspecies somatic cell hybrids had shown that the genes for complementation groups I and II were located on the mammalian X chromosome (21).
One of the mutants in our collection, CCL16-B2, recently has been shown to be complemented by the matrix NADH dehydrogenase of yeast, a single peptide encoded by the NDI1 gene (22). Although the yeast Ndi1 protein cannot pump protons out of the mitochondrial matrix, it can oxidize NADH and pass electrons via ubiquinone to the downstream mammalian electron transport chain. It therefore releases the feedback inhibition of the Krebs cycle with high NADH levels and makes the transfected cells once again capable of respiration and oxidative phosphorylation. This result also proves that the complex I defect is the only defect in this mutant.
The growing interest in complex I deficiencies related to human “mitochondrial diseases” (23, 24) has been the impetus in recent years to clone cDNAs and to map many of the nuclear complex I genes. As part of this effort, Zhuchenko et al. (25) have isolated and mapped the human NDUFA1 gene on the X chromosome. Because X linkage is largely conserved in mammals and because it is the only X-linked gene known so far, a complementation test with this gene seemed promising.
The NDUFA1 gene is a small gene (≈5 kb) with two introns (1.5 kb and 3 kb), and it encodes a peptide of 70 amino acids. We use NDUFA1 when referring to the gene, mRNA, or cDNA but use MWFE when referring to the peptide or protein. The MWFE protein is imported into mitochondria and is associated with complex I apparently without requiring proteolytic processing (14). It is listed among the ≈28 accessory peptides for which no function has been established so far. We show in this communication that the defect in one of the complex I mutants in our collection is in the NDUFA1 gene. The results show that the MWFE protein is essential for a functional complex I in mammalian mitochondria.
MATERIALS AND METHODS
Cell Lines and Cell Culture.
The parental Chinese hamster cell lines and res− mutants derived from them have been described (see ref. 26 for a review). They were cultured routinely in DMEM with 5 mg/ml glucose, 10% (vol/vol) FCS, nonessential amino acids, gentamycin (100 μg/ml), and fungizone (2.5 μg/ml). Under these conditions, even the res− cell lines grow normally. To distinguish res− from res+ cell lines, the same medium was used, except that glucose was replaced by 1 mg/ml galactose. This mixture is referred to as DMEM/Gal (27). At low glucose or with galactose substituted for glucose, only res+ cells remain viable.
Plasmids and Genes.
The human NDUFA1 gene has been cloned and characterized by Zhuchenko et al. (25). The ORF has been defined, and the mouse and bovine cDNA sequences are also available in the GenBank database (accession nos. Y07708 and X63222, respectively). A comparison shows a high degree of conservation of sequence. Oligonucleotides for cloning the corresponding cDNAs from human, hamster, and mouse cells were designed from regions where the sequence was identical. The oligonucleotide used for the 3′ rapid amplification of cDNA ends (RACE; ref. 28) overlaps the start codon, and the oligonucleotide for the 5′ RACE is further downstream in the ORF. Their sequences are, for NDUFA1 forward, AACGGTGCGGAGATGTGGTTCG, and for NDUFA1 reverse, TAATCAACCAGGAAAATGCTTC. A cDNA containing the entire ORF could be obtained by the 3′ RACE protocol and was cloned into the expression vector pBK-CMV (Stratagene) for sequencing and transfection into the hamster mutant cell lines. All cDNAs in the expression vector were sequenced before use in transfections. Northern analyses with total RNA from various cell lines were performed as described (17) by using the hamster NDUFA1 cDNA for the preparation of the probe.
Complementation Tests.
The cells were transfected by incubating ≈1 × 106 to ≈4 × 106 cells per plate with 10 μg of plasmid and 20 μl of Lipofectin (Life Technologies, Grand Island, NY). Controls included the vector pBK-CMV with an unrelated (dummy) cDNA. After a 48-h incubation in DMEM (glucose), the cells were treated with trypsin and distributed (1:10 dilution) to multiple plates. G418 was added to some plates (selection for neomycin resistance), and in others, the medium was changed to DMEM/Gal for a direct selection for the res+ phenotype as described (27). The neomycin-resistant colonies were marked on the plate after about 2 weeks and tested for their res−/res+ phenotype by exposure to DMEM/Gal.
Measurement of Respiratory Activities.
The respiratory chain activities of various cells (n = 3 × 107 to 8 × 107) were measured by using the method developed by Seo et al. (22). The cells were harvested by treatment with trypsin and resuspended in 1 ml of 20 mM Hepes, pH 7.1/250 mM sucrose/10 mM MgCl2. The cells were treated with 50–150 μg of digitonin until more than 90% of the cells were stained by trypan blue. The digitonin-treated cells were washed with the same medium. Oxygen consumption (respiration) was measured polarographically in 0.6 ml of 20 mM Hepes, pH 7.1/250 mM sucrose/10 mM MgCl2 by using a Clark electrode in a water-jacketed chamber maintained at 37°C.
Isolation of Mitochondria and Mitochondrial Fractions.
Intact and sonicated mitochondria were isolated from freshly harvested cells essentially according to Trounce et al. (29). Mitochondrial fractions were prepared as follows. CCL16 (wild-type), CCL16-B2, and NDUFA1-transfected cells (≈1 × 109 cells) were washed twice with PBS and harvested by treatment with trypsin. The pellets were suspended in 5 ml of buffer [210 mM mannitol/70 mM sucrose/1 mM EDTA/5 mM Hepes (pH 7.2)/0.2 mM PMSF/0.5% fatty acid free BSA (isolation buffer)]. The cell suspensions were treated with 1–2 mg/ml digitonin for 1 min on ice. The suspension was diluted 10-fold with the isolation buffer and centrifuged at 3,000 × g for 5 min to remove excess detergent. The cell pellet was resuspended with the isolation buffer and homogenized by using a tight-fit Dounce homogenizer (15–20 up/down strokes). The homogenate was centrifuged at 625 × g for 5 min at 4°C to remove unbroken cells and nuclei. The supernatant was centrifuged at 10,000 × g for 20 min at 4°C. The pellet was suspended in 0.1 ml of the isolation buffer. This fraction is designated as the intact mitochondrial fraction.
The mitochondrial suspension was subjected to sonication for 2 min in an ice-water bath by using a Branson sonifier at output 3 and 50% pulse mode. The sonication was repeated three times after allowing the sample to cool down for 2 min in the ice-water bath. The suspension was centrifuged at 150,000 × g for 30 min at 4°C, and the pellets were resuspended in 0.1 ml of the isolation buffer and used as the mitochondrial membrane fraction.
Immunochemical Assays.
SDS/12% polyacrylamide gels were prepared and loaded with 40–150 μg of mitochondria or various amounts of mitochondrial membranes isolated from CCL16, CCL16-B2, and NDUFA1-cDNA-transfected CCL16-B2 cells. After electrophoresis and transfer of the proteins to nitrocellulose membranes, the membranes were blotted with affinity-purified antibodies against the 51-kDa subunit of bovine flavoprotein (FP; ref. 30), the 24-kDa subunit of bovine FP (30), the 49-kDa subunit of bovine iron–sulfur protein (IP; ref. 11), the 30-kDa subunit of bovine IP (11), the 13-kDa subunit of bovine IP (11), Paracoccus NQO6 (homologue to bovine PSST; ref. 31), Paracoccus NQO3 (homologue to the 75-kDa subunit of bovine IP; ref. 32), and Paracoccus NQO9 (homologue to bovine TYKY; ref. 1) as described (30, 31, 33). Alkaline phosphatase and an ECL kit (Amersham Pharmacia) were used for the visualization of signals on the immunoblots. Antisera specific to bovine complex I subunits and to IP fractions were generous gifts from Youssef Hatefi (The Scripps Research Institute). Protein concentration was determined by bicinchoninic acid protein assay (Pierce).
RESULTS
Mammalian Cell Mutants and Complementation by the NDUFA1 cDNA.
The mutant cell lines CCL16-B2 and V79-G4 represent the two complementation groups of interest because of X linkage. Previous studies have shown also that these mutations could be complemented by X-linked genes from the hamster or the mouse but apparently not by a human X chromosome. Therefore, we cloned the NDUFA1 cDNA from hamster cells, and we sought to establish whether it can complement the mutations in either the CCL16-B2 or V79-G4 mutant cells.
Successful complementation results in the restoration of the cells’ ability to grow in medium with very low glucose or in DMEM/Gal. The selection can be made directly in DMEM/Gal; alternatively, selection for the neomycin-resistance marker on the transfecting expression vector can occur first, before the cells are tested in DMEM/Gal. In all the tests, permanently transfected cell lines were established, and their phenotype was verified by prolonged growth under conditions that would be nonpermissive for the mutant cells. It had been noted in one of our earlier publications that complementation is not instantaneous, because time is required not only for the expression of the gene product but also for the generation of an active complex I and res+ mitochondria. Thus, a direct selection in DMEM/Gal cannot be imposed immediately but can be several days after transfection. By employing G418 in the first selection, more than 100 clones were obtained on a plate. When clearly visible to the eye, they were marked and observed again for several days after a change to DMEM/Gal. Cells in mutant colonies (mock-transfected) disintegrated visibly within 1 to 2 days. Colonies in which the mutation had been complemented successfully continued to grow. The direct selection in DMEM/Gal also yielded many (>100) viable and growing colonies, indicative of complementation. A small number of those were isolated for further characterization. NDUFA1 cDNA from normal hamster cells restored the ability to grow in DMEM/Gal in the CCL16-B2 cells but not in V79-G4 cells, confirming the earlier distinction between CCL16-B2 and V79-G4 cells and their assignment to separate complementation groups
Identification of the Mutation in the NDUFA1 Gene of Chinese Hamster CCL16-B2 Mutants.
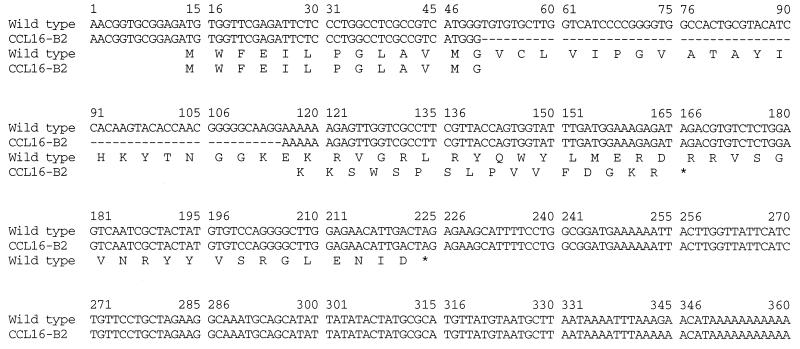
The results obtained prompted us to clone the NDUFA1 cDNAs from the mutant cell lines CCL16-B2 and V79-G4. The RACE protocol described in Materials and Methods with oligomers designed from the hamster or human cDNA sequences yielded clones from both cell lines that were sequenced. It should be noted that the transcripts are derived from an X-linked gene (the parental cell lines originated from a male hamster), and, therefore, only one type of transcript is expected. The V79-G4 mutant cells yielded cDNA with the wild-type sequence, confirming that the mutation must be in another X-linked gene. A mutation was found in the cDNA clone from the CCL16-B2 mutant cells (Fig. 1). The mutation consists of a 65-nt deletion in the cDNA, creating a premature stop codon in the translated mRNA, and, hence, the mutation would cause the production of a truncated MWFE peptide. The mutant peptide has 13 amino acids from the wild-type sequence at the N terminus, followed by 16 amino acids from a different reading frame. It should be mentioned that the structure of the hamster gene is not known, and the number of exons and introns may differ from that in humans; however if the human and hamster genes are similar, the deletion would be entirely within exon I. From the comparison of the hamster amino acid sequence with the human (25), mouse (GenBank accession no. Y07708), and bovine (14) sequences, it is clear that this peptide is highly conserved in evolution (see Fig. 2). A major difference between species is observed in the nonconservative substitutions at positions 41 and 42 (Fig. 2). The substitutions distinguishing human and hamster sequences in this region appeared quite dramatic and possibly could explain our previous failure to complement the hamster mutants with the human X chromosome. To test this idea directly, the human and mouse cDNAs were cloned from human HeLa cells and from mouse C2C12 myoblasts. The human NDUFA1 cDNA failed to complement the CCL16-B2 hamster mutant, in agreement with previous results from hamster–human somatic cell hybrids and the failure to observe complementation in human–hamster hybrid cell lines. As expected, the mouse cDNA complemented the hamster mutations in the CCL16-B2 cells, although the growth rate was lower, possibly because of a less efficient complex I containing a heterologous peptide. A more quantitative definition of differences caused by these species-specific sequence differences remains to be measured.
Figure 1.
Alignment of the wild-type and mutant CCL16-B2 cDNA sequences. The wild-type hamster sequence has been submitted to GenBank (accession no. AF100706). The deletion in the mutant is indicated by the dashed line. Translation in the mutant yields a truncated peptide, starting with a wild-type sequence (first 13 amino acids), followed by 16 amino acids from a different reading frame.
Figure 2.
Alignment of the available mammalian MWFE amino acid sequences emphasizing the very significant conservation (identity indicated in black) and a striking set of changes around positions 39–44. This polypeptide is not processed proteolytically on import into mitochondria (14).
Northern Analyses.
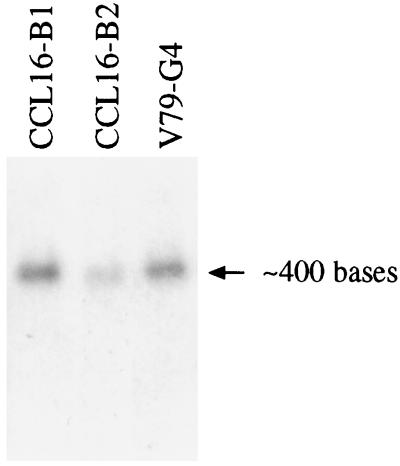
Northern analysis identified a small transcript of ≈400 nt in wild-type cells and in a variety of mutant cells (Fig. 3). The hamster mRNA has a 5′ untranslated region of 48 nt and a 3′ untranslated region of 141 nt. Together with a coding sequence of 210 nt, the mRNA has a length of 399 nt. The mRNA is slightly smaller in CCL16-B2 cells because of the deletion, and it also seems to be less abundant compared with the other Chinese hamster cell lines. It is possible that its stability is reduced by the deletion and premature stop codon, although an effect on transcription is not excluded completely.
Figure 3.
Northern blot of total RNA from wild-type cells, CCL16-B1, and the two mutant cell lines described here, CCL16-B2 and V79-G4. Equal loading of the gels was verified by both staining and labeling of ribosomal RNAs. From molecular mass markers run in an adjoining lane, the size of the mRNA was estimated to be ≈400 nt. The CCL16-B2 mRNA is slightly smaller and reduced in amount (see Results).
Complex I Activities in Wild-Type, Mutant, and Transfected Cell Lines.
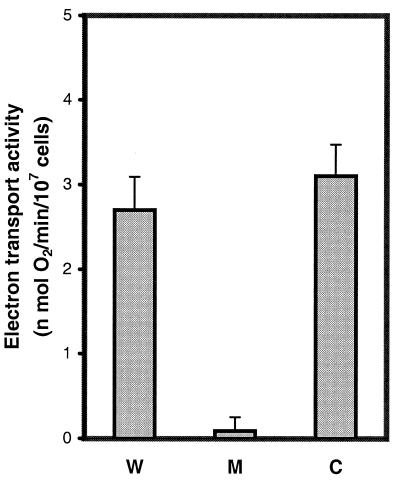
The respiratory chain activities of wild-type, mutant, and transfected complemented cell lines were investigated. As described above, when CCL16-B2 mutant cells were transfected with NDUFA1 cDNA, they can survive in a medium containing low glucose or galactose. These results suggest that NDUFA1 cDNA restores the respiration in CCL16-B2 cells. To confirm these results, the NADH-dependent, rotenone-sensitive respiration of NDUFA1 cDNA-transfected CCL16-B2 cells was examined and compared with wild-type and mutant cells. Because NADH cannot enter mitochondria directly, the combination of malate plus glutamate was used as the substrate; both malate and glutamate enter mitochondria and are metabolized readily with the concomitant production of NADH. Fig. 4 shows the complex I-dependent respiration rates of nontransfected and NDUFA1-cDNA-transfected mutants. In addition, as a control, the respiration rate of the parent cells was measured. The activity in the mutant is less than 10% of the control (see also the results in ref. 20) or close to the background measurement in wild-type cells inhibited with rotenone. It is clear that NDUFA1-cDNA-transfected CCL16-B2 cells are restored to the level of the parental cells with respect to rotenone-sensitive respiration.
Figure 4.
Comparison of rotenone-sensitive NADH-quinone oxidoreductase activities in wild-type (W), mutant CCL16-B2 (M), and a complemented mutant clone transfected with the NDUFA1-cDNA expression vector (C). Determinations were made by measuring oxygen consumption (respiration) that was stimulated by malate plus glutamate and inhibited by rotenone in cells that had been made permeable with digitonin (see Materials and Methods). Bars represent the average of five such measurements, and the standard deviation is indicated.
Immunochemical Assays of Complex I Subunits.
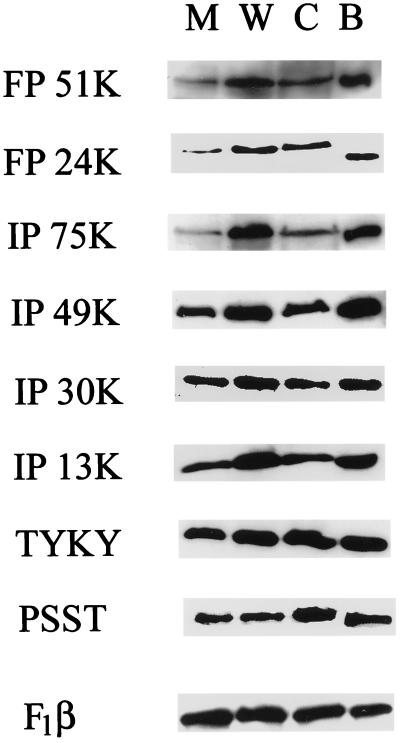
In previous studies establishing complex I deficiency in these mutants, the biochemical analyses were restricted to identifying the lack of NADH-dependent, rotenone-sensitive respiration in isolated mitochondria. In the meantime, specific antisera have become available to examine the state of complex I in the mutant mitochondria. Immunoblotting of mitochondria and mitochondrial membranes isolated from the parental cells and from nontransfected and transfected CCL16-B2 mutant cells was performed with antibodies specific to the 51-kDa and 24-kDa subunits of FP, the 75-kDa, 49-kDa, 30-kDa, and 13-kDa subunits of IP, the TYKY subunit, and the PSST subunit. Mitochondrial membranes from bovine heart were used as controls. As seen in Fig. 5, all subunits examined exist in the mitochondrial membranes of CCL16-B2 cells with somewhat reduced levels of the 75-kDa subunit of IP and the 51-kDa and 24-kDa subunits of FP compared with wild-type mitochondrial membranes. The lower abundance of the 75-kDa, 51-kDa, and 24-kDa peptides was also apparent when whole mitochondrial proteins were examined. After transfection, the 51-kDa subunit of FP and the 75-kDa subunit of IP, but not the 24-kDa subunit of FP, are recovered in whole mitochondria, although the relative amounts of the three peptides are still somewhat lower in the comparison of mitochondrial membranes (Fig. 5)
Figure 5.
Immunoblotting analysis of several peripheral membrane proteins of complex I in wild-type (W), mutant CCL16-B2 (M), and transfected (complemented) cells (C). A preparation of bovine heart mitochondrial membranes (B) also is included for comparison. Mitochondrial membranes were prepared as described in Material and Methods. Gels were loaded to obtain comparable signals for the β-subunit of the F1-ATPase.
DISCUSSION
Major findings from these studies are the identification of the precise defect in a res− Chinese hamster cell mutant, CCL16-B2, and the identification of an essential function of the MWFE protein in NADH:ubiquinone oxidase activity. The mitochondrial complex I deficiency results from mutations in an X-linked gene, NDUFA1, encoding a small peptide of 70 amino acids. This peptide has a highly hydrophobic N-terminal domain and a hydrophilic, positively charged C-terminal domain. It is likely to be associated with the integral membrane fraction of the complex.
Of the ≈42 peptides in the mammalian complex I, 14, including the 7 encoded by the mtDNA, have homologues in the bacterial complex. The MWFE peptide belongs to the group of 28 polypeptides to which an unknown accessory function has been attributed. The mutation in the CCL16-B2 (frameshift) mutant abolished complex I activity almost completely, as measured by rotenone-sensitive respiration stimulated by malate plus glutamate. This activity could be restored completely by the wild-type NDUFA1 cDNA transfected into the mutant cells. Measuring complex I activity by itself would require the measurement of the reduction of ubiquinone by NADH. However, the indirect experiments point quite convincingly to a specific defect in NADH-ubiquinone oxidoreductase in the CCL16-B2 mutant. First, succinate- and α-glycerolphosphate-stimulated respiration is near normal in CCL16-B2 mitochondria (20), indicative of an intact electron transport chain from ubiquinone to oxygen. Some reduction in activity of downstream complexes may be caused by a compromised protein import in these res− mitochondria (reduced ΔΨ; ref. 17). Second, in a recent report, we describe the restoration of respiration stimulated by malate plus glutamate in CCL16-B2 cells by the yeast NADH dehydrogenase, Ndi1p (22). This single polypeptide, associated with the mitochondrial membrane on the matrix side, can oxidize NADH and reduce ubiquinone. Complementation and restoration of respiration by the NDI1 gene, therefore, also required an intact downstream electron transport chain. The Ndi1 protein substitutes for the complex I function, except that it cannot pump protons out of the matrix.
Immunoblotting experiments with whole mitochondrial proteins and mitochondrial membrane fractions were performed with antisera against 8 polypeptides from the peripheral, hydrophilic domain of the complex. All peptides were detectable in the membrane fraction from sonicated mitochondria. When the loading in each lane was adjusted to obtain comparable signals for the β-subunit of F1-ATPase, comparable band intensities were observed for bovine heart, wild-type hamster, and most but not all complex I polypeptides in the CCL16-B2 mutant. In particular, two polypeptides (75 kDa and 51 kDa), and a possible third polypeptide (24 kDa) appeared to be present in reduced quantities, but this partial deficiency also is observed in the complemented cells. Parallel observations were made in the examination of intact (whole) mitochondria.
The immunoblotting experiments might suggest a problem in assembly. However, if MWFE is an integral membrane protein, it is not obvious why or how it could cause the failure of assembly, specifically of the 75-kDa and 51-kDa proteins. Furthermore, these two peptides are not completely absent from the CCL16-B2 mutant mitochondrial membranes, and one might have expected to see partial activity. However, the loss in activity seems almost total (Fig. 3). It is also possible that the MWFE protein may be involved in other aspects of complex I integrity, such as formation or integration of cofactors. One could even speculate that the protein might have a role in the import of some subunits, as is the case with the core proteins of complex III, which serve as processing enzymes (34). Whether the lack of MWFE protein affects other respiratory complexes remains to be seen. The effect could be either direct or indirect because of the reduced membrane potential (ΔΨ) in these mutants.
Deficiencies in complex I have been observed in human patients with encephalomyopathies and related mitochondrial diseases. Mutations in mitochondrial DNA have been identified, and patients with nuclear mutations also have been described (35, 36). In one patient, the investigators found a mutation in the gene encoding the 18-kDa subunit (AQDQ; ref. 36). A strong male preponderance among the patients in the study prompted speculations about mutations in an X-linked gene; thus, the NDUFA1 gene in 17 patients (14 of them male) was examined. No mutations in this gene were found (35). The authors also described highly variable expression of this gene in different tissues, but this variation was found in normal individuals as well.
Human patients with complex I deficiency caused by nuclear mutations must have leaky mutations to be viable. Regulatory mutations resulting in a lower level of expression or missense mutations affecting the activity are possibilities. In contrast, cells in tissue culture can be completely res− and still grow normally in the presence of abundant glucose (26). Thus, null mutants like the CCL16-B2 mutant described here can be isolated, and it is expected that such null mutants will have useful applications in the study of other specific mutations that can be introduced with modified complementing cDNAs.
Acknowledgments
We thank Dr. Youssef Hatefi for generously providing all the antisera specific to the various subunits of complex I tested in this study, Ms. Deena Ream-Robinson who provided expert technical assistance, and Ms. Negin Iranfar from the laboratory of Dr. W. Loomis for DNA sequencing. This work was supported by a grant from the American Cancer Society (to I.E.S.) and by U.S. Public Health Service Grant R01DK53244 (to A.M.-Y. and T.Y.).
ABBREVIATIONS
- res−
respiration-deficient
- res+
respiration-competent
- RACE
rapid amplification of cDNA ends
- FP
flavoprotein complex
- IP
iron–sulfur protein complex
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF100706).
References
- 1.Yagi T, Yano T, Di Bernardo S, Matsuno-Yagi A. Biochim Biophys Acta. 1998;1364:125–133. doi: 10.1016/s0005-2728(98)00023-1. [DOI] [PubMed] [Google Scholar]
- 2.Rasmusson A G, Heiser V, Zabaleta E, Brennicke A, Grohmann L. Biochim Biophys Acta. 1998;1364:101–111. doi: 10.1016/s0005-2728(98)00021-8. [DOI] [PubMed] [Google Scholar]
- 3.Guénebaut V, Schlitt A, Weiss H, Leonard K, Friedrich T. J Mol Biol. 1998;276:105–112. doi: 10.1006/jmbi.1997.1518. [DOI] [PubMed] [Google Scholar]
- 4.Walker J E. Biochim Biophys Acta. 1995;1271:221–227. doi: 10.1016/0925-4439(95)00031-x. [DOI] [PubMed] [Google Scholar]
- 5.Schulte U, Weiss H. Methods Enzymol. 1995;260:3–14. doi: 10.1016/0076-6879(95)60126-0. [DOI] [PubMed] [Google Scholar]
- 6.Finel M. Biochim Biophys Acta. 1998;1364:112–121. doi: 10.1016/s0005-2728(98)00022-x. [DOI] [PubMed] [Google Scholar]
- 7.Yagi T. Biochim Biophys Acta. 1993;1141:1–17. doi: 10.1016/0005-2728(93)90182-f. [DOI] [PubMed] [Google Scholar]
- 8.Yagi T, Yano T, Matsuno-Yagi A. J Bioenerg Biomembr. 1993;25:339–345. doi: 10.1007/BF00762459. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi M, Hatefi Y. Biochemistry. 1993;32:1935–1939. doi: 10.1021/bi00059a008. [DOI] [PubMed] [Google Scholar]
- 10.Finel M, Majander A S, Tyynelä J, De Jong A M P, Albracht S P J, Wikström M. Eur J Biochem. 1994;226:237–242. doi: 10.1111/j.1432-1033.1994.tb20046.x. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi M, Belogrudov G I, Hatefi Y. J Biol Chem. 1998;273:8094–8098. doi: 10.1074/jbc.273.14.8094. [DOI] [PubMed] [Google Scholar]
- 12.Ohnishi T. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 13.Walker J E. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 14.Walker J E, Arizmendi J M, Dupuis A, Fearnley I M, Finel M, Medd S M, Pilkington S J, Runswick M J, Skehel J M. J Mol Biol. 1992;226:1051–1072. doi: 10.1016/0022-2836(92)91052-q. [DOI] [PubMed] [Google Scholar]
- 15.Soderberg K, Mascarello J T, Breen G A M, Scheffler I E. Somatic Cell Genet. 1979;5:225–240. doi: 10.1007/BF01539163. [DOI] [PubMed] [Google Scholar]
- 16.Burnett K G, Scheffler I E. J Cell Biol. 1981;90:108–115. doi: 10.1083/jcb.90.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Au H C, Scheffler I E. Somatic Cell Mol Genet. 1997;23:27–35. doi: 10.1007/BF02679953. [DOI] [PubMed] [Google Scholar]
- 18.Soderberg K, Ditta G S, Scheffler I E. Cell. 1977;10:697–702. doi: 10.1016/0092-8674(77)90103-9. [DOI] [PubMed] [Google Scholar]
- 19.Oostveen F G, Au H C, Meijer P-J, Scheffler I E. J Biol Chem. 1995;270:26104–26108. doi: 10.1074/jbc.270.44.26104. [DOI] [PubMed] [Google Scholar]
- 20.Breen G A M, Scheffler I E. Somatic Cell Genet. 1979;5:441–451. doi: 10.1007/BF01538879. [DOI] [PubMed] [Google Scholar]
- 21.Day C, Scheffler I E. Somatic Cell Genet. 1982;8:691–707. doi: 10.1007/BF01543012. [DOI] [PubMed] [Google Scholar]
- 22.Seo B B, Kitajima-Ihara T, Chan E K L, Scheffler I E, Matsuno-Yagi A, Yagi T. Proc Natl Acad Sci USA. 1998;95:9167–9171. doi: 10.1073/pnas.95.16.9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schapira A H. Biochim Biophys Acta. 1998;1364:261–270. doi: 10.1016/s0005-2728(98)00032-2. [DOI] [PubMed] [Google Scholar]
- 24.Robinson B H. Biochim Biophys Acta. 1998;1364:271–286. doi: 10.1016/s0005-2728(98)00033-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhuchenko O, Wehnert M, Bailey J, Sun Z S, Lee C C. Genomics. 1996;37:281–288. doi: 10.1006/geno.1996.0561. [DOI] [PubMed] [Google Scholar]
- 26.Scheffler I E. In: Biochemical Genetics of Respiration-Deficient Mutants of Animal Cells. Morgan M J, editor. London: Plenum; 1986. pp. 77–109. [Google Scholar]
- 27.Ditta G, Soderberg K, Scheffler I E. Somatic Cell Genet. 1976;2:331–344. doi: 10.1007/BF01538838. [DOI] [PubMed] [Google Scholar]
- 28.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trounce I A, Kim Y L, Jun A S, Wallace D C. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 30.Han A L, Yagi T, Hatefi Y. Arch Biochem Biophys. 1988;267:490–496. doi: 10.1016/0003-9861(88)90055-0. [DOI] [PubMed] [Google Scholar]
- 31.Takano S, Yano T, Yagi T. Biochemistry. 1996;35:9120–9127. doi: 10.1021/bi9605853. [DOI] [PubMed] [Google Scholar]
- 32.Yano T, Yagi T, Sled V D, Ohnishi T. J Biol Chem. 1995;270:18264–18270. doi: 10.1074/jbc.270.31.18264. [DOI] [PubMed] [Google Scholar]
- 33.Hekman C, Tomich J M, Hatefi Y. J Biol Chem. 1991;266:13564–13571. [PubMed] [Google Scholar]
- 34.Schulte U, Arretz M, Schneider H, Tropschug M, Wachter E, Neupert W, Weiss H. Nature (London) 1989;339:147–149. doi: 10.1038/339147a0. [DOI] [PubMed] [Google Scholar]
- 35.Leoffen J, Smeets R, Smeitink J, Ruitenbeek W, Janssen A, Mariman E, Sengers R, Trijbels F, van den Heuvel L. J Inherit Metab Dis. 1998;21:210–215. doi: 10.1023/a:1005339332062. [DOI] [PubMed] [Google Scholar]
- 36.van den Heuvel L, Ruitenbeek W, Smeets R, Gelman-Kohan Z, Elpeleg O, Loeffen J, Trijbels F, Mariman E, de Bruijn D, Smeitink J. Am J Hum Genet. 1998;62:262–268. doi: 10.1086/301716. [DOI] [PMC free article] [PubMed] [Google Scholar]