Abstract
Acyl homoserine lactones (acyl-HSLs) are important intercellular signaling molecules used by many bacteria to monitor their population density in quorum-sensing control of gene expression. These signals are synthesized by members of the LuxI family of proteins. To understand the mechanism of acyl-HSL synthesis we have purified the Pseudomonas aeruginosa RhlI protein and analyzed the kinetics of acyl-HSL synthesis by this enzyme. Purified RhlI catalyzes the synthesis of acyl-HSLs from acyl–acyl carrier proteins and S-adenosylmethionine. An analysis of the patterns of product inhibition indicated that RhlI catalyzes signal synthesis by a sequential, ordered reaction mechanism in which S-adenosylmethionine binds to RhlI as the initial step in the enzymatic mechanism. Because pathogenic bacteria such as P. aeruginosa use acyl-HSL signals to regulate virulence genes, an understanding of the mechanism of signal synthesis and identification of inhibitors of signal synthesis has implications for development of quorum sensing-targeted antivirulence molecules.
Many Gram-negative bacteria synthesize acyl-homoserine lactone (acyl-HSL) signal molecules that serve in a cell-to-cell communication system termed quorum sensing. Quorum sensing enables population density control of gene expression (for recent reviews of quorum sensing see refs. 1–4). Because quorum sensing has been implicated as an important factor in the expression of virulence genes in animal and plant pathogens (2, 5–7), understanding the mechanism of acyl-HSL synthesis is of importance. Although all acyl-HSLs possess an HSL ring, the length of the acyl side chain and the substitutions on the side chain differ and are specificity determinants for different quorum-sensing systems. In most systems, acyl-HSL signal synthesis requires a member of the LuxI family of proteins. LuxI family members occur in a number of different bacterial genera; all LuxI proteins direct the synthesis of specific acyl-HSLs and show sequence similarity (2–4, 8).
There are three reports of in vitro catalysis of acyl-HSL synthesis by LuxI family members. The Vibrio fischeri LuxI protein was purified as a maltose-binding protein fusion (9) and the Agrobacterium tumefaciens TraI protein as a His-tagged fusion (10). Both of these proteins functioned as acyl-HSL synthases when provided with S-adenosylmethionine (SAM) as the amino donor and an appropriate acyl–acyl carrier protein (acyl-ACP) as an acyl donor. Subsequently, the Pseudomonas aeruginosa RhlI protein was purified from recombinant Escherichia coli in the form of insoluble inclusion bodies. In vivo, RhlI directs the synthesis of N-butyryl-HSL and small amounts of N-hexanoyl-HSL (11). The purified protein was reported to catalyze the synthesis of butyryl-HSL when provided with butyryl-CoA, HSL, and NADPH (12). The activity of the RhlI preparation was substantially lower than the activity of the LuxI or TraI preparations (10−6), thus raising concerns as to whether butyryl-CoA and HSL are relevant substrates for acyl-HSL synthesis by RhlI.
It has been hypothesized that by analogy to fatty acid biosynthesis, the first step in acyl-HSL synthesis involves formation of a covalent acyl-enzyme intermediate (3, 10), and that active-site cysteine residues or serine residues are acyl acceptors for such a mechanism (2, 3, 10). Analyses of the effects of site-specific mutations in LuxI and RhlI on acyl-HSL synthesis in recombinant E. coli suggest that the proposed mechanism for signal synthesis may be incorrect (8, 13). None of the cysteine residues in LuxI or RhlI are essential for acyl-HSL synthesis, and in RhlI, a conserved serine residue within the proposed active site is also not essential for enzyme activity. Perhaps acyl-HSL synthesis proceeds via a mechanism that does not involve an acyl-enzyme intermediate or perhaps the acyl group acceptor has not been targeted in the site-specific mutagenesis studies.
In addition to site-specific mutagenesis, RhlI and LuxI function has been studied by analyzing random single amino acid substitution mutants that retain little or no activity in recombinant E. coli (8, 13). These studies have revealed a conserved region containing a number of specific residues required for both RhlI and LuxI activity. This region corresponds to residues 25–104 of the 193-aa LuxI polypeptide, and it has been proposed to represent the active site for formation of the amide bond between the acyl group and the amino donor, SAM (8, 13). The fact that analogous mutations in RhlI and LuxI inactivate both enzymes raises further doubt concerning the proposal that the substrates for RhlI are HSL and acyl-CoA rather than SAM and acyl-ACPs, which are the substrates for LuxI and TraI.
To probe the mechanism of acyl-HSL synthesis, we purified a native, soluble form of RhlI from P. aeruginosa. RhlI was chosen as a model because the activated acyl substrates butyryl-ACP and butyryl-CoA are relatively easy to prepare. We report that the enzyme shows greatest activity with butyryl-ACP and SAM as substrates. There is low activity when butyryl-CoA is provided in place of butyryl-ACP, and even in the presence of NADPH there is no detectable activity when HSL or homoserine are provided in place of SAM. Inhibitor studies suggested that RhlI produces butyryl-HSL by using a sequential ordered reaction mechanism initiated by SAM binding. This is inconsistent with previous proposals of acyl–enzyme intermediate formation as the first step in acyl-HSL synthesis.
MATERIALS AND METHODS
Plasmid Construction and Transformation.
The RhlI expression vector, pRhlI-2, was constructed by ligation of an 899-bp ptac-rhlI BamHI-HindIII fragment from pRhlI-1 (8) with BamHI-HindIII-digested pBBR1MCS5 (14). Standard procedures were used for all molecular genetic manipulations (15), and E. coli XL1-Blue was used as a cloning vehicle. P. aeruginosa PAO-JP1, a LasI− strain, was transformed with pRhlI-2 by electroporation as described below, and transformants were selected by plating on peptone-trypticase-soy (PTS) agar (16) supplemented with gentamicin (25 μg/ml) and tetracycline (50 μg/ml).
Electroporation was performed as follows: P. aeruginosa was grown for 5 h in 50 ml of PTS broth with tetracycline at 37°C with shaking. The inoculum was 0.5 ml of an overnight culture. Cells were harvested by centrifugation and suspended in 25 ml of ice-cold 300 mM sucrose. After centrifugation at 4°C, the cells were suspended in 1 ml of 300 mM sucrose. This cell suspension was incubated with plasmid DNA (about 500 ng per 100 μl) on ice for 5 min and then subjected to electroporation (100 μl at 2.5 V, 25 μFd, 200 Ω). Immediately after electroporation, 700 μl of PTS broth was added to the cell suspension. After 1 h at 37°C, transformants were selected as described above.
Purification of RhlI.
P. aeruginosa PAO-JP1 with pRhlI-2 was grown at 30°C in 10 liters of PTS broth containing gentamicin to an optical density of 0.85 at 660 nm in a Biostat B fermentor (B. Braun, St. Louis, MO). The cells were harvested by centrifuging at 4°C for 10 min at 5,000 × g, washed in 25 ml of cold buffer I [10% (vol/vol) glycerol/0.1 mM DTT/0.1 mM EDTA/0.1 mM PMSF in 20 mM potassium phosphate buffer, pH 7.5], suspended in 70 ml of cold buffer I, and lysed in a French Pressure cell (twice at 8,000 psi; 1 psi = 6.89 kPa). The resulting cell extract was centrifuged at 12,000 × g for 30 min at 4°C, and the soluble fraction was clarified by centrifuging at 74,000 × g for 60 min. RhlI was purified by using HiTrap Q, HiTrap S, and Superdex 75 column chromatography (Amersham Pharmacia). The HiTrap Q column (5 ml bed volume) was eluted with a 0–1 M gradient of NaCl in buffer I. Fractions with RhlI activity were pooled, and the buffer was exchanged with buffer II (10% glycerol/0.1 mM DTT/0.1 mM EDTA/0.1 mM PMSF in 20 mM potassium phosphate buffer, pH 7.2). RhlI was then further purified by using HiTrap S column chromatography (5 ml column bed volume) in a 0–1 M NaCl gradient in buffer II. The RhlI-containing fractions from HiTrap S column chromatography were pooled, and the buffer was exchanged with buffer III (10% glycerol/0.1 mM DTT/0.1 mM EDTA/0.1 mM PMSF/ 500 mM NaCl in 20 mM potassium phosphate buffer, pH 7.2). Protein was concentrated, and the concentrate was fractionated by Superdex 75 column chromatography in buffer III (75 ml bed volume). The Superdex 75-purified RhlI was stored in buffer III with 20% (vol/vol) glycerol at −70°C. Protein concentrations were determined by using the Bradford method (Bio-Rad). SDS/PAGE was as described (17). Western immunoblotting with RhlI antiserum was by procedures described elsewhere (8).
RhlI Activity Assays.
Unless otherwise specified, we measured RhlI activity by using the following rapid and sensitive analysis: RhlI was added to the indicated acyl-ACP or acyl-CoA and 25 nCi (1 Ci = 37 GBq) carboxy-14C-labeled SAM at the indicated SAM concentrations (Amersham Pharmacia) in a buffer containing 2 mM DTT, 200 mM NaCl, and 20 mM Tris (pH 7.8; final volume, 100 μl in siliconized 2-ml microcentrifuge tubes). After 10 min at 37°C, the reactions were stopped by addition of 4 μl of 1 M hydrochloric acid. Reaction mixtures were extracted twice with 100 μl of ethyl acetate, and the amount of 14C label in the pooled ethyl acetate extracts was determined by using scintillation counting. Acyl-HSLs readily partition into ethyl acetate, whereas SAM remains in the aqueous phase. Thus, the amount of label in the organic phase is a measure of the amount of SAM converted to acyl-HSL. The amount of acyl-HSL synthesized was calculated from the amount of label in the solvent phase and the partition coefficient of the reaction product. This assay was validated by comparison to results obtained with the bioassays described below.
Standard reaction mixtures contained 61 μM SAM and 40 μM butyryl-ACP, and reactions with purified RhlI contained 72 ng of protein. The rate of butyryl-HSL synthesis by clarified cell extracts or purified RhlI was linear over time for at least 20 min and was directly proportional to the amount of protein added.
RhlI activity was also assessed by measuring acyl-HSLs in ethyl acetate extracts from reaction mixtures as described above with bioassays. The production of butyryl or hexanoyl-HSL was measured with the P. aeruginosa Rhl bioassay (8) and octanoyl-HSL was measured with the Ralstonia solanacearum bioassay (18). Standard curves were generated with synthetic acyl-HSLs.
We used HPLC to identify reaction products. Ethyl acetate extracts were dried under a stream of nitrogen and then dissolved in 250 μl of 20% methanol in water. C18-reverse-phase HPLC in a 20–100% methanol gradient was as described (19, 20).
Chemicals.
With the exception of acyl-ACPs, all chemicals were purchased from commercial sources. Butyryl-ACP was synthesized chemically (21). Longer chain acyl-ACPs were synthesized from fatty acids and holo-ACP by using purified Vibrio harveyi acyl-ACP synthetase (22). Holo-ACP was purified from an E. coli strain that overproduces the protein by methods described elsewhere (23). Butyryl-SAM was synthesized from the N-hydrosuccinimide of butyric acid and SAM by the procedure for synthesis of hexanoyl-SAM from the N-hydrosuccinimide of hexanoic acid and SAM described previously (9). The butyryl-SAM had the expected mass as determined by using electrospray mass spectrometry.
RESULTS
Purification of RhlI.
A series of preliminary studies revealed that when overexpressed in E. coli or in P. aeruginosa PAO-JP1, RhlI was in the form of insoluble inclusion bodies (data not shown). This is consistent with a previous report of RhlI overexpression in E. coli (12). When we used P. aeruginosa PAO-JP1 (pRhlI-2) grown at 30°C the majority of RhlI was soluble as judged by immunoblotting with antiserum against RhlI. When cultures were incubated at 37°C, most of the RhlI was insoluble. We chose to use strain PAO-JP1 because it carries an insertion mutation in lasI, the other P. aeruginosa gene known to direct the synthesis of acyl-HSLs (24), and we purified RhlI from cells grown at 30°C.
A three-step purification procedure was used to obtain soluble, active RhlI from clarified P. aeruginosa (pRhlI-2) extracts (≈3.6 mg of RhlI from a 10-liter culture). RhlI was monitored throughout the purification by activity assays (Fig. 1, Table 1) and by immunoblotting (data not shown). Fractions containing enzyme activity correlated with fractions containing immunoreactive protein.
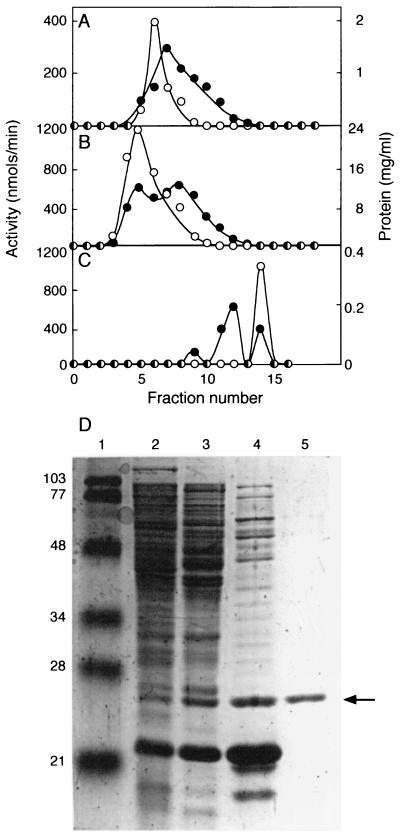
Figure 1.
Purification of RhlI from clarified cell extracts. (A) HiTrap Q column chromatography. (B) HiTrap S column chromatography. (C) Superdex 75 column chromatography. ○, RhlI activity (nmol of butyryl-HSL produced min−1 in each fraction); ●, protein levels. (D) SDS/PAGE of RhlI activity peaks. Lane 1, molecular mass standards (prestained low-range markers, Bio-Rad); lane 2, 30 μg of cell extract; lane 3, 35 μg of protein from pooled HiTrap Q fractions 3–8; lane 4, 30 μg of protein from pooled HiTrap S fractions 5–8; lane 5, 1.4 μg of protein from pooled Superdex 75 fractions 13–15. The numbers to the left indicate the molecular mass of the standard. The arrow indicates RhlI.
Table 1.
Purification of RhlI
| Step | Total activity* | Total protein, mg | Fold purification | Specific activity, units/mg | Recovery, % |
|---|---|---|---|---|---|
| Crude extract | 18 | 840 | 1.0 | 0.022 | — |
| HiTrap Q | 11 | 315 | 1.6 | 0.035 | 60 |
| HiTrap S | 1.6 | 27 | 2.9 | 0.060 | 9 |
| Superdex 75 | 7.4 | 3.6 | 95 | 2.07 | 40 |
Activity given in μmol of product formed per minute.
The apparent Mr of RhlI was ≈26,000 (Fig. 1). This is greater than the Mr predicted from the sequence of rhlI, ≈22,000. Similar observations have been reported for other LuxI family members (10, 25). In Superdex 75 size exclusion column chromatography (Fig. 1), RhlI behaved as a protein with a Mr of 25,000. This indicates RhlI is probably monomeric. Of interest, the last step in the purification resulted in a 4- to 5-fold increase in the total RhlI activity (Table 1), presumably because of the removal of an inhibitor of the reaction.
Substrates and Conditions for Acyl-HSL Synthesis by Purified RhlI.
A previous report suggested that in the presence of NADPH, HSL and butyryl-CoA served as substrates for the synthesis of butyryl-HSL by RhlI (12). When supplied with these substrates under our standard reaction conditions (Table 2) or under conditions identical to those described in the previous report (12), the amount of butyryl-HSL detected was not above the background level in the absence of added RhlI. Butyryl-CoA served as an acyl donor when SAM was a substrate, but activity was only ≈10% of the activity observed when butyryl-ACP was provided as a substrate together with SAM. The acyl-HSL from reactions containing either butyryl-ACP or butyryl-CoA and SAM coeluted with authentic butyryl-HSL in HPLC. Hexanoyl- and octanoyl-HSL were produced when hexanoyl- or octanoyl-ACP were provided as substrates, but the rates of synthesis were lower than the rate of butyryl-HSL synthesis from butyryl-ACP (Table 2). There was no detectable activity when sodium butyrate was provided in place of butyryl-ACP. This indicates that the thioester bond of the acyl donor is required for activity.
Table 2.
Substrate requirements for synthesis of acyl-HSLs by RhlI
| Substrates added | Acyl-HSL produced, nmol⋅min−1⋅mg−1 RhlI |
|---|---|
| Butyryl-ACP and SAM | 650 (530) |
| Butyryl-ACP, SAM, and NADPH | 720 |
| Butyryl-CoA, HSL, and NADPH | <0.02 |
| Butyryl-CoA and SAM | 80 (40) |
| Butyrate and SAM | <0.02 |
| Butyryl-SAM | (80) |
| Hexanoyl-ACP and SAM | 260 |
| Hexanoyl-CoA and SAM | <0.02 |
| Octanoyl-ACP and SAM | 30 |
| Decanoyl-ACP and SAM | <0.02 |
| Butyryl-ACP and S-adenosylhomocysteine | (<0.5) |
| Butyryl-ACP and S-adenosylcysteine | (<0.5) |
| Butyryl-ACP and HSL | (<0.5) |
| Butyryl-ACP and homocysteine | (1) |
| Butyryl-ACP and homoserine | (<0.5) |
| Butyryl-ACP and methionine | (<0.5) |
Each reaction mixture contained 72 ng of purified RhlI plus the indicated amino donor at 60 μM and the indicated acyl donor at 40 μM. For butyryl-SAM, the concentration was 200 μM. Where indicated, 500 μM NADPH was included in the reaction mixture. As described in Materials and Methods, the specific activity of RhlI was determined by measuring the production of 14C-labeled acyl-HSL. The values in parenthesis are obtained by using bioassays to measure acyl-HSLs.
The reaction required SAM. Activity was not detected when this amino donor was replaced by other molecules including S-adenosylhomocysteine, S-adenosylcysteine, or homoserine (Table 2). This analysis supports the view that RhlI and other members of the LuxI family of proteins catalyze the synthesis of acyl-HSLs from acyl-ACPs and SAM. Interestingly, butyryl-SAM served as a substrate for butyryl-HSL synthesis, although the activity was low relative to the activity with SAM and butyryl-ACP (Table 2). This suggests that butyryl-SAM could be an intermediate in the synthesis of butyryl-HSL from SAM and butyryl-ACP.
RhlI catalyzed synthesis of butyryl-HSL from butyryl-ACP and SAM over a pH range of 6–11 with optimum activity at pH 7.8–8.0. The enzyme was active at temperatures up to ≈60°C and activity at 50°C was about twice that at 37°C.
Kinetics of Acyl-HSL Synthesis by RhlI.
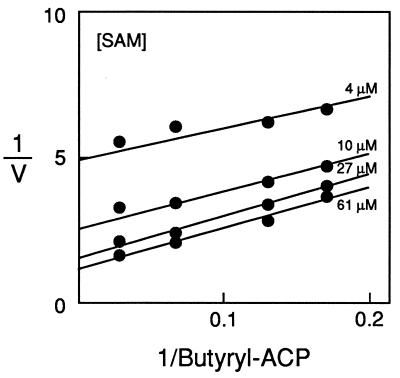
Km and Vmax for SAM and for several different acyl group donors were determined (Table 3). The Vmax for butyryl-HSL synthesis from butyryl-ACP and SAM was 16 mol⋅min−1⋅mol−1 of RhlI. This is >10-fold greater than the rates of acyl-HSL synthesis by either of the previously purified autoinducer synthase fusion proteins. The Km for butyryl-CoA was ≈40-fold greater than the Km for butyryl-ACP. This suggests that the natural substrate for RhlI is butyryl-ACP. A substrate initial velocity plot (Fig. 2) showed a pattern of nearly parallel lines, an expected result for a ping-pong reaction mechanism. This is consistent with suggestions that a covalent acyl–enzyme intermediate is formed during the synthesis of butyryl-HSL (3, 10). However, a more detailed analysis is required to draw conclusions about the reaction mechanism (see below).
Table 3.
Kinetics of acyl-HSL synthesis by RhlI
| Substrate | Km (μM) | Vmax (mol of products⋅min−1⋅mol−1 RhlI) |
|---|---|---|
| SAM | 14 | 16 |
| butyryl-ACP | 6 | 16 |
| butyryl-CoA | 230 | 2 |
| hexanoyl-ACP | 8 | 10 |
| octanoyl-ACP | 43 | 2 |
| butyryl-SAM | 38 | 2 |
Kinetic constants were calculated with the appropriate FORTRAN programs (35). SAM was tested over a range of 4–95 μM at a butyryl-ACP concentration of 60 μM with 72 ng of RhlI. For the fatty acyl substrates, the SAM concentration was 95 μM. Butyryl-ACP was tested over a range of 4–60 μM with 72 ng of RhlI, hexanoyl-ACP over a range of 8.6–690 μM with 144 ng of RhlI, octanoyl-ACP over a range of 4.4–264 μM with 1.4 μg of RhlI, and butyryl-CoA over a concentration range of 28–780 μM with 3.6 μg of RhlI. Butyryl-SAM was tested over a concentration range of 10 μM–1 mM with 72 ng of RhlI. For SAM and butyryl-ACP, true Km values are given; for the other substrates, apparent Km values are indicated.
Figure 2.
Substrate initial velocity patterns with SAM and butyryl-ACP. A double reciprocal plot with butyryl-ACP as the varied substrate at SAM concentrations of 61, 27, 10, and 4 μM.
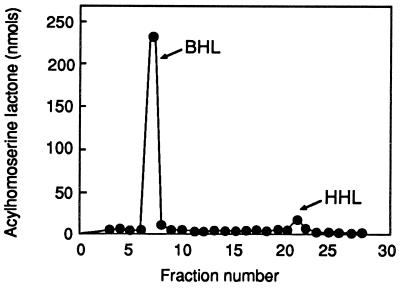
The kinetics of hexanoyl-HSL synthesis from hexanoyl-ACP and SAM were not remarkably different from the kinetics observed with butyryl-ACP and SAM. Thus, the kinetics analysis does not provide an explanation as to why the ratio of butyryl-HSL to hexanoyl-HSL produced by P. aeruginosa is ≈15:1 (11). Perhaps cellular levels of hexanoyl-ACP are limited relative to butyryl-ACP levels. To test this hypothesis, we determined the ratio of butyryl- to hexanoyl-HSL produced by RhlI in vitro in the presence of saturating levels of both butyryl-ACP and hexanoyl-ACP. The ratio of butyryl-HSL to hexanoyl-HSL was 20:1 (Fig. 3). Thus, RhlI has the ability to discriminate between butyryl- and hexanoyl-ACP. Our data fail to support the hypothesis that differences in the amounts of butyryl- and hexanoyl-HSLs produced in vivo are due to the differences in the cellular hexanoyl- and butyryl-ACP pools.
Figure 3.
HPLC analysis of acyl-HSLs synthesized by purified RhlI. Reaction mixtures contained saturating levels of SAM and 40 μM of both butyryl-ACP and hexanoyl-ACP. Synthetic butyryl-HSL was eluted in peak 7 and synthetic hexanoyl-HSL in peaks 21 and 22.
Analysis of the Mechanism of Butyryl-HSL Synthesis.
The initial velocity kinetics (Fig. 2) suggest a ping-pong reaction mechanism, but this sort of analysis cannot be considered conclusive. Therefore, we studied the kinetics of inhibition by reaction products and substrate analogs. First, we tested a number of different molecules for their ability to inhibit synthesis of butyryl-HSL (Table 4). Of the compounds tested, only the end products 5′-methylthioadenosine (MTA) and more weakly, holo-ACP, as well as the SAM analogs, S-adenosylhomocysteine, S-adenosylcysteine and sinefungin inhibited RhlI activity. The possible reaction intermediate, butyryl-SAM also inhibited the reaction. One of the products, butyryl-HSL, did not inhibit the reaction.
Table 4.
Influence of reaction products, substrate analogs, and other compounds on RhlI activity
| Inhibitors* | Inhibition, % |
|---|---|
| Holo-ACP (50 μM) | 2 |
| Holo-ACP (500 μM) | 55 |
| MTA (50 μM) | 67 |
| MTA (500 μM) | 91 |
| l-S-adenosylhomocysteine (50 μM) | 62 |
| l-S-adenosylhomocysteine (500 μM) | 91 |
| d-S-adenosylhomocysteine (50 μM) | 43 |
| d-S-adenosylhomocysteine (500 μM) | 88 |
| l-S-adenosylcysteine (50 μM) | 77 |
| l-S-adenosylcysteine (500 μM) | 97 |
| Sinefungin (100 μM) | 58 |
| Butyryl-SAM (50 μM) | 24 |
| Butyryl-SAM (500 μM) | 65 |
Reactions were performed under standard conditions except the substrate concentrations were 10 μM SAM and 9 μM butyryl-ACP. Percent inhibition was calculated based on activity without added inhibitors.
Compounds that did not inhibit the reaction: Butyryl-HSL (1 mM), CoA (575 μM), NADH (750 μM), methionine (200 μM), homocysteine (200 μM), HSL (200 μM), pantothenate (420 μM), homoserine (200 μM), butyryic acid (1 mM), cerulenin (224 μM), ATP (1 mM), ADP (1 mM), 4-hydroxybutyrate (1 mM), butyryl-CoA (500 μM), apo-ACP (155 μM). The concentrations shown in parenthesis represent the highest concentration of each compound tested.
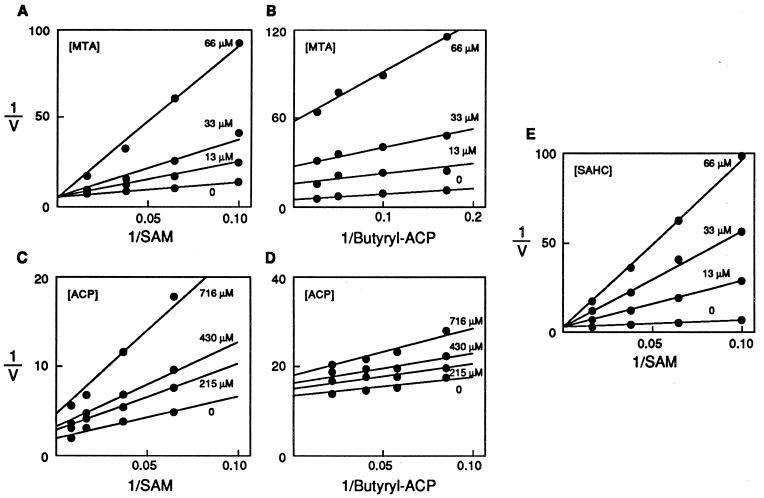
We determined the types of inhibition by measuring butyryl-HSL synthesis from butyryl-ACP and SAM and varying the inhibitor concentration and the concentration of one of the two substrates, at a fixed concentration of the other substrate (26). MTA was a competitive inhibitor of SAM at saturating and nonsaturating butyryl-ACP concentrations (Fig. 4). MTA was a noncompetitive inhibitor of butyryl-ACP at nonsaturating concentrations of SAM, but did not inhibit when SAM was saturating (data not shown). ACP was a noncompetitive inhibitor of SAM and butyryl-ACP under all conditions tested (Fig. 4). We also examined the SAM analog, S-adenosylhomocysteine. S-adenosylhomocysteine was a competitive inhibitor of SAM at both saturating (data not shown) and nonsaturating levels of butyryl-ACP. However, it was a noncompetitive inhibitor of butyryl-ACP at both saturating and nonsaturating levels of SAM (Fig. 4).
Figure 4.
Analysis of inhibition kinetics. Double-reciprocal plots of product and dead-end inhibitors vs. the substrates butyryl-ACP and SAM. (A) Inhibition pattern with the product inhibitor MTA vs. varied concentrations of the substrate SAM with butyryl-ACP held at a nonsaturating concentration. (B) Inhibition pattern with the product inhibitor MTA vs. butyryl-ACP with SAM held at a nonsaturating concentration. (C) Inhibition pattern with the product inhibitor ACP vs. the substrate SAM with butyryl-ACP held at a nonsaturating concentration. (D) Inhibition pattern with the product inhibitor ACP vs. the substrate butyryl-ACP with SAM held at a nonsaturating concentration. (E) The inhibition pattern with differing amounts of the dead-end inhibitor S-adenosylhomocysteine vs. varied concentrations of the substrate SAM with butyryl-ACP held at nonsaturating levels.
The data are consistent with a bi ter (two substrate, three product) sequential ordered enzymatic reaction mechanism. The pattern of MTA inhibition of SAM indicates that these two molecules bind the same form of the enzyme, that SAM is the first substrate that binds the enzyme, and that MTA is the last product released. The MTA inhibition of butyryl-ACP is consistent with butyryl-ACP as the second substrate that binds the enzyme with an irreversible step (release of butyryl-HSL) between butyryl-ACP binding and MTA release. The ACP and S-adenosylhomocysteine inhibition data are consistent with a bi ter sequential ordered mechanism.
The data are inconsistent with a ping-pong mechanism, which would be observed if, as previously suggested, a covalent acyl–enzyme intermediate were formed in the first step of the reaction (3, 10). With a ping-pong mechanism involving formation of a covalent acyl–enzyme intermediate, MTA would be a noncompetitive inhibitor of SAM because SAM would bind the acylated form of the enzyme, whereas MTA would bind free enzyme. If SAM binding were the first step in a ping-pong mechanism, the released MTA would be a noncompetitive inhibitor of SAM, not a competitive inhibitor.
DISCUSSION
We used a strategy in which RhlI was mildly overexpressed in its native environment, P. aeruginosa, to obtain sufficient quantities of this enzyme for detailed kinetic analysis. A lasI mutant strain was used to eliminate the complication of a second acyl-HSL synthase. This strategy, coupled with direct radiochemical acyl-HSL synthase assays, allowed monitoring of the purification procedure. The purified enzyme catalyzed the synthesis of acyl-HSLs at a rate >10-fold greater than the rates obtained with purified LuxI and TraI fusion proteins (9, 10). The substrates for butyryl-HSL synthesis by RhlI in vivo are likely to be butyryl-ACP and SAM (Table 2). With these substrates, purified RhlI synthesized butyryl-HSL at a rate 107 times greater than that obtained with acyl-CoA and HSL as substrates (12). Therefore, we believe that previous conclusions that RhlI differs from the other LuxI homologs in substrate specificity (12) are unfounded. Rather, our evidence indicates that RhlI is a suitable model for studies of acyl-HSL synthesis by members of the LuxI family of enzymes, all of which catalyze the synthesis of acyl-HSLs from acyl-ACPs and SAM. The development of a convenient radioactive assay for RhlI activity has facilitated our work and should be useful in future studies of acyl-HSL synthesis.
Butyryl-CoA was a poor substitute for butyryl-ACP as the acyl donor for RhlI, and the enzyme showed no detectable activity with sodium butyrate (Table 2). This suggests that the reaction requires a thioesterified acyl group and that the enzyme preferentially recognizes butyryl-ACP. The enzyme is most active when provided with butyryl- or hexanoyl-ACP (Table 2), but it shows a strong preference for butyryl-ACP in the presence of both acyl donors (Fig. 3). This indicates that the acyl group preference observed in vivo (11) is defined by the substrate specificity of the enzyme rather than by the supply of acyl substrates available in the cytoplasm.
The best amino donor for RhlI-catalyzed acyl-HSL synthesis was SAM (Table 2). As discussed above, this is consistent with previous studies of LuxI and TraI, which indicated SAM was the amino donor. The Km for SAM was 14 μM (Table 3). Because SAM levels in bacteria are thought to be above 300 μM (27), SAM should not limit RhlI activity in vivo.
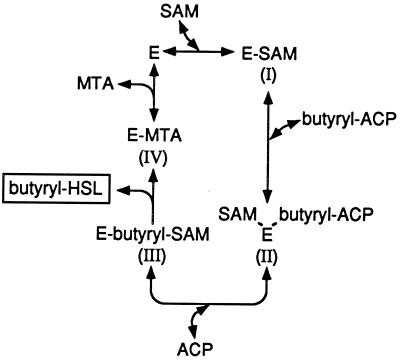
The availability of sufficient quantities of active, pure RhlI, and the substrates for RhlI activity, together with a convenient and quantitative activity assay allowed our kinetic analysis of end-product and dead-end inhibitors of RhlI activity (Table 4 and Fig. 4). The most effective inhibitors were the reaction product MTA and the SAM analogs. The reaction product holo-ACP was a weak inhibitor, and we did not observe inhibition of RhlI by butyryl-HSL. The lack of end-product inhibition by butyryl-HSL might be explained if there is an acyl-SAM intermediate that irreversibly cyclizes to form MTA and butyryl-HSL. The fact that butyryl-SAM serves as both a substrate and an inhibitor of RhlI is consistent with the idea that there is an enzyme-acyl-SAM intermediate. We propose a bi ter sequential ordered mechanism for RhlI-catalyzed butyryl-HSL synthesis (Fig. 5). There is more to be learned from studies of butyryl-SAM interactions with the enzyme.
Figure 5.
The proposed enzymatic reaction mechanism for autoinducer synthesis by RhlI. Bound substrates and products are in parentheses. This is a bi ter sequential ordered reaction in which the substrates bind in a defined order and the products are released in a defined order.
Because pathogenic bacteria such as P. aeruginosa involve acyl-HSL signals in the regulation of virulence genes (7, 24, 28–30) there has been a growing interest in blocking this system with inhibitors. Although some effort has been made to develop acyl-HSL signal analogs that block signal reception (31–34), our lack of understanding of the mechanism of signal synthesis has limited efforts to target this step in quorum sensing. We have proposed that the initial step in synthesis of butyryl-HSL by RhlI is SAM binding, and we showed that several SAM analogs were inhibitors of RhlI activity (Table 4). Furthermore, acyl-HSL synthases are unlike most other SAM-utilizing enzymes in that the SAM is an amino donor rather than a methyl donor, and not surprisingly, we cannot identify motifs in the acyl-HSL synthases that characterize other enzymes with SAM-binding sites. Thus, screening efforts could reveal SAM analogs that inhibit autoinducer synthases specifically. We know less about acyl group transfer from acyl-ACP to RhlI, but the evidence against formation of a covalent acyl–enzyme intermediate suggests that acyl group transfer also may be a target for development of specific acyl-HSL synthase inhibitors.
Acknowledgments
We thank Bryce Plapp for his help in the design and interpretation of enzyme kinetics and inhibitor studies. This work was supported by a grant from the National Science Foundation (MCB 9808308) and the National Institutes of Allergy and Infectious Diseases (AI15650). M.R.P. is a National Institutes of Health Postdoctoral Fellow (GM 18740-01A1), and B.L.H. has been supported by U. S. Public Health Service Training Grant 732 GM8365.
ABBREVIATIONS
- ACP
acyl carrier protein
- HSL
homoserine lactone
- SAM
S-adenosylmethionine
- MTA
5′-methylthioadenosine
References
- 1.Fuqua C, Greenberg E P. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 2.Fuqua C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 3.Sitnikov D M, Schineller J B, Baldwin T O. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 4.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck von Bodman S, Farrand S K. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 8.Parsek M R, Schaefer A L, Greenberg E P. Mol Microbiol. 1997;26:301–310. doi: 10.1046/j.1365-2958.1997.5741935.x. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moré M I, Finger D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 11.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P C, Bycroft B W, et al. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, Camara M, Chhabra S R, Hardie K R, Bycroft B W, Lazdunski A, Salmond G P C, Stewart G S A B, Williams P. Mol Microbiol. 1998;28:193–203. doi: 10.1046/j.1365-2958.1998.00789.x. [DOI] [PubMed] [Google Scholar]
- 13.Hanzelka B L, Stevens A M, Parsek M R, Greenberg E P. J Bacteriol. 1997;179:4882–4887. doi: 10.1128/jb.179.15.4882-4887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovach M E, Phillips R W, Elzer P H, Roop R M, II, Petersen K M. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Ohman D E, Cryz S J, Iglewski B H. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kropinski A M, Parr T R, Jr, Angus B L, Hancock R E W, Ghiorse W C, Greenberg E P. J Bacteriol. 1987;169:172–179. doi: 10.1128/jb.169.1.172-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer, A. L., Hanzelka, B. L., Parsek, M. R. & Greenberg, E. P. (1999) Methods Enzymol., in press. [DOI] [PubMed]
- 19.Puskas A, Greenberg E P, Kaplan S, Schaefer A L. J Bacteriol. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cronan J E, Jr, Klages A L. Proc Natl Acad Sci USA. 1981;78:5440–5444. doi: 10.1073/pnas.78.9.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Z, Fice D, Byers D M. Anal Biochem. 1992;204:34–39. doi: 10.1016/0003-2697(92)90135-t. [DOI] [PubMed] [Google Scholar]
- 23.Keating D H, Carey M R, Cronan J E., Jr J Biol Chem. 1995;270:22229–22235. doi: 10.1074/jbc.270.38.22229. [DOI] [PubMed] [Google Scholar]
- 24.Pearson J P, Pesci E C, Iglewski B H. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engebrecht J, Silverman M. Proc Natl Acad Sci USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleland W W. Biochim Biophys Acta. 1963;67:188–196. doi: 10.1016/0006-3002(63)91816-x. [DOI] [PubMed] [Google Scholar]
- 27.Val D L, Cronan J E., Jr J Bacteriol. 1998;180:2644–2651. doi: 10.1128/jb.180.10.2644-2651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pesci E C, Iglewski B H. Trends Microbiol. 1997;5:132–135. doi: 10.1016/S0966-842X(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 29.Pesci E C, Pearson J P, Seed P C, Iglewski B H. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Beaber J W, Moré M I, Fuqua C, Eberhard A, Winans S C. J Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passador L, Tucker K D, Guertin K R, Journet M P, Kende A S, Iglewski B H. J Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer A L, Hanzelka B L, Eberhard A, Greenberg E P. J Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eberhard A, Widrig C A, McBath P, Schineller J B. Arch Microbiol. 1986;146:35–40. doi: 10.1007/BF00690155. [DOI] [PubMed] [Google Scholar]
- 35.Cleland W W. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]