Abstract
Apolipophorin III (apoLp-III) from the sphinx moth, Manduca sexta, is a helix bundle protein that interacts reversibly with lipoproteins. Its five elongated amphipathic α-helices are organized in an antiparallel fashion, with helices 3 and 4 connected by a short 6-residue (PDVEKE) linker helix, termed helix 3′. Upon interaction with lipoproteins, apoLp-III opens to expose a continuous hydrophobic interior. It was postulated that helix bundle opening is preceded by an initiation step wherein helix 3′ serves to recognize available lipoprotein surface binding sites. To test this hypothesis, helix 3′ was replaced by residues that have a propensity to form a type I β-turn, NPNG. This mutant apoLp-III was defective in lipoprotein binding assays. To define a more precise mode of interaction, the relevance of the presence of the hydrophobic Val-97 flanked by Asp-96 and Glu-98 was investigated by site-directed mutagenesis. V97N and D96N/V97N/E98Q apoLp-III were unable to compete with wild-type apoLp-III to initiate an interaction with lipoproteins, whereas D96N/E98Q apoLp-III was as competent as wild-type apoLp-III. The results suggest that Val-97 is critical, whereas Asp-96 and Glu-98 are irrelevant for initiating binding to lipoproteins. A model of binding is presented wherein apoLp-III is oriented with the helix 3′ end of the molecule juxtaposed to the lipoprotein surface. Recognition of lipoprotein surface hydrophobic defects by Val-97 triggers opening of the helix bundle and facilitates formation of a stable binding interaction.
A recurring theme in exchangeable apolipoproteins is a preponderance of amphipathic α-helical segments that display lipid-binding capability (1). Exchangeable apolipoproteins play a vital role in regulating the dynamics of lipoprotein interconversions (2). Apolipophorin III (apoLp-III) from the sphinx moth, Manduca sexta, serves as a model exchangeable apolipoprotein to study structure–function relationships. Association and dissociation of apoLp-III to/from lipophorins is dependent on, and proportional to, the particle content of the neutral lipid diacylglycerol (DAG) (3–5). apoLp-III and other exchangeable apolipoproteins are characterized by an inherent conformational flexibility (6), which permits their alternate existence in lipid-free and lipid-bound states. However, the molecular basis of apolipoprotein recruitment to lipoprotein surfaces is not clear.
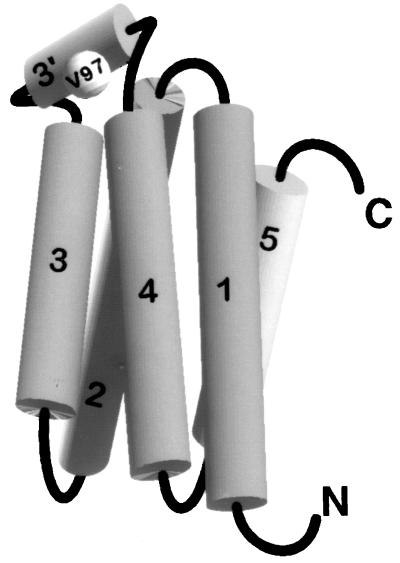
The molecular architecture of M. sexta apoLp-III is currently available and provides the solution structure of a full-length apolipoprotein in the lipid-free state (7). It is organized as a five-helix bundle (Fig. 1) in which the amphipathic α-helices are arranged in an antiparallel manner, an organization that is similar to that observed in Locusta migratoria apoLp-III (8). The hydrophobic faces of the helices are oriented toward the protein interior, whereas the hydrophilic faces are directed to the aqueous environment. An examination of M. sexta apoLp-III structure reveals the presence of a short helix, termed helix 3′, connecting helices 3 and 4. This linker helix is composed of 6 residues (95PDVEKE100) and resides outside the helix bundle (7).
Figure 1.
Schematic representation of M. sexta apoLp-III. The tertiary fold of apoLp-III reveals a five-helix bundle, where the hydrophobic faces of the amphipathic helices are oriented inwards. Helix 3′ (95PDVEKE100) connects helices 3 and 4 and lies outside the context of the bundle.
By using structure-guided disulfide bond engineering, it was demonstrated that, during interaction with lipoprotein surfaces, apoLp-III displays a preferred opening of the helix bundle about putative hinge loops located between helices 2 and 3 and helices 4 and 5 (9). During this event, the hydrophobic faces of the amphipathic helices are exposed, thereby facilitating binding to the lipid surface. Opening of the apoLp-III molecule permits helices 3 and 4 to move away from helices 1, 2, and 5. This mode of opening suggests that helix 3′ may serve as a site of recognition of exposed hydrophobic surfaces on lipoproteins, a step that precedes opening of the helix bundle and formation of a stable binding interaction (7).
In this paper, we address the above hypothesis experimentally by using site-directed mutagenesis in conjunction with lipoprotein binding assays. We have determined that Val-97, located in the middle of helix 3′, is a critical residue for lipoprotein binding and dictates the initial interaction of apoLp-III with a lipoprotein surface. We suggest that helix 3′ is responsible for oriented presentation of apoLp-III to the hydrophobic surface.
EXPERIMENTAL PROCEDURES
Materials.
3H-labeled amino acid mixture was from Amersham, and the QuikChange mutagenesis kit was purchased from Stratagene. All other chemicals and media were reagent grade. Low density lipoprotein (LDL) was isolated from fresh human plasma (10).
Design of Mutants and Site-Directed Mutagenesis.
Four different mutants were designed at the helix 3′ end of the molecule: (i) a substitution mutation wherein 10 residues (93–102) encompassing helix 3′ (95–100) were replaced by residues NPNG, which are predicted to form a type I β-turn (11). Because the distance between Pro-95 and Glu-100 (10.5 Å) is greater than that of a β-turn (5.3 Å), residues from Ala-93 to Asn-102, whose backbone atoms are separated by 6.3 Å, were included in this substitution mutation. The new sequence conformed to standard criteria for defining a β-turn: the distance between Cα(i) and Cα(i + 3) was <7 Å (11), and φ, ψ angles were in a region that is permissible in a Ramachandran plot (12); (ii) single point mutation, V97N; (iii) a double point mutation, D96N/E98Q; and (iv) a triple point mutation, D96N/V97N/E98Q. Site-directed mutagenesis was carried out in accordance with Stratagene’s instructions. The mutations were confirmed by sequencing (13).
Expression of apoLp-III.
Wild-type (WT) and mutant apoLp-IIIs were expressed in Escherichia coli cultured in M9 minimal medium (14) supplemented with 2 mM MgSO4, 0.1 mM CaCl2, and 13.3 mM glucose at 37°C, followed by induction with 2 mM isopropyl β-d-thiogalactoside (IPTG) (15) at 30°C for 5 h. apoLp-III, the only major protein to accumulate in the culture medium, was concentrated about 30-fold by ultrafiltration and dialyzed against 0.1 M sodium phosphate, pH 7.5.
In the case of 3H-labeled WT apoLp-III, the expression protocol was modified as follows: 3H-labeled amino acid mixture (150 μCi; 1 μCi = 37 kBq) was added to 1 liter of medium 30 min after induction (5-h expression period at 30°C). 3H-labeled apoLp-III recovered in the culture medium was concentrated and dialyzed extensively. The retentate was used without further purification (15). The specific activity of 3H-labeled WT apoLp-III was 3–4 × 105 dpm/mg of protein.
Purification and Characterization of Unlabeled apoLp-III.
Unlabeled WT and mutant apoLp-IIIs were purified by HPLC (15). The purity of the pooled fractions was established by analytical reversed-phase HPLC. Electrospray ionization mass spectrometric analysis (VG Biotech, Fisons Instruments, Loughborough, U.K.) revealed the following masses: Δ93–102/NPNG apoLp-III, 17,677 ± 4 Da (expected, 17,672 Da); V97N apoLp-III, 18,399 ± 6 Da (expected, 18,396 Da); D96N/E98Q apoLp-III, 18,388 ± 13 Da (expected, 18,379 Da), and D96N/V97N/E98Q apoLp-III, 18,398 ± 10 Da (expected, 18,394 Da). Circular dichroism (CD) analysis and guanidine hydrochloride (Gdn⋅HCl) denaturation of WT and mutant apoLp-IIIs were carried out as described (16). Δ93–102/NPNG apoLp-III was examined by 1H–15N heteronuclear single quantum correlation (HSQC) NMR spectroscopy and compared with WT apoLp-III (7).
Binding of apoLp-III to Phospholipase C (PL-C)-Treated LDL.
Incubation of exchangeable apolipoproteins with LDL does not result in formation of a stable binding interaction because the particle surface is already stabilized by the nonexchangeable apolipoprotein B-100 and amphiphilic phospholipids. Destruction of the surface “coat” by enzymatic (PL-C) conversion of phosphatidylcholine into DAG leads to particle aggregation (17). Although this aggregation is irreversible, it can be prevented by coincubation with exchangeable apolipoproteins (18). Apolipoprotein binding to hydrophobic sites created by phospholipolysis constitutes the basis of binding assays employed.
A typical binding assay contained LDL (50 μg of protein) in 50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/2 mM CaCl2 and 50 μg of apoLp-III at 37°C (total volume of 250 μl); phospholipolysis was initiated by the addition of 160 milliunits (mU) of PL-C (1,420 U/mg of protein; 1 U liberates 1 μmol of water-soluble organic phosphate from egg yolk l-α-phosphatidylcholine per min at pH 7.3 and 37°C). The absorbance at 340 nm was measured at given time points. In specified incubations, PL-C activity was inhibited after 30 min and turbidity development was compared with that in parallel assays containing active PL-C. All assays were performed in triplicate with three different preparations of the proteins.
Competition Binding Assays.
In the binding assay described above, if two distinct apolipoproteins are present in the incubation mixture, the one that more effectively senses developing hydrophobic sites on a lipoprotein surface will be able to initiate binding more efficiently. In the present assay, a specific mutant apoLp-III competes with WT apoLp-III for binding sites created on the surface of LDL. LDL (250 μg of protein) in 50 mM Tris⋅HCl, pH 7.5, containing 150 mM NaCl and 2 mM CaCl2, was coincubated with Δ93–102/NPNG apoLp-III and WT apoLp-III in the presence of 880 mU of PL-C (1 ml total assay volume). Control incubation mixtures, containing either WT apoLp-III or Δ93–102/NPNG apoLp-III alone, were included. After 40 min, LDL was reisolated to separate unbound apoLp-III (18) and analyzed by SDS/PAGE (10–22% acrylamide gradient). From the difference in electrophoretic mobility of WT and Δ93–102/NPNG apoLp-III, the relative amounts of each protein were determined by densitometry and normalized with respect to apolipoprotein B-100. However, SDS/PAGE analysis was not suitable for point mutants, as their size did not differ significantly from that of WT apoLp-III. In this case, mutant apoLp-IIIs were allowed to compete with 3H-labeled WT apoLp-III, which was prepared as described above. The strategy was to determine the extent of decrease in LDL-bound 3H-labeled WT apoLp-III caused by the presence of unlabeled competitor apolipoprotein.
A typical radioactive competition assay consisted of LDL (250 μg of protein), 3H-labeled WT apoLp-III (250 μg of total culture medium protein), and unlabeled competitor apolipoprotein (either WT or a mutant apoLp-III, 250 μg of protein) in a total volume of 1 ml. The reaction was initiated by the addition of 880 mU of PL-C, followed by incubation at 37°C. After 40 min, EDTA and KBr were added (final concentrations, 3.5 mM and 2.5 M, respectively), LDL was reisolated, and the amounts of radioactivity and protein were determined. A control incubation with no added PL-C gave an indication of nonspecific binding, while another control with no unlabeled competitor apolipoprotein indicated maximal 3H-labeled apoLp-III binding (taken as 100%).
RESULTS
Helix 3′ is composed of 6 residues, PDVEKE, with the prominent presence of a valine (Val-97) in the middle of the helix, flanked by Asp-96 and Glu-98 (7). The design of the first mutant, Δ93–102/NPNG apoLp-III, was based on evaluating the effect of helix 3′ on initiation of protein–lipid interactions. After evaluating the functionality of Δ93–102/NPNG apoLp-III, point mutants in helix 3′ were designed to further define the role of the Val-97, Asp-96, and Glu-98.
Physical and Structural Characterization.
Mass spectrometric analyses of the different protein preparations indicated the presence of the respective mutations, a result independently confirmed by DNA sequencing. 1H–15N heteronuclear single quantum correlation NMR analysis of Δ93–102/NPNG apoLp-III gave rise to a spectrum that was very similar to that of WT apoLp-III (7), with minor changes in the substituted region, indicating that the global fold of this mutant has not been disrupted. CD spectroscopy of the mutants indicated that their respective secondary structure contents were similar to that of WT apoLp-III (16). Also, Gdn⋅HCl denaturation indicated that the mutant proteins have stability properties similar to WT apoLp-III (midpoint of Gdn⋅HCl denaturation was ≈0.4 M).
Functional Characterization: Competition between WT and Δ93–102/NPNG apoLp-III for Binding to PL-C-Treated LDL.
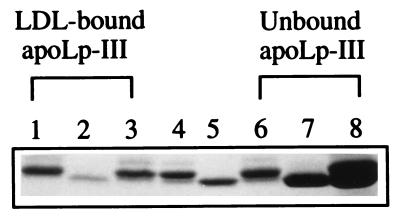
In principle, the competition assay employed can discriminate the relative ability of two exchangeable apolipoproteins to initiate a binding interaction with hydrophobic sites on a lipoprotein particle created by PL-C-mediated phospholipolysis. WT- and/or Δ93–102/NPNG apoLp-III were incubated with LDL in the absence or presence of PL-C. To determine the extent of binding, LDL was reisolated and analyzed by 10–22% acrylamide gradient SDS/PAGE (Fig. 2). Because WT apoLp-III (lane 4) and Δ93–102/NPNG apoLp-III (lane 5) migrate differently, this analysis permitted determination of the relative amount of LDL-bound and unbound apoLp-IIIs. In the absence of PL-C, no binding occurs. When present alone with LDL and PL-C under the conditions employed, WT apoLp-III distributes approximately equally between LDL (lane 1) and the infranatant fractions (lane 6). By contrast, under the same conditions, only small amounts of Δ93–102/NPNG apoLp-III were recovered in association with LDL (lane 2), with a corresponding larger amount recovered in the infranatant (lane 7). In a competition assay containing equal amounts of WT and Δ93–102/NPNG apoLp-IIIs, the latter was a poor competitor for hydrophobic surface binding sites, with a majority of the LDL-bound protein corresponding to WT apoLp-III (≈70%, as estimated by densitometric quantitation, lane 3). As expected, a majority of the unbound apolipoprotein was Δ93–102/NPNG apoLp-III (lane 8).
Figure 2.
Competition between WT and Δ93–102/NPNG apoLp-III for binding to PL-C-treated LDL. LDL (250 μg of protein) was incubated with 880 mU of PL-C in the presence of WT (250 μg), Δ93–102/NPNG (250 μg), or both WT and Δ93–102/NPNG apoLp-III (250 μg of each) at 37°C. After 40 min, unbound apoLp-III was separated from LDL-bound apoLp-III. The LDL-containing (lanes 1, 2, and 3), and the bottom fractions (lanes 6, 7, and 8) were analyzed by SDS/PAGE. The lanes contained PL-C-treated LDL plus the following: WT apoLp-III (lanes 1 and 6); Δ93–102/NPNG apoLp-III (lanes 2 and 7); and WT and Δ93–102/NPNG apoLp-III (lanes 3 and 8). Standard WT and Δ93–102/NPNG apoLp-III were in lanes 4 and 5, respectively.
These results support the premise that helix 3′ is involved in lipid binding and that, while it is able to weakly initiate a binding interaction with LDL, Δ93–102/NPNG apoLp-III is unable to compete with WT apoLp-III. This result led us to examine helix 3′ in greater detail, using point mutants to define the role(s) of Val-97, Asp-96, and Glu-98. Similar SDS/PAGE analysis, however, was not adequate because a difference in electrophoretic mobility between WT apoLp-III and the point mutants could not be discerned. To circumvent this, 3H-labeled WT apoLp-III was prepared. The extent of decrease in LDL-bound 3H-labeled WT apoLp-III caused by the presence of unlabeled competitor apolipoprotein was taken as an indication of the efficiency of the mutant apoLp-III in initiating a binding interaction.
Competition Between WT apoLp-III and Point Mutants for an Available Hydrophobic Surface.
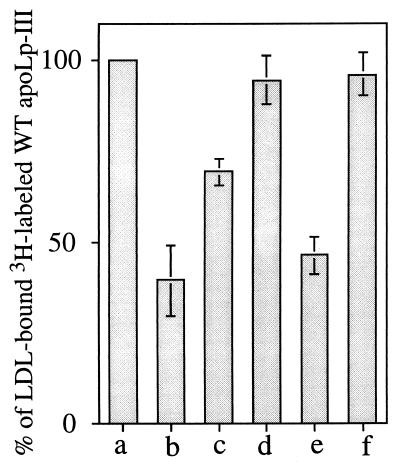
As shown in Fig. 3, in the absence of a competitor apolipoprotein, PL-C-treated LDL binds only 3H-labeled WT apoLp-III (bar a, taken as 100%). If the competitor is as effective as the WT protein, then the modified LDL surface is covered by both WT and competitor apolipoprotein, which decreases LDL-bound radioactivity. With unlabeled WT apoLp-III (bar b) as competitor, a 60% decrease in LDL-associated radioactivity was observed. V97N apoLp-III (bar d), on the other hand, induced a mere 6% decrease in LDL-bound radioactivity, suggesting that it is unable to compete with 3H-labeled WT apoLp-III for available hydrophobic sites created on the surface of LDL. By contrast, D96N/E98Q apoLp-III (bar e) caused a 54% decrease in LDL-bound radioactivity, indicating that it competes efficiently with 3H-labeled WT apoLp-III. When a third substitution, V97N, was introduced into D96N/E98Q apoLp-III (bar f), only a 4% decrease in LDL-bound radioactivity occurred, indicating that, like V97N apoLp-III, it is an ineffective competitor of 3H-labeled WT apoLp-III. Δ93–102/NPNG apoLp-III (bar c) decreased the amount of LDL-bound radioactivity by about 31%, indicating it is partially effective in competing with 3H-labeled WT apoLp-III, a result that corroborates the SDS/PAGE analysis (Fig. 2). Contributions from nonspecific interactions were negligible (<3%) as evidenced from incubation of 3H-labeled WT apoLp-III with LDL in the absence of PL-C (not shown).
Figure 3.
Competition between WT apoLp-III and point mutants for initiation of lipoprotein binding. LDL (250 μg of protein) was incubated at 37°C with 3H-labeled WT apoLp-III (250 μg of culture medium protein) and 880 mU of PL-C in the absence (bar a) or presence of unlabeled competitor apoLp-III: WT (bar b), Δ93–102/NPNG (bar c), V97N (bar d), D96N/E98Q (bar e), or D96N/V97N/E98Q (bar f) (250 μg each). After 40 min of incubation, unbound apoLp-III was separated from LDL-bound apoLp-III by density gradient ultracentrifugation, and the radioactivity in the bound fraction was determined. The bars represent percentage of LDL-bound 3H-labeled WT apoLp-III in the presence of competitor compared with that in the absence of competitor apolipoprotein (taken as 100%). A control incubation, with no added PL-C, was included to estimate nonspecific binding of 3H-labeled WT apoLp-III (<3%; not shown).
Functional Ability of Mutant apoLp-IIIs to Bind to PL-C-Treated LDL.
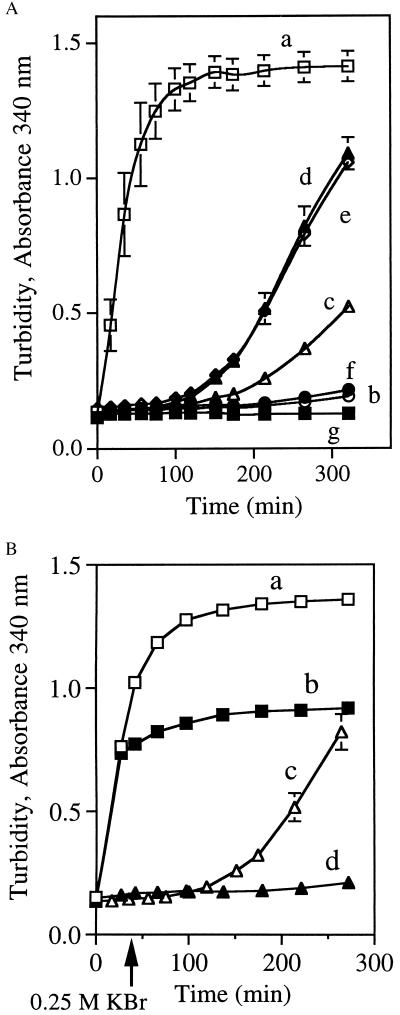
The competition assay described above examined interaction of apoLp-III with lipolyzing LDL after a 40-min incubation. Since some mutants showed essentially no binding, we evaluated the intrinsic ability of these proteins to bind to PL-C-treated LDL in the absence of competing WT apoLp-III. Fig. 4A depicts a comparative evaluation of the binding ability of WT and mutant apoLp-IIIs. A plot of PL-C-induced turbidity as a function of time revealed that LDL aggregates rapidly in the absence of added apolipoprotein (curve a). This aggregation, however, is prevented by inclusion of WT apoLp-III (curve b). Δ93–102/NPNG apoLp-III afforded protection against aggregation at early time points but appeared to be defective after 100 min (curve c). A similar lag in PL-C-induced aggregation was observed with V97N-apoLp-III, with or without the negatively charged flanking residues (V97N and D96N/V97N/E98Q apoLp-III, curves d and e, respectively). By contrast, when the residues flanking Val-97 were replaced by Asn and Gln, this mutant was as efficient as WT apoLp-III in preventing LDL aggregation (curve f, D96N/E98Q apoLp-III). A control incubation with no added PL-C was included (curve g).
Figure 4.
Binding and stabilization of PL-C-treated LDL by apoLp-III. (A) LDL (50 μg of protein) was incubated at 37°C with 160 mU of PL-C in the absence (curve a) or presence (50 μg each) of apolipoproteins, WT (curve b), Δ93–102/NPNG (curve c), V97N (curve d), D96N/V97N/E98Q (curve e), or D96N/E98Q (curve f). Control incubation containing no PL-C (curve g) was included. Sample absorbance was measured at 340 nm. (B) Incubations were the same as for A, except incubations for curves b and d, in which KBr (0.25 M, final concentration) was added after 30 min (see arrow), to inhibit PL-C activity (19). LDL was treated with PL-C in the absence (curves a and b) or the presence of V97N apoLp-III (curves c and d).
In the case of V97N and D96N/V97N/E98Q apoLp-III, their failure to protect PL-C-treated LDL against aggregation after 100 min could be due to either defective initiation or an inability to maintain a stable binding interaction. To distinguish between these two processes, assays were conducted wherein PL-C activity was inhibited by KBr (19) after 30 min (Fig. 4B). In designing this experiment, we reasoned that, if poor stability of binding of mutant apoLp-III was responsible for the delayed onset of turbidity, the mutant apoLp-III-LDL complex, once formed, would not maintain a stable interaction. Specifically, if V97N apoLp-III (or D96N/V97N/E98Q apoLp-III, data not shown) was unable to maintain a stable binding interaction with the LDL surface, it would dissociate from the particle, leading to an increase in sample turbidity. However, it was noted that after inhibition of PL-C activity in incubation mixtures containing LDL and V97N apoLp-III (curve d), no further change in sample turbidity occurred. Similarly, after inhibition of PL-C activity in incubations of LDL and PL-C in the absence of apolipoprotein, further turbidity development was prevented (curve b). Taken together, these results suggest that the initiation process has been affected by the mutations, not the stability of mutant apoLp-III–LDL complex. Furthermore, in denaturation studies of lipid-associated WT and Δ93–102/NPNG apoLp-III (as dimyristoyl phosphatidylcholine/apoLp-III disc complexes), no significant differences in their respective midpoints of Gdn⋅HCl denaturation were noted (2.5 M versus 2.7 M, respectively), suggesting that the stability of binding has not been altered.
DISCUSSION
The molecular mechanism of the initiation of exchangeable apolipoprotein binding to lipoprotein surfaces has been investigated. M. sexta apoLp-III exists as a lipid-free, globular, five-helix bundle hemolymph protein in resting insects. During flight, upon stimulation by adipokinetic hormone, apoLp-III is recruited onto preexisting lipophorin (insect lipoprotein) particles, concomitant with uptake of DAG from fat body tissue (3–5, 20). Evidence suggests that the presence of DAG in the phospholipid monolayer of lipophorin induces apoLp-III to bind and stabilize the swelling macromolecular structure (20–22). However, questions remain about how apoLp-III recognizes lipophorin surface hydrophobic defects.
A brief survey of lipid-free M. sexta apoLp-III structure reveals that the elongated helix bundle contains a short helix present at one end of the molecule (7). The outside of the helix bundle is dominated by polar and charged residues. Given this architecture, the molecular mechanism by which DAG accumulation on the lipophorin surface induces apoLp-III binding is intriguing. On the basis of our earlier observation that apoLp-III undergoes an oriented conformational opening during interaction with PL-C-treated LDL (9), it was suggested that the feature responsible for initiation of binding is located at the end of the molecule bearing helix 3′ (7). The strategic location of helix 3′, the presence of a hydrophobic residue (Val-97), and its relatively solvent-exposed nature suggest it may mediate the initial interaction between apoLp-III and the lipoprotein particle.
Alignment of the primary sequence of M. sexta apoLp-III with the sequences available from other lepidopteran species indicates that the residues constituting helix 3′, particularly P95, D96, V97, and E98, are highly conserved (PDVEKN in Bombyx mori P50 strain, National Center for Biotechnology Information Entrez no. 1381798; PDVERQ in Spodoptera litura, no. 3719453; PEVEKE in Agrius convolvuli, no. 2197055; and PDVERQ in Galleria mellonella, no. 3426083). Likewise, in the N-terminal domain of human apolipoprotein E (apoE), which is a four-helix bundle (23), a 9-residue helix connects helices 1 and 2. Significantly, sequence analysis suggests that this region of apoE is the most conserved among species (24). Thus, while lipid-binding capability is conferred by amphipathic helical segments in apolipoproteins, it is possible that the ability to recognize available hydrophobic sites on lipoprotein particles is provided by a cognate hydrophobic site at one end of the apolipoprotein molecule.
Mutations in apoLp-III could affect functionality of the protein either by directly blocking protein–lipid interactions or by disrupting the structural integrity of the protein. The latter possibility may be eliminated in all the mutants used here on the basis of CD and denaturation data which indicate that, in comparison to WT apoLp-III, overall structural integrity is maintained. The substituted amino acids in the point mutants are not helix-breaking residues and, therefore, should not disrupt the helical nature of helix 3′. Furthermore, their location outside the helix bundle precludes any possibility that they interrupt the lipid-binding ability of the amphipathic helices. In the case of the more aggressive mutation, NMR studies of Δ93–102/NPNG apoLp-III revealed that the overall global fold is retained, with minimal perturbation of the structure. As expected, the 15N/1H amide chemical shifts were different in and around the site of the mutation.
Initial experiments with Δ93–102/NPNG apoLp-III helped focus the issue of initiation to the helix 3′ end of the molecule. It was necessary to design further mutations in this site to evaluate the role of individual residues in the initiation/recognition process. Since we are examining the interaction between apoLp-III and hydrophobic surfaces on a lipoprotein particle, we hypothesized that Val-97 plays a significant role in recognition and initiation of binding. V97N and D96N/V97N/E98Q apoLp-III were unable to compete with WT apoLp-III for available sites on PL-C-treated LDL. On the other hand, D96N/E98Q apoLp-III retained its functionality and was demonstrated to compete effectively with WT apoLp-III. In assays with PL-C, LDL, and V97N or D96N/V97N/E98Q apoLp-III (Fig. 4A), we noted a delay in the onset of turbidity development, compared with assays conducted in the absence of apolipoprotein. This delay is likely because of a residual ability of these mutant apoLp-IIIs to interact with modified LDL surface. However, as evidenced by the development of sample turbidity in these assays as a function of time, as well as the competition assays with WT apoLp-III, it is apparent that these mutants are defective. These mutants appear to be able to repair hydrophobic defects created in the initial stages by PL-C, but are subsequently unable to keep pace with the generation of binding sites. It is noteworthy that a similar delay in onset of LDL aggregation was observed in an earlier study (9). In this case, a disulfide bond was engineered to tether neighboring helical segments and prevent helix bundle opening. Under oxidizing, but not reducing, conditions this mutant was unable to prevent PL-C-induced LDL aggregation, although there was a lag in the onset of turbidity development, compared with the control incubation lacking apoLp-III. Insofar as the stability of the lipid-binding interaction of the V97N or D96N/V97N/E98Q apoLp-III was not affected by the mutations in the present study (Fig. 4B), we conclude that the ability of these proteins to initiate binding to PL-C-treated LDL has been compromised. Furthermore, since the underlying commonality between the nonfunctional mutants is loss of the hydrophobic residue, the data strongly suggest that Val-97 functions in initiation of apoLp-III–lipoprotein binding interactions.
In terms of apoLp-III-mediated transformation of phospholipid vesicles, charge–charge interactions have been shown to play a role in the binding process (25). Alternatively, the concept that hydrophobic interactions determine binding of apoLp-III to lipoprotein surfaces has been proposed (8, 20, 21, 26). On the basis of the crystal structure of L. migratoria apoLp-III (8), it was suggested that the hydrophobic residues in the loop regions of the five-helix bundle play a role in initiating lipid-binding interaction. Using progressive sequence alignment algorithms, Smith et al. (27) reiterated the significance of leucine residues at or near positions 32 and 93 of L. migratoria apoLp-III, located in the loop regions at one end of the elongated molecule, in mediating the initial recognition process. Recent site-directed mutagenesis experiments in L. migratoria apoLp-III suggest an important role for these residues in initiation of lipid binding (P. Weers, V.N., C.M.K., and R.O.R., unpublished results). By using surface plasmon resonance spectroscopy (26), a two-step sequential binding hypothesis was evoked, with an initial recognition process relying on a putative “hydrophobic sensor” site on M. sexta apoLp-III, followed by opening of the helix bundle. Alternatively, a recent report suggested that a partially folded molten globule conformation represents the lipid binding-competent form of apoLp-III (28). Using a combination of structure-aided site-directed mutagenesis and a sensitive lipoprotein binding assay, we provide here experimental evidence for an initiation process taking place at the helix 3′ end of M. sexta apoLp-III.
It is envisaged that the elongated apoLp-III molecule is oriented on the lipoprotein surface with helix 3′ juxtaposed to the phospholipid monolayer of the lipoprotein surface (its axis parallel to the plane of the monolayer), possibly attracted by weak charge interactions (Fig. 5). Perturbation of the monolayer surface by appearance of DAG (29, 30) appears to be the event that “triggers” initiation of binding of apoLp-III leading to rotation of helix 3′, thereby exposing the otherwise buried Val-97. A signal from the lipoprotein particle is thus a mandatory step in initiating binding of lipid-free apoLp-III. The proposal of helix rotation is analogous to the “lid-opening” of hepatic and lipoprotein lipases (31). The “lids” in lipases cover the catalytic site and are considered to be organized as amphipathic helices that come in contact with lipase substrates and dictate substrate specificity (31). In M. sexta apoLp-III, the recognition event may trigger opening of the helix bundle, leading to replacement of helix–helix contacts by helix–lipid contacts at the lipoprotein surface. Given the increased stability of lipid-bound apoLp-III (6), this conformation will be retained in situ, until metabolic processes eliminate lipoprotein surface hydrophobicity, resulting in apoLp-III release and return to the helix-bundle conformation. A question then arises: where does the “helix-to-turn” mutant fit into this interpretation? One possible explanation is that removal of helix 3′ exposes other hydrophobic residue(s) from the helix bundle interior, which substitutes for the role played by Val-97. This mutant displays intermediate functionality in terms of its ability to compete for and bind to lipid surfaces. This observation is crucial, as it leads to a model of apolipoprotein initiation and binding interactions, wherein helix 3′ acts as a scaffold, presenting the appropriate residues to potential binding sites on the lipoprotein particle. It is possible that a similar mode of lipid interaction takes place with other exchangeable apolipoproteins, involving an initial recognition step followed by a lipid-binding step accompanied by large conformational alterations.
Figure 5.
Model of apoLp-III binding interaction (not to scale). In the lipid-free state, apoLp-III exists as a closed five-helix bundle, poised to bind to lipophorin. Appearance of DAG in the lipoprotein surface monolayer induces apoLp-III binding at its helix 3′ end, which triggers helix-bundle opening. This opening ultimately leads to replacement of helix–helix contacts in the bundle conformation by helix–lipid contacts in the lipoprotein-bound state.
Acknowledgments
We thank Kim Oikawa for the biophysical studies and Dr. P. Weers and C. Fisher for useful discussions and critical reading of the manuscript. We also thank P. d’Obrenan for preparation of the model figures. This work was supported by grants from the Medical Research Council of Canada to R.O.R.
ABBREVIATIONS
- apoLp-III
apolipophorin III
- DAG
diacylglycerol
- LDL
low density lipoprotein
- PL-C
phospholipase C
- U
unit
- WT
wild type
- Gdn⋅HCl
guanidine hydrochloride
References
- 1.Segrest J P, Garber D W, Brouillette C G, Harvey S C, Anantharamaiah G M. Adv Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- 2.Fielding C J, Fielding P E. J Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 3.Van der Horst D J. Biochim Biophys Acta. 1990;1047:195–211. doi: 10.1016/0005-2760(90)90518-3. [DOI] [PubMed] [Google Scholar]
- 4.Ryan R O. Curr Opin Struct Biol. 1994;4:499–506. [Google Scholar]
- 5.Soulages J L, Wells M A. Adv Protein Chem. 1994;45:371–415. doi: 10.1016/s0065-3233(08)60644-0. [DOI] [PubMed] [Google Scholar]
- 6.Wientzek M, Kay C M, Oikawa K, Ryan R O. J Biol Chem. 1994;269:4605–4612. [PubMed] [Google Scholar]
- 7.Wang J, Gagné S M, Sykes B D, Ryan R O. J Biol Chem. 1997;272:17912–17920. doi: 10.1074/jbc.272.29.17912. [DOI] [PubMed] [Google Scholar]
- 8.Breiter D R, Kanost M R, Benning M M, Wesenberg G, Law J H, Wells M A, Rayment I, Holden H M. Biochemistry. 1991;30:603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- 9.Narayanaswami V, Wang J, Kay C M, Scraba D G, Ryan R O. J Biol Chem. 1996;271:26855–26862. doi: 10.1074/jbc.271.43.26855. [DOI] [PubMed] [Google Scholar]
- 10.Schumaker V N, Puppione D L. Methods Enzymol. 1986;128:155–170. doi: 10.1016/0076-6879(86)28066-0. [DOI] [PubMed] [Google Scholar]
- 11.Wilmot C M, Thornton J M. J Mol Biol. 1988;203:221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 12.Richardson J S. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 13.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Vol. 3. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Ryan R O, Schieve D, Wientzek M, Narayanaswami V, Oikawa K, Kay C M, Agellon L B. J Lipid Res. 1995;36:1066–1072. [PubMed] [Google Scholar]
- 16.Ryan R O, Oikawa K, Kay C M. J Biol Chem. 1993;268:1525–1530. [PubMed] [Google Scholar]
- 17.Suits A G, Chait A, Aviram M, Heinecke J W. Proc Natl Acad Sci USA. 1989;86:2713–2717. doi: 10.1073/pnas.86.8.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Scraba D G, Ryan R O. FEBS Lett. 1993;316:27–33. doi: 10.1016/0014-5793(93)81730-n. [DOI] [PubMed] [Google Scholar]
- 19.Aakre S-G, Little C. Biochem J. 1982;203:799–801. doi: 10.1042/bj2030799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells M A, Ryan R O, Kawooya J K, Law J H. J Biol Chem. 1987;262:4172–4176. [PubMed] [Google Scholar]
- 21.Wang J, Liu H, Sykes B D, Ryan R O. Biochemistry. 1995;34:6755–6761. doi: 10.1021/bi00020a021. [DOI] [PubMed] [Google Scholar]
- 22.Soulages J L, van Antwerpen R, Wells M A. Biochemistry. 1996;35:5191–5198. doi: 10.1021/bi952794d. [DOI] [PubMed] [Google Scholar]
- 23.Wilson C, Wardell M R, Weisgraber K H, Mahley R W, Agard D A. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 24.Weisgraber K H. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Lewis R N A H, McElhaney R N, Ryan R O. Biochemistry. 1993;32:3942–3952. doi: 10.1021/bi00066a014. [DOI] [PubMed] [Google Scholar]
- 26.Soulages J L, Salamon Z, Wells M A, Tollin G. Proc Natl Acad Sci USA. 1995;92:5650–5654. doi: 10.1073/pnas.92.12.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith A F, Owen L M, Strobel L M, Chen H, Kanost M R, Hanneman E, Wells M A. J Lipid Res. 1994;35:1976–1984. [PubMed] [Google Scholar]
- 28.Soulages J L, Bendavid O J. Biochemistry. 1998;37:10203–10210. doi: 10.1021/bi980622l. [DOI] [PubMed] [Google Scholar]
- 29.Das S, Rand P R. Biochemistry. 1986;26:2882–2889. doi: 10.1021/bi00358a022. [DOI] [PubMed] [Google Scholar]
- 30.Siegel D P, Banschbach J, Alford D, Ellens H, Lis L J, Quinn P J, Yeagle P L, Bentz J. Biochemistry. 1989;28:3703–3709. doi: 10.1021/bi00435a012. [DOI] [PubMed] [Google Scholar]
- 31.Dugi K A, Dichek H L, Santamarina-Fojo S. J Biol Chem. 1995;270:25396–25401. doi: 10.1074/jbc.270.43.25396. [DOI] [PubMed] [Google Scholar]