Abstract
mRNA decay in prokaryotic cells involves the action of both endo- and exoribonucleases. In Escherichia coli, degradation of RNA to the mononucleotide level was thought to depend on RNase II and polynucleotide phosphorylase. Here, we show that the enzyme oligoribonuclease is an essential part of this process as well. Thus, inactivation of the orn gene encoding oligoribonuclease leads to a cessation of cell growth. Moreover, although pulse-labeled RNA decays normally in orn mutant cells under nonpermissive conditions, a large fraction of the resulting products is small oligoribonucleotides rather than the mononucleotides generated in wild-type cells. The oligoribonucleotides that accumulate are 2–5 residues in length; longer molecules disappear during the decay process. These data indicate that oligoribonuclease is required to complete the degradation of mRNA to mononucleotides and that this process is required for cell viability. Inasmuch as close homologues of the orn gene are found in a wide range of eukaryotes, extending up to humans, these findings raise the possibility that oligoribonuclease also participates in mRNA degradation in these organisms.
It now is appreciated that mRNA degradation is an important contributor to the overall level of gene expression in both prokaryotic and eukaryotic cells (1, 2). Nevertheless, there is still relatively little information about the enzymes that participate in this process or the factors that might influence their mode of action. For Escherichia coli, the system that probably is understood best, it is thought that mRNA decay is initiated by endonucleolytic cleavages, mainly catalyzed by the enzyme RNase E, followed by exonucleolytic breakdown of the resultant products to the mononucleotide level (1, 3). The principal enzymes found to be involved in this latter step are RNase II, a 3′-to-5′ hydrolytic exoribonuclease, and polynucleotide phosphorylase, a 3′-to-5′ phosphorolytic nuclease (1, 4); in fact, mutant strains devoid of these two RNases are inviable (5), suggesting that degradation of the mRNA fragments to mononucleotides is required for normal cell growth.
However, there are several inconsistencies with this currently accepted scheme for mRNA decay. One difficulty is that in E. coli mRNA, degradation is largely hydrolytic, generating nucleoside monophosphates as products (6, 7); yet, it is polynucleotide phosphorylase, which produces nucleoside diphosphates, that is associated with RNase E and other proteins in a complex called the degradosome, which has been implicated in the mRNA decay process (8, 9). A second problem with the proposed scheme, which is the subject of this paper, is that it is not understood how RNase II or polynucleotide phosphorylase could complete the degradation of mRNA because both are known to be inefficient in degrading small oligoribonucleotides (10, 11). Consequently, because mRNA represents as much as 60% of overall transcription at any given time and also is degraded very rapidly (1, 12), much of the nucleotide content of the cell soon would accumulate as small oligoribonucleotides if they were not removed rapidly. It has been suggested that a possible solution to this problem may lie with the enzyme oligoribonuclease (13, 14).
Oligoribonuclease originally was identified as an RNase specific for small oligoribonucleotides (15). Our laboratory subsequently demonstrated that oligoribonuclease is distinct from the other exoribonucleases known in E. coli and that it is the only one able to degrade small oligoribonucleotides (14). Recently, we identified the orn gene encoding oligoribonuclease and showed that it has close homologs in other organisms, extending up to humans (16). In this paper we report that orn is an essential gene, the only one of the eight known exoribonuclease genes in E. coli to be required for cell viability. Moreover, we demonstrate that upon loss of oligoribonuclease activity, cells accumulate short oligoribonucleotides that are derived from pulse-labeled RNA. Based on these data, we propose that oligoribonuclease is an essential component of the mRNA-decay pathway in E. coli and, possibly, in higher organisms as well.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids. All strains used were E. coli K-12 derivatives. Strains UT481[Δ(lac-pro) hsdS (r−m−) lacIq lacZ] (17) and JM109 (Promega) were used for plasmid transformation. Strain CF881 (recB xthA rna) was used as the recipient for linear transformation and as the wild-type control for enzyme assays. Plasmid pUC4K (Pharmacia) provided the kanamycin-resistance (Kanr) cassette used to interrupt the orn gene. Plasmid pMAK705, a kind gift from S. Kushner (University of Georgia), contains a temperature-sensitive (ts) replicon and a chloramphenicol-resistance (Camr) marker (18). Strain CF881/pYJ19–1, which carries plasmid pYJ19–1 (see below), was used as the wild-type control for most experiments.
Plasmid Constructions.
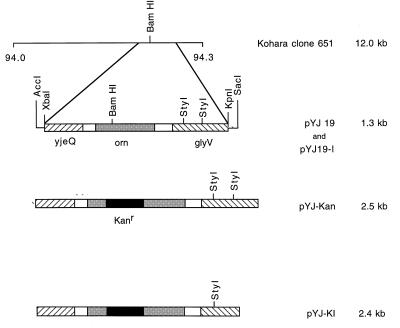
Plasmid pYJ19, carrying the E. coli orn gene, and plasmid pYJ-Kan, in which the orn gene is interrupted with a Kanr cassette, have been described (16) (Fig. 1). Downstream of orn in this plasmid is the glyV operon comprising 3 tRNAGly genes. The presence of these genes interfered with linear transformation of orn. Consequently, pYJ-Kan was digested with StyI to eliminate 113 bp from the glyV operon. The resulting large fragment was self-ligated with T4 DNA ligase (NEB, Beverly, MA) to generate plasmid pYJ-K1 (Fig. 1). The orn-containing fragment of pYJ19 was excised with SacI and AccI and also subcloned into pMAK705 to generate pYJ19–1, which carries wild-type orn on a plasmid with a ts replicon.
Figure 1.
Restriction map of E. coli genomic clone 651 and subclones derived from it containing orn. Depicted is the 94-min region of the E. coli chromosome present on clone 651 from the Kohara library (24). The three chromosomal genes subcloned from clone 651 are represented by shaded or hatched boxes. The Kanr fragment inserted into orn is shown as a solid box, but is not drawn to scale. The names of the various subclones and the sizes of their insert DNA are shown on the right. A small amount of vector DNA and restriction sites in the vector are shown for pYJ19 because these were used to prepare pYJ19–1. Construction of the clones is described under Experimental Procedures.
Interruption of Chromosomal orn.
Plasmid pYJ-K1, containing a Kanr interruption in orn, was linearized with ScaI and introduced into strains CF881 and CF881/pYJ19–1 by linear transformation and selection for kanamycin resistance. Whether interruption of the chromosomal copy of orn had occurred was determined by Southern blot analysis (see Results).
Growth Conditions.
Cells were grown in yeast extract/tryptone (YT) medium (19). For solid medium, 1.6% agar was added. Antibiotics, when present, were at the following concentrations: 50 μg/ml ampicillin, 25 μg/ml kanamycin, 34 μg/ml chloramphenicol, 500 μg/ml rifampicin, and 20 μg/ml nalidixic acid.
Materials.
Restriction endonucleases, T4 DNA ligase, T4 RNA ligase, and Klenow fragment DNA polymerase I were obtained from New England Biolabs. Calf intestine alkaline phosphatase and Prime-a-Gene Labeling system were from Promega. Bacterial alkaline phosphatase was a product of Worthington. [α-32P]dATP (3,000 Ci/mmol; 1 Ci = 37 GBq), [5′-32P]pCp (3,000 Ci/mmol), and [5,6-3H]uridine were obtained from NEN. Uridine, ApC, and ApCpC were from Sigma. GeneScreen-Plus membrane was an NEN product. All other chemicals were reagent grade.
Preparation of Cell Extracts.
Cell extracts were prepared as described previously (14).
Preparation of Oligoribonuclease Substrates.
ApCpC[32P]pC was prepared by ligating [5′-32P]pCp to ApCpC, followed by treatment with bacterial alkaline phosphatase. The ApCpC[5′-32P]pC product was separated from the substrates by paper chromatography on Whatman 3MM paper by using 1 M ammonium acetate/95% ethanol (60:40, vol/vol) as the solvent. The radioactive tetranucleotide product was eluted from the paper with H2O and used directly for the oligoribonuclease assay.
Assay of Oligoribonuclease Activity.
The activity of oligoribonuclease was determined by monitoring the release of [32P]CMP from ApCpC[32P]pC. The product was separated by chromatography on Whatman 3MM paper by using 1 M ammonium acetate/95% ethanol (60:40, vol/vol) as the solvent. Strips corresponding to [32P]CMP were cut out and counted directly. Assays were carried out in 100 μl of reaction mixtures containing 100 mM Tris⋅Cl at pH 8.0, 5 mM MnCl2, and 6 pmol of substrate. Cell extract was added, and the sample was incubated at 37°C for 10 min. Two micromoles of EDTA, pH 8.0, was added to stop the reaction, and a portion of it was analyzed by paper chromatography. All assays were done in duplicate, and extracts were prepared at least three times. One unit of enzyme activity was defined as the amount that produced 1 nmol of [32P]CMP from ApCpC[32P]pC under the assay conditions described. The specific activity was defined as units per mg of protein. Protein was estimated by the method of Bradford (20).
Labeling of RNA and Measurement of RNA Decay.
Cells were grown at 31°C to an OD550 ≈ 0.2 and then shifted to 44°C and allowed to grow for another 90 min. Cells then were pulse-labeled for 2 min with [3H]uridine (5 μCi/ml). Incorporation was terminated by the addition of uridine (200 μg/ml), rifampicin (500 μg/ml), and nalidixic acid (20 μg/ml) (21). Portions (0.5 ml) of the culture were removed at different times after the labeling period. Cells then were washed three times with 0.9% NaCl to remove unincorporated label and suspended in 100 μl of buffer containing 50 mM Tris⋅Cl at pH 7.6, 10% sucrose, and 1 mM DTT. Three hundred microliters of 0.5% yeast RNA and 400 μl of 20% trichloroacetic acid then were added, and the samples were left for 10 min on ice. Samples were centrifuged for 10 min at 10,000 × g to remove acid-precipitable RNAs and large oligoribonucleotides. Small oligoribonucleotides remain in the acid-soluble fraction under these conditions. This fraction was extracted three times with ether to remove trichloroacetic acid and evaporated to dryness, and the pellet was dissolved in water for further analysis.
Determination of the Size Distribution of Small Oligonucleotides.
The solution containing the acid-soluble fraction was treated with bacterial alkaline phosphatase and applied to a 0.5-ml column of AG-1-X2 (Bio-Rad). The nucleoside fraction, derived from the mononucleotides in the acid-soluble fraction by phosphatase action, first was washed off the column with seven 1-ml portions of H2O. The remaining oligonucleotides then were eluted with seven 1-ml portions of 0.1 M HCl. The HCl eluant was neutralized and fractionated by paper chromatography by using 1 M ammonium acetate/95% ethanol (70:30, vol/vol). Oligoribonucleotides of different sizes were identified with appropriate standards, cut out of the paper, and counted directly in a scintillation counter.
All experiments presented were carried out at least twice with essentially identical results.
RESULTS
Interruption of Chromosomal orn.
Plasmid pYJ-K1, carrying the orn gene with a Kanr insertion (Fig. 1), was linearized with ScaI and introduced into strain CF881 to place the orn mutation in the chromosome. Cells transformed with the linear DNA fragment first were selected for kanamycin resistance and then screened for ampicillin sensitivity to eliminate any cells containing intact plasmid. Under these conditions, less than one Kanr ampicillin-sensitive (Amps) transformant was isolated per microgram of linearized plasmid DNA in each of two successive experiments (Table 1). Chromosomal DNA from each of the three transformants isolated in these experiments was subjected to Southern blot analysis using probes specific for the orn gene and the Kanr cassette. These analyses showed that the orn gene was not interrupted in any of the strains and that integration had occurred elsewhere in the chromosome (data not shown). Our inability to disrupt chromosomal orn raised the possibility that it may be an essential gene in E. coli.
Table 1.
Transformation efficiency and properties of strains transformed with orn∷kan
| Recipient strain | Relevant genotype | No. of transformants/ μg DNA* | Viable colonies
|
|
|---|---|---|---|---|
| 31°C | 44°C | |||
| CF881 | Wild type | <1 (total = 3) | 3/3 | 3/3 |
| CF881/pYJ19-1 | ts plasmid- borne orn | ≈25 (total = 170) | 50/50 | 6/50 |
Cells were linearly transformed with pYJ-K1, which carries orn∷kan.
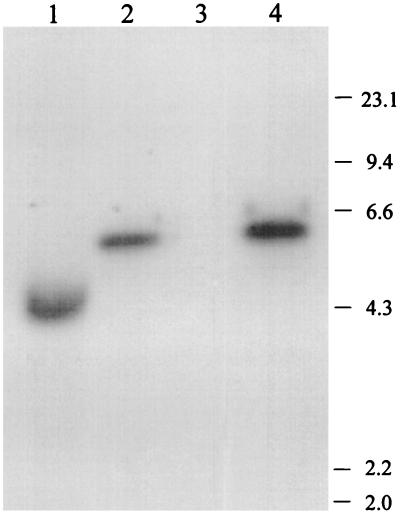
To examine this possibility, linear transformation was repeated with the same DNA fragment, but using strain CF881/pYJ19–1 as the recipient. Plasmid pYJ19–1, present in this strain, contains a ts replicon, a Camr marker, and the wild-type allele of orn (Fig. 1). If orn were essential, we would expect that the plasmid would serve to maintain cell viability even in the absence of an intact chromosomal copy of the gene; however, the resulting cells would have a ts phenotype (22). With this recipient strain a much higher yield of transformants was obtained. Thus, approximately 25 transformants (Kanr Camr Amps) per μg of linearized plasmid were recovered, and 44 of 50 transformants examined were found to be ts and unable to grow at 44°C (Table 1). Seven of these were analyzed further by Southern blot hybridization using a probe specific for the orn gene. All seven gave rise to a larger, 5.5-kb band (one example is shown in Fig. 2, lane 2) instead of the 4.3-kb band originally present in the recipient strain (lane 1), as would be expected if the interrupted orn gene had replaced the wild type. Using the Kanr cassette as a probe confirmed this conclusion, as it hybridized with the same band (land 4), but gave no signal with the strain before transformation (lane 3). These data indicate that the chromosomal orn gene had been interrupted by the 1.2-kb Kanr cassette. In contrast, when one of the temperature-resistant transformants was analyzed, only the original 4.3-kb band was detected with the orn probe (data not shown), indicating that, in this case, the Kanr cassette had integrated elsewhere on the chromosome. The data show that the chromosomal copy of orn can be interrupted only when an additional wild-type gene is present in the cell, in this case on a ts plasmid, and they support the conclusion that orn is an essential gene. These cells, carrying orn+ on a ts plasmid and a Kanr interruption in chromosomal orn, were used for all the subsequent experiments and are referred to as orn mutants.
Figure 2.
Southern blot analysis of chromosomal DNA. Samples of chromosomal DNA from wild-type strain and the orn mutant strain, prepared as in ref. 25, were digested with BglI and fractionated on a 0.8% agarose gel. Lanes 1 and 2 were loaded with digested chromosomal DNA from the wild type and mutant, respectively, and were probed with an EcoRI–HindIII fragment from pYJ19 containing orn. Lanes 3 and 4 were loaded with wild type and mutant, respectively, and were probed with Kanr cassette DNA. The migration positions of the HindIII-digested λ DNA size standards (in kb) are shown on the right.
Growth Properties of orn Mutant Cells.
Studies were carried out to ascertain the phenotype of the orn mutant cells under various growth conditions. Mutant cells grew normally on YT plates at 31°C. However, when plated on YT at 44°C, only microcolonies appeared and only after 36–48 h, indicating that the mutant cells are ts. Upon transfer to liquid YT medium at 44°C, such colonies grew only slightly over a period of 24 h, but when replated on YT medium at 31°C, many colonies appeared. Two thousand of these colonies were tested, and all were Kan- and Cam-resistant, indicating that only those cells retaining the plasmid carrying orn+ survive at 44°C. This finding strongly suggests that the orn gene is indispensable for E. coli viability.
The growth properties of the mutant cells in liquid culture also were examined. Cells were grown at 31°C to an OD550 ≈ 0.2, and a portion of the culture then was shifted to 44°C (Fig. 3A). Cells initially grew more rapidly after the temperature shift. However, within 1 h their growth slowed, and it almost ceased after 90 min. In contrast, at 31°C, mutant cells continued to grow at the same rate. Likewise, wild-type cells (CF881/pYJ19–1) shifted to 44°C continued to grow normally (data not shown). These data again show that mutant cells display a ts phenotype, consistent with the fact that their only source of a wild-type orn gene is a plasmid with a ts replicon.
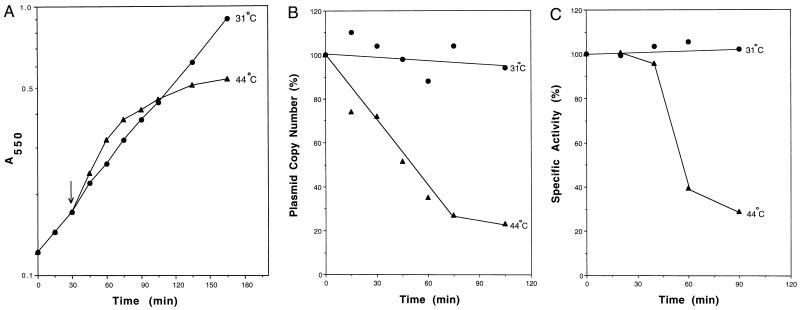
Figure 3.
Properties of orn mutant cells after temperature shift. (A) Growth. orn mutant cells were grown in YT medium at 31°C until an OD550 ≈ 0.2. Half of the culture then was shifted to 44°C, and the remaining half continued growth at 31°C. Growth was followed by measurement of OD550. The arrow indicates the time of the temperature shift. (B) Plasmid copy number. Duplicate cultures of orn mutant cells were grown at 31°C until an OD550 ≈ 0.2, at which point one culture was shifted to 44°C. After the shift, portions of each culture were removed and plasmid DNA was prepared. DNA was blotted on a GeneScreenPlus membrane and hybridized according to the manufacturer’s protocol with a 3.5-kb plasmid-specific probe obtained by digesting pMAK705 with EcoRI and HindIII. The signal was quantitated with a phosphoimager. (C) Oligoribonuclease activity. Duplicate cultures of orn mutant cells were grown at 31°C until an OD550 ≈ 0.2, at which point one culture was shifted to 44°C. After the shift, portions of each culture were removed, cell lysates were prepared, and oligoribonuclease activity was measured as described in Experimental Procedures. In B and C, 0 min represents the time of the temperature shift.
Plasmid Copy Number in orn Mutant Cells.
To examine this point in more detail, we determined the amount of plasmid in the mutant cells after the temperature shift. Duplicate cultures of mutant cells were grown at 31°C until they reached an OD550 ≈ 0.2, after which one culture was shifted to a higher (44°C) temperature. At various times after the temperature shift, portions of each culture were removed and plasmid was isolated. The relative amount of plasmid present was quantitated by DNA slot-blot analysis by using a probe specific for pYJ19–1. Fig. 3B shows that the plasmid copy number per cell decreased almost immediately after shifting the culture to the nonpermissive temperature, whereas the copy number of the plasmid in cells grown at 31°C remained essentially constant. These data support the conclusion that the slowdown in growth of mutant cells at 44°C is due to loss of the plasmid carrying the wild-type orn gene.
Thermolability of Oligoribonuclease Activity in orn Mutant Cells.
To ascertain the effect of the loss of plasmid on the level of oligoribonuclease activity, enzyme assays were carried out at various times after the temperature shift. As before, mutant cells initially were grown at 31°C until an OD550 ≈ 0.2, and a portion was shifted to 44°C. As can be seen in Fig. 3C, the specific activity of oligoribonuclease began to decrease by 40 min after the temperature shift and was reduced by 75% by 90 min after the shift. In contrast, when mutant cells were maintained at 31°C, oligoribonuclease specific activity remained constant. Likewise, in CF881 wild-type cells, oligoribonuclease specific activity was unaffected for at least 90 min after a temperature shift (data not shown). The initial level of oligoribonuclease activity in mutant cells carrying the orn+ plasmid was 1.4-fold that in CF881 wild-type cells. Thus, at 90 min after the shift, the level of activity in the mutant would be reduced to only 35% of that normally present in a wild-type cell. These data demonstrate that oligoribonuclease activity is lost upon elevating the temperature of mutant cells; moreover, this loss occurs after the decrease in plasmid copy number, but before the slowdown in growth of the mutant cells, supporting the contention that oligoribonuclease is essential for normal cell growth and probably not present in much excess.
mRNA Decay in orn Mutant Cells.
The information presented to this point demonstrated that oligoribonuclease is an essential enzyme in E. coli. To ascertain what this essential role in RNA metabolism might be, we focused on the one process in which small oligoribonucleotides were known to be generated, mRNA decay. Accordingly, wild-type and mutant cells were shifted to 44°C, and, after 90 min at this temperature to eliminate most of the oligoribonuclease from the mutant cells (Fig. 3C), each was pulse-labeled with [3H]uridine for 2 min, as described in Experimental Procedures; incorporation was terminated by the addition of uridine, rifampicin, and nalidixic acid. After various periods of chase, the cellular levels of high-molecular-weight mRNA and small oligoribonucleotides and mononucleotides derived from mRNA were determined.
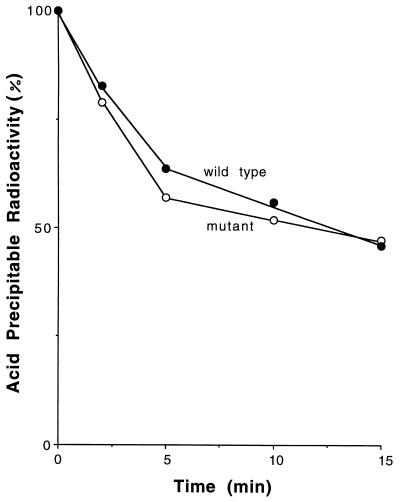
The time course of reduction in acid-precipitable radioactivity is shown in Fig. 4. No difference in mRNA decay rates between wild-type and orn mutant cells was seen. Both types of cells lost approximately 40% of their pulse-labeled RNA in 5 min, followed by a slower rate of decay over the next 10 min of an additional 15–20% of the RNA. The RNA remaining after 15 min presumably represents the stable species, rRNA and tRNA, which were synthesized during the 2-min pulse period. These data indicate that the reduction in oligoribonuclease activity in the mutant cells has no effect on the decay of mRNA to acid-soluble fragments.
Figure 4.
Decay of pulse-labeled acid-precipitable RNA in wild-type and orn mutant cells. Cells were grown for 90 min at 44°C and pulse-labeled with [3H]uridine as described in Experimental Procedures. Portions of each culture were removed at various times during the chase period, and acid-precipitable radioactivity was determined. Incorporation at 0 time: wild type, 7,642 cpm; mutant, 4,129 cpm.
Accumulation of Oligoribonucleotides in orn Mutant Cells.
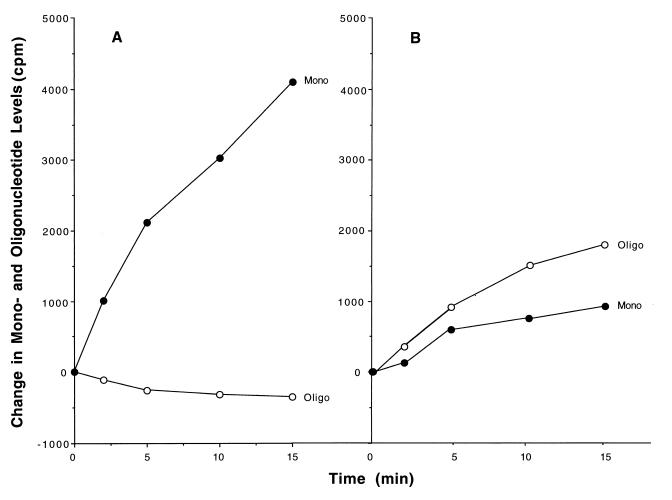
Concomitant with the decrease in acid-precipitable radioactivity was an increase in radioactivity in the acid-soluble fraction and in the medium (data not shown). This material was isolated and analyzed. First, mononucleotides and oligonucleotides were separated by fractionation on AG-1-X2 columns as described in Experimental Procedures. As shown in Fig. 5A, in wild-type cells the amount of radioactive mononucleotides increased continually during the 15-min chase period, consistent with the fact that the pulse-labeled RNA is undergoing degradation. Oligoribonucleotides also slowly disappear as they are converted to mononucleotides. In orn mutant cells, on the other hand (Fig. 5B), mononucleotides increased at a considerably slower rate, and, most significantly, oligoribonucleotide levels also increased dramatically. Thus, in cells deficient in oligoribonuclease, oligoribonucleotides derived from pulse-labeled RNA are converted poorly to mononucleotides and, consequently, accumulate as the mRNA decays.
Figure 5.
Generation of mononucleotides and oligonucleotides from pulse-labeled RNA. The acid-soluble fractions of the samples analyzed in Fig. 4 were treated with alkaline phosphatase and separated into mononucleotide and oligonucleotide fractions by chromatography on AG-1-X2 as described in Experimental Procedures. Radioactivity in the medium at each time point, which was derived from intracellular mononucleotide (see Discussion), was added to that in the mononucleotide fraction to obtain the total mononucleotide radioactivity. Data are presented as the change in radioactivity in each fraction compared with that present at 0 time. At 0 time, wild-type radioactivity in mononucleotides was 1,334 cpm and, in oligonucleotides, was 882 cpm; in mutant, radioactivity in mononucleotides was 1,176 cpm and, in oligonucleotides, was 782 cpm.
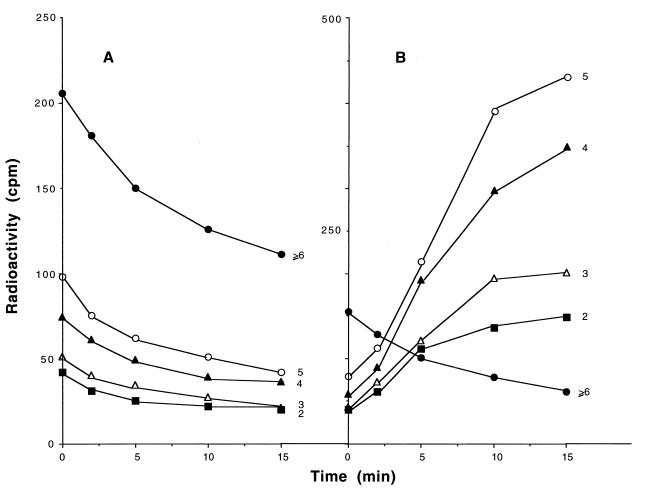
To examine in more detail the pattern of oligoribonucleotide accumulation, the oligoribonucleotide fraction was analyzed by paper chromatography to separate di-, tri-, tetra-, and pentanucleotides and those ≥6 residues in length (Fig. 6). In wild-type cells, the amount of each of the oligoribonucleotide species decreased during the 15-min chase period (Fig. 6A). In contrast, in the orn mutant, while the largest oligoribonucleotides (≥6) decreased during the chase period, those 2–5 residues in length increased considerably (Fig. 6B). The greatest accumulation was in oligoribonucleotides 5 residues long, and the level of accumulation decreased with the size of the oligonucleotide. These data show that the enzyme oligoribonuclease is essential for the removal of small oligoribonucleotides generated during the course of mRNA decay and that, in its absence, molecules 2–5 residues in length accumulate.
Figure 6.
Size distribution of oligonucleotides. The radioactive oligonucleotide samples from Fig. 5 were fractionated by paper chromatography and counted as described in Experimental Procedures. The numbers at the right of each curve indicate the chain length of the oligonucleotide.
DISCUSSION
The data presented here demonstrate that the orn gene encoding the enzyme oligoribonuclease is essential for E. coli viability. Many attempts to incorporate an interrupted orn gene into the chromosome by linear transformation were unsuccessful (16). The few Kanr transformants that could be obtained were shown by Southern blot analysis to retain an intact orn gene, indicating a low level of integration at other sites. Successful interruption of the chromosomal orn gene could be accomplished only in the presence of a low-copy-number plasmid carrying a wild-type copy of the gene. Because this plasmid also carried a ts replicon, it enabled us to verify that orn is essential for growth. Thus, upon elevation of the temperature, there is a decrease in plasmid copy number, oligoribonuclease activity is reduced, and cell growth slows dramatically in liquid culture. On plates, only microcolonies are obtained at elevated temperatures. Moreover, all cells that do grow were shown to harbor the plasmid carrying wild-type orn. Based on this information, it is clear that oligoribonuclease serves an essential function in E. coli.
Interestingly, oligoribonuclease is the only one of eight known E. coli exoribonucleases found to be essential for cell viability. For the other exoribonucleases there is generally sufficient functional redundancy that removal of a single RNase does not alter cell proliferation (4). In fact, as many as four exoribonucleases (RNases II, D, BN, and T) may be eliminated without much effect on cell growth (23). However, in the case of oligoribonuclease, the finding that it is essential for cell viability is consistent with its in vitro specificity, because it is the only exoribonuclease able to hydrolyze small oligoribonucleotides, and these are its only known substrates in vitro (13, 14). Thus, in attempting to elucidate its in vivo function, we assumed that its essential role would be in a process requiring the degradation of small oligoribonucleotides.
This led us to an examination of mRNA metabolism. Using mutant cells in which oligoribonuclease activity is reduced dramatically upon elevation of temperature, we now have shown that oligoribonucleotides derived from pulse-labeled RNA accumulate, whereas they are converted to mononucleotides in the wild type. This observation supports the conclusion that oligoribonuclease is required to complete the breakdown of mRNA and is consistent with the known inability of RNase II and polynucleotide phosphorylase to act on small oligonucleotides (10, 11). In view of the fact that elimination of oligoribonuclease leads to inviability, it appears that this enzyme plays an important role in mRNA decay and that removal of small oligoribonucleotides is an important process. Furthermore, because close homologues of the orn gene have been identified in many eukaryotes, extending from yeast to humans (16), there is a strong possibility that oligoribonuclease participates in mRNA decay in these organisms as well.
Of course, a question that remains to be answered is why the accumulation of small oligoribonucleotides would lead to a cessation of cell growth. One possibility is that as oligoribonucleotides accumulate, there is a depletion of the cell’s mononucleotide pool. A second possibility is that the accumulated oligoribonucleotides may inhibit certain enzymes and interfere with an essential metabolic process. Finally, it is also possible that oligoribonuclease has an additional, as yet unknown, function and that it is the elimination of this latter process that actually is responsible for the growth defect. Further work will be needed to sort out these various possibilities.
Finally, an interesting sidelight to these experiments that is independent of the action of oligoribonuclease is that a substantial portion of the mononucleotides released from pulse-labeled RNA at 44°C is secreted into the culture medium in the form of nucleosides (unpublished observation). This release of nucleoside occurs in both wild-type and mutant cells at 44°C, but is not found at 31°C. It is not due to the presence of rifampicin during the chase period. To our knowledge, the loss of nucleotide material from cells at elevated temperatures has not been examined, and its physiological significance remains unclear.
Acknowledgments
This work was supported by Grant GM16317 from the National Institutes of Health.
ABBREVIATIONS
- Kanr
kanamycin resistance
- Camr
chloramphenicol resistance
- ts
temperature-sensitive
- YT
yeast extract/tryptone
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kushner S R. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 849–860. [Google Scholar]
- 2.Beelman C A, Parker R. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 3.Ehretsmann C P, Carpousis A J, Krisch H M. FASEB J. 1992;6:3186–3192. doi: 10.1096/fasebj.6.13.1397840. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher M P. J Bacteriol. 1993;175:4577–4583. doi: 10.1128/jb.175.15.4577-4583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donovan W P, Kushner S R. Proc Natl Acad Sci USA. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaney S G, Boyer P D. J Mol Biol. 1972;64:581–591. doi: 10.1016/0022-2836(72)90084-8. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher M P, Reuven N B. Proc Natl Acad Sci USA. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpousis A J, Van Houwe G, Ehretsmann C, Krisch H M. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 9.Blum E, Py B, Carpousis A J, Higgins C F. Mol Micro. 1997;26:387–396. doi: 10.1046/j.1365-2958.1997.5901947.x. [DOI] [PubMed] [Google Scholar]
- 10.Shen V, Schlessinger D. In: The Enzymes. Boyer P D, editor. 15, Part B. New York: Academic; 1982. pp. 501–515. [Google Scholar]
- 11.Littauer U Z, Soreq H. In: The Enzymes. Boyer P D, editor. 15, Part B. New York: Academic; 1982. pp. 518–553. [Google Scholar]
- 12.Bremer H, Dennis P R. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1553–1569. [Google Scholar]
- 13.Datta A K, Niyogi S K. J Biol Chem. 1975;250:7313–7319. [PubMed] [Google Scholar]
- 14.Yu D, Deutscher M P. J Bacteriol. 1995;177:4137–4139. doi: 10.1128/jb.177.14.4137-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niyogi S K, Datta A K. J Biol Chem. 1975;250:7307–7312. [PubMed] [Google Scholar]
- 16.Zhang X, Zhu L, Deutscher M P. J Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Deutscher M P. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S K. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. pp. 431–435. [Google Scholar]
- 20.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Kuwano M, Ono M, Endo H, Katsuji H, Nakamura K, Hirota Y, Ohnishi Y. Mol Gen Genet. 1977;153:279–285. doi: 10.1007/BF00571283. [DOI] [PubMed] [Google Scholar]
- 22.Jasin M, Schimmel P. J Bacteriol. 1984;159:783–786. doi: 10.1128/jb.159.2.783-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly K O, Deutscher M P. J Bacteriol. 1992;174:6682–6684. doi: 10.1128/jb.174.20.6682-6684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohara Y, Akiyama K, Isono K. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Kuo T. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]