Abstract
Transcription factor UAF (upstream activation factor) is required for a high level of transcription, but not for basal transcription, of rDNA by RNA polymerase I (Pol I) in the yeast Saccharomyces cerevisiae. RRN9 encodes one of the UAF subunits. We have found that rrn9 deletion mutants grow extremely slowly but give rise to faster growing variants that can grow without intact Pol I, synthesizing rRNA by using RNA polymerase II (Pol II). This change is reversible and does not involve a simple mutation. The two alternative states, one suitable for rDNA transcription by Pol I and the other favoring rDNA transcription by Pol II, are heritable not only in mitosis, but also in meiosis. Thus, S. cerevisiae has an inherent ability to transcribe rDNA by Pol II, but this transcription activity is silenced in normal cells, and UAF plays a key role in this silencing by stabilizing the first state.
There are many examples of epigenetic repression of gene expression. In Drosophila, genes placed near heterochromatin by chromosomal rearrangements are often transcriptionally repressed without undergoing mutation. The repressed states are heritable during mitotic growth, but are not completely stable and switch between inactive and active states. This phenomenon is called position effect variegation or silencing (1, 2). Similarly, in the yeast Saccharomyces cerevisiae, silencing of genes at the silent mating type loci (the HM loci) and at or near telomeres has been studied extensively (3). These studies deal with on and off states of a particular gene or group of genes that are normally transcribed by RNA polymerase II (Pol II). Silencing is thought to be caused by special repressive chromatin structures (heterochromatin structures in higher eukaryotes).
Here we describe another case of gene silencing. The genes for rRNA (rDNA) in all eukaryotes are transcribed by RNA polymerase I (Pol I). We have discovered that the yeast S. cerevisiae has an inherent ability to transcribe rDNA by Pol II, but this transcription activity is silenced in normal yeast cells. In mutants defective in transcription factor UAF (upstream activation factor), two alternative reversible states exist for rDNA transcription: one favoring transcription by Pol II (the polymerase-switched state or PSW) and the other suitable for Pol I transcription (non-PSW). The presence of UAF in normal cells appears to stabilize the second state, thus achieving a stringent silencing of rDNA transcription by Pol II.
Like other eukaryotic rDNA promoters, the yeast rDNA promoter for the 35S precursor rRNA gene consists of two cis elements, the upstream element and the core promoter. The upstream element is required for high level transcription, but is dispensable for basal transcription, whereas the core promoter is essential for accurate transcription initiation (4–6). Initiation of yeast rDNA transcription by Pol I uses four factors in addition to Pol I: UAF, core factor, TATA-binding protein, and Rrn3p (7). UAF is a multiprotein transcription factor containing three Pol I-specific proteins, Rrn5p, Rrn9p, and Rrn10p (encoded by RRN5, RRN9, and RRN10, respectively), histones H3 and H4, and uncharacterized protein p30 (8, 9). In vitro, UAF interacts with the upstream element of the rDNA promoter, forming a stable UAF–template complex and committing the template to transcription. UAF is required for high level transcription mediated by the upstream element but not for basal transcription from the core promoter in vitro (7, 8). Consistent with this finding, the genes encoding Pol I-specific subunits of UAF, RRN5, RRN9, and RRN10, are not absolutely required for cell growth (8). This was demonstrated in two ways. First, mutants in these genes were originally isolated as galactose-dependent mutants by using strains carrying a “helper” plasmid pNOY103, which contains the rRNA coding region (35S rDNA) fused to the GAL7 promoter (8, 10, 11). We found that, in contrast to mutants defective in essential Pol I subunit genes or core factor subunit genes, these UAF mutants actually showed extremely slow growth on glucose that was detectable after long incubation. Second, mutants deleted for these genes individually (8) or in combination (unpublished data) formed tiny colonies in the absence of helper plasmids. We have now found that such mutant cultures growing in the absence of helper plasmid produce, with a high frequency, variants that grow faster. Similarly, we have found that deletion mutants carrying helper plasmid produce variants that can grow faster on glucose. Analyses of these variants led to the discovery of rDNA transcription by Pol II and a reversible switching between the two states, non-PSW and PSW.
MATERIALS AND METHODS
Media, Strains and Plasmids.
Strains used in this study are listed in Table 1, and plasmids are listed in Table 2. To construct NOY900, strain NOY898 was first constructed by crossing Pol II temperature-sensitive strain Y260 (MATa ura3–52 rpb1; ref. 12) with strain NOY556 and screening haploid segregants for temperature-sensitive growth and other desired genotypes. NOY898 was then crossed with NOY703 to obtain NOY899, which is galactose-dependent for growth and temperature-sensitive. NOY900 was isolated from NOY899 as a PSW variant able to grow on glucose.
Table 1.
Yeast strains used
| Strain | Description |
|---|---|
| NOY505 | MATa ade2–1 ura3–1 trp1–1 leu2–3,112 his3–11 can1–100 |
| NOY556 | Matα ade2–1 ura3–1 his3–11 trp1–1 leu2–3,112 can1–100 pNOY103 |
| NOY678 | MATa/MATα ade2–1/ade2–1 ura3–1/ura3–1 his3–11/his3–11 trp1–1/trp1–1 leu2–3,112/leu2–3,112 can1–100/can1–100 RRN9/rrn9Δ∷HIS3 (Ref. 8) |
| NOY684 | MATα ade2–1 ura3–1 his3–11 trp1–1 leu2–3,112 can1–100 rrn9Δ∷HIS3 (a haploid segregant derived from NOY678); Non–PSW (unstable) |
| NOY703 | MATa ade2–1 ura3–1 his3–11 trp1–1 leu2–3,112 can1–100 rrn9Δ∷HIS3 pNOY103; Non–PSW |
| NOY794 | Same as NOY852, but rpa135Δ∷LEU2 |
| NOY852 | Same as NOY684; PSW |
| NOY877 | Same as NOY852, but carries pNOY199; PSW |
| NOY878 | Same as NOY703, but PSW |
| NOY896 | Same as NOY703 (non–PSW), but rap135Δ∷LEU2 |
| NOY897 | Same as NOY878 (PSW), but rpa135Δ∷LEU2 |
| NOY898 | MATα ade2–1 ura3 his3–11 rpb1–1 pNOY103 |
| NOY899 | MATα ade2–1 ura3 his3–11 leu2–3,112 rpb1–1 rrn9Δ∷HIS3 pNOY103; Non–PSW |
| NOY900 | MATα ade2–1 ura3 his3–11 leu2–3,112 rpb1–1 rrn9Δ∷HIS3 pNOY103; PSW |
Table 2.
Yeast plasmids used
| Plasmid | Description |
|---|---|
| pRS316 | Escherichia coli–yeast shuttle vector carrying CEN6 ARSH4 URA3 (Ref. 36) |
| PNOY102 | High–copy-number plasmid carrying GAL7-35S rDNA, URA3, 2μ, amp (Ref. 11) |
| pNOY103 | High-copy-numer plasmid carrying GAL7-35S rDNA, ADE3, URA3, 2μ, amp (Ref. 10) |
| pNOY199 | High-copy-number plasmid carrying GAL7-352S rDNA, TRP1, 2μ, amp |
| pNOY446 | A derivative of CEN URA3 vector pRS316 carrying RRN9 |
| pNOY3237 | pBluescript KS+ containing the SmaI-XbaI fragment of rDNA (−210 to +4,470 with respect to the Pol I start site), constructed by J. Keener |
YEP-glucose (YEPD), YEP-galactose, synthetic galactose (SGal), and synthetic glucose (SD) media were described previously (10, 13). The following supplements were added to the synthetic media as appropriate to satisfy nutritional requirements: casamino acids (5 mg/ml), tryptophan (20 μg/ml), adenine (20 μg/ml), and uracil (20 μg/ml).
Spot Test for PSW and Non-PSW Phenotypes.
Individual colonies (diameter ≈1 mm) formed on galactose media were picked, suspended in 100 μl of H2O, and 5-μl aliquots of 10-fold serial dilutions were spotted on YEP-galactose and YEPD plates. They were usually incubated at 30°C for 7 days.
Analysis of 5′ Ends of Precursor rRNAs by Primer Extension.
RNA was prepared as described (7) from 25 ml of culture (A600 0.4–0.6) growing exponentially in YEPD or from cells after temperature shift-up. The primer (Genosys, The Woodlands, TX) used for the analysis was 5′-ACACGCTGTATAGAGACTAGGC-3′, which hybridizes to 35S precursor rRNA 130 nt downstream of the published Pol I start site (14). Total RNA (1 μg for NOY556, NOY898, NOY505, and NOY794 and 4 μg for NOY878 and NOY900) was subjected to primer extension analysis as described (7).
RESULTS
RNA PSW Variants.
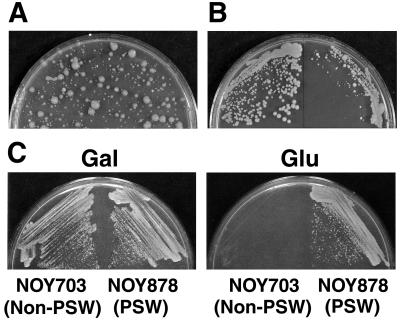
Fig. 1A shows the heterogeneous population of a culture of haploid strain NOY684 carrying the rrn9Δ∷HIS3 (“rrn9Δ”) allele that was freshly constructed from diploid strain NOY678 (see Table 1). On restreaking, larger colonies produced mostly larger (but smaller than the wild-type) colonies, whereas small colonies produced some larger colonies among mostly small colonies (Fig. 1B). Thus, strains carrying rrn9Δ (and rrn5Δ or rrn10Δ; data not shown) were not stable. However, by introducing helper plasmid pNOY102 (or pNOY103 or pNOY199; Table 2) and maintaining the cells on galactose medium, strains carrying these deletion alleles could be maintained much more stably (8). Strain NOY703 is such a rrn9Δ strain and can grow fairly well on YEP-galactose (doubling time 6–7 hr at 30°C) by synthesizing rRNA from the GAL7-35S rDNA fusion gene on helper plasmids. However, these strains grow very poorly on YEPD because of repression of the fusion gene by glucose (Fig. 1C). Variant colonies that grow faster in the absence of intact UAF without helper plasmids (e.g., NOY852 derived from NOY684; Fig. 1B) appear to grow by transcribing native chromosomal rDNA using Pol II rather than Pol I (see below). We call this phenotype PSW (polymerase switch), and the phenotype without such a switch non-PSW. PSW variants also were obtained by plating 104–105 cells of rrn9Δ strains carrying a helper plasmid directly on glucose (NOY878; Fig. 1C).
Figure 1.
RNA polymerase switch variants. (A) An unstable rrn9Δ haploid segregant (NOY684) derived from RRN9/rrn9Δ diploid strain NOY678. A heterogeneous cell population recovered from a tiny rrn9Δ colony formed on a tetrad-dissection plate was suspended in H2O, spread on a YEPD plate, and incubated at 30°C for 7 days. (B) An isolated larger colony (left) and an isolated small colony (right) from the plate shown in A were restreaked on YEPD and incubated at 30°C for 7 days. (C) Non-PSW strain NOY703 and PSW strain NOY878 derived from NOY703 were streaked on YEP-galactose (Gal) and YEPD (Glu) plates and incubated at 30°C for 5 days.
To examine whether growth of a PSW strain is Pol I-independent, we disrupted the gene (RPA135) for the second largest subunit (A135) of Pol I. PSW strain NOY878 and its parent non-PSW strain NOY703 were transformed with DNA carrying the disrupted gene (rpa135Δ∷LEU2; ref. 15) in the presence of galactose. All transformants (e.g., NOY897) derived from NOY878 grew on glucose as well as on galactose, indicating a polymerase switch for rDNA transcription. In contrast, no transformants (e.g., NOY896) derived from control strain NOY703 grew on glucose. Southern analysis of DNA confirmed disruption of RPA135 (data not shown) in both NOY897 and NOY896, demonstrating that the good growth displayed by PSW strain NOY878 is, in fact, achieved by a switch to a new RNA polymerase system.
Identification of 5′ Ends of Primary rRNA Transcripts and Evidence for Transcription by Pol II in PSW Strains.
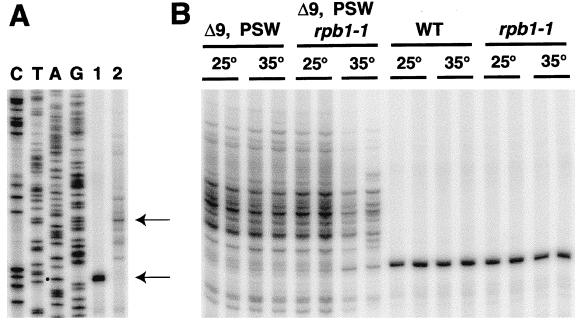
We first examined the 5′ start site of rDNA transcription in PSW strains. RNA was isolated from PSW strain NOY794 (rrn9Δ rpa135Δ) and control strain NOY505, and 5′ ends were analyzed by using primer extension. RNA from the control strain showed the start site (+1) expected from the previously published work (14) (Fig. 2A, lane 1). In contrast, RNA from the PSW strain showed many different 5′ ends that ranged from −9 to −95. A major start site was located at approximately position −29 (Fig. 2A, lane 2). Although we cannot exclude the possibility that these apparent start sites arise from RNA degradation or pausing of reverse transcriptase, it is clear that transcription initiates upstream from the normal Pol I start site. We then used this primer extension analysis to examine the possibility of rDNA transcription by Pol II in PSW strains.
Figure 2.
Identification of 5′ ends of primary rRNA transcripts and evidence for transcription by Pol II in PSW strains. (A) RNA samples from control strain NOY505 (lane 1) and PSW strain NOY794 (lane 2) were analyzed by primer extension using a 32P-labeled primer. The sequence ladder was obtained by dideoxy sequencing using the same primer and pNOY3237 DNA as template. Major start sites are indicated by ←. A dot marks the band in lane A corresponding to the published Pol I start site. (B) RNA samples were prepared from NOY878 (Δ9, PSW), NOY900 (Δ9, PSW, rpb1–1), NOY556 (WT), and NOY898 (rpb1–1) with and without temperature shift from 25° to 35° as indicated. Primer extension was carried out in duplicate as in A. Quantification of the sum of all Pol II-specific bands from −9 to −95 was done for NOY878 and NOY900 and that of the Pol I-specific band at +1 was done for NOY556 and NOY898 by using PhosphorImager analysis. A significant decrease (≈3-fold) of rRNA synthesis after temperature shift-up was observed for NOY900 but not for the other strains (see below). Gels shown are those visualized by a PhosphorImager. Four to seven reverse transcription analyses similar to that shown in the figure were done by using two independent RNA preparations for each strain. The values obtained for the ratio of rRNA synthesis at 35°C to that at 25°C are: NOY900, 0.33 ± 0.07; NOY878, 1.01 ± 0.29; NOY898, 0.80 ± 0.09; and NOY556, 0.83 ± 0.19.
We constructed an rrn9Δ PSW strain carrying a temperature-sensitive mutation (rpb1–1) in the largest subunit of Pol II (12). This strain (NOY900) and three others—an rrn9Δ PSW strain without this mutation (NOY878) and RRN9 strains with (NOY898) and without (NOY556) this mutation—were grown at 25°C, shifted to 35°C, and incubated for 1 hr. RNA then was isolated from these cultures as well as control cultures without temperature shift, and the amounts of unstable precursor rRNA, which is expected to reflect the rRNA synthesis rate, were analyzed by using primer extension. As shown in Fig. 2B, temperature shift-up caused a significant decrease (≈3-fold; see the legend) of rRNA synthesis in the PSW strain with the Pol II temperature-sensitive mutation, but not in the PSW strain without this mutation or in the control strains synthesizing rRNA by Pol I. Because rRNA synthesis by Pol I was not affected by the Pol II temperature-sensitive mutation in the non-PSW strain, the observed decrease in NOY900 cannot be explained by an indirect effect of the Pol II mutation, such as a decrease in ribosomal protein synthesis leading to instability of rRNAs. Thus, the results demonstrate that PSW strains synthesize rRNA by Pol II.
We also constructed a reporter fusion gene consisting of the ADE2 protein-coding region fused to the rDNA promoter, and integrated it into the chromosomal rDNA repeats in the PSW strain (NOY852) and into those in a control strain (NOY505). For the PSW strain, the phenotype of the resultant strain was Ade+, demonstrating the synthesis of functional ADE2 mRNA. In contrast, the control strain remained as Ade−, confirming the expectation that transcription of the ADE2 coding region by Pol I does not yield a functional ADE2 mRNA (data not shown). From all of these experiments, we conclude that PSW strains use Pol II to synthesize rRNA.
Reversibility of Polymerase Switch.
PSW variants arise from rrn9Δ strains more often than the usual frequency of mutation, on the order of approximately 10−4 per division (unpublished data), suggesting an epigenetic event rather than a simple mutation. (See Discussion for the term “epigenetic change” used here.) The following experiments confirm this suggestion.
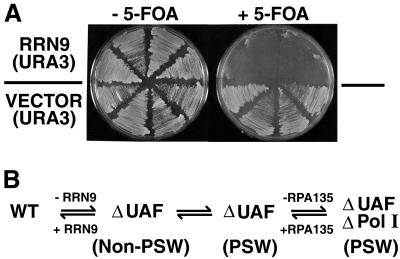
NOY852 is an rrn9Δ strain with the PSW phenotype and grows well without a helper plasmid. We asked whether the PSW phenotype is stably maintained after reintroduction of RRN9 to this strain. A URA3 based CEN plasmid carrying RRN9 (pNOY446) was introduced into this strain by transformation by using URA3 for selection. As a control, the vector plasmid without RRN9 was introduced in parallel into NOY852. Both types of Ura+ transformants were grown on YEPD to facilitate segregation of the introduced plasmid, then streaked on 5-fluoroorotic acid (5-FOA)-containing medium, which does not allow growth of Ura+ cells (16). As shown in Fig. 3A, the control transformants grew well on 5-FOA, whereas cells carrying RRN9 on the plasmid were mostly unable to grow. This shows that most of the cells, which originally employed Pol II-mediated rDNA transcription to grow well in the absence of an intact RRN9 gene, lost this ability (the PSW phenotype) after growth with the reintroduced RRN9.
Figure 3.
(A) Loss of the PSW phenotype on reintroduction of the missing RRN9 into an rrn9Δ PSW strain. Plasmid pNOY446 carrying RRN9 (and URA3) or control URA3 plasmid pRS316 was introduced into NOY852 (rrn9Δ, PSW) by transformation. Four of the transformants isolated as Ura+ from each transformation were streaked on YEPD medium and then streaked on SD with and without 5-FOA (1 mg/ml). The plates were incubated at 30°C for 4 days. (B) A summary of experiments showing reversible switching between the non-PSW and PSW states.
In another experiment, PSW strain NOY794 (rrn9Δ rpa135Δ PSW) carrying pNOY199 was grown on galactose, and the effect of introducing RRN9 (on the URA3 plasmid, pNOY446) to this strain was examined by comparing the transformants carrying RRN9 with two control transformants: one receiving the vector URA3 plasmid and another receiving a URA3 plasmid carrying both RRN9 and RPA135. After growth on YEP-galactose, the three kinds of transformants were examined for their ability to grow on glucose containing 5-FOA. Most transformants carrying both RRN9 and RPA135 failed to grow on glucose containing 5-FOA, i.e., lost the PSW phenotype. However, the transformants carrying RRN9 alone retained the ability to grow on glucose containing 5-FOA as did the control, which received the vector (data not shown). This experiment demonstrates that, in the absence of intact Pol I, UAF neither inhibits Pol II transcription of rDNA in the PSW strain nor stimulates switch back to the non-PSW state.
The conclusion that the switch from the non-PSW to PSW state is reversible also was demonstrated by using rrn9Δ PSW strain NOY878, which was derived from non-PSW strain NOY703. Both strains carry a helper plasmid and can grow on galactose, but the PSW strain forms slightly larger colonies on galactose medium than does the non-PSW strain. This PSW strain, which had been grown on glucose, was streaked on YEP-galactose repeatedly. By testing smaller colonies, we identified some clones that lost the ability to grow on glucose, that is, the PSW phenotype. The results show that the switch from the non-PSW to the PSW state can be reversed without introduction of the RRN9 gene. Fig. 3B summarizes these experimental results, indicating reversibility of switching between the non-PSW and PSW states.
Crosses Between PSW and Non-PSW strains.
We crossed a non-PSW strain (NOY703, carrying URA3 on plasmid pNOY103) with a PSW strain (NOY877, carrying TRP1 on plasmid pNOY199) by plate-mating on galactose medium that allowed the growth of both strains. Diploids were then selected as Ura+Trp+ colonies by streaking the mating mixture on SGal(−U,−T) medium. PSW phenotypes of 12 diploid colonies obtained in this way, as well as meiotic haploid segregants derived from these clones, were examined by spot test on galactose and glucose plates.
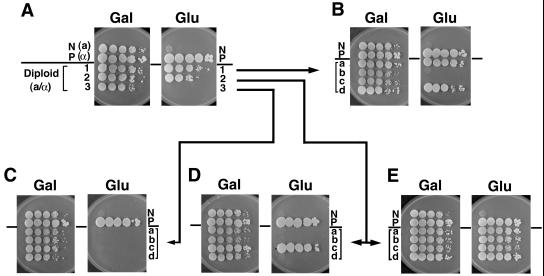
Some diploid clones showed the apparent PSW phenotype (Fig. 4A, diploid clone 1), some showed the non-PSW phenotype (Fig. 4A, diploid clone 3) and others showed a “mixed phenotype” (Fig. 4A, diploid clone 2; this clone was a mixture of ≈10% PSW cells and ≈90% non-PSW cells). By repeating mating in liquid culture and analyzing individual diploid cells formed at 7 hr after the start of mating, we found that the original diploid cells show the PSW phenotype, and that clones with the non-PSW phenotype appear during subsequent growth on galactose with high frequency. On sporulation and tetrad dissection, diploid clones showing the non-PSW phenotype produced tetrads in which all of the haploid segregants were non-PSW (Fig. 4C). However, in the case of diploid clones showing the PSW phenotype or the mixed phenotype, some tetrads produced four PSW haploid segregants without any non-PSW segregants (Fig. 4E), some tetrads produced both PSW and non-PSW haploid segregants (Fig. 4 B and D), and others produced four non-PSW haploid segregants (not shown). It is clear that the PSW phenotype is not a result of simple mutation. First, diploid clones that should have the same genotype showed several different phenotypes: non-PSW, PSW, and mixed phenotypes. Second, some diploid cells gave rise to tetrad segregants, all of which were PSW, whereas some other diploid cells produced tetrad segregants, all of which were non-PSW. Thus, the original PSW phenotype must be produced by a reversible change that appears to be an epigenetic change. In addition, the results indicate that the PSW state can be maintained through meiosis.
Figure 4.
Analysis of diploid clones from a cross between non-PSW and PSW strains. Strain NOY703 (non-PSW; N) and NOY877 (PSW; P) were crossed by plate-mating. Three diploid clones together with haploid parents (N and P) were analyzed for PSW phenotype by spotting aliquots of 10-fold serial dilutions of colonies on YEP-galactose (Gal) and YEPD (Glu), and the results are shown in A. Clones 1, 2, and 3 were sporulated, and tetrads were dissected on YEP-galactose. Colonies from tetrads showing four viable ones were analyzed for their PSW phenotype by spot test. The two original haploid parents (N and P) also were analyzed as controls. One tetrad from clone 1 (B), one tetrad from clone 3 (C), and two tetrads from clone 2 (D and E) are shown.
DISCUSSION
[Rhoo] Yeast System.
Transcription of yeast rDNA by Pol II was previously reported by Conrad-Webb and Butow (17) for respiratory-deficient mitochondrial [rhoo] strains. Our PSW strains can grow well on glycerol and thus are not mitochondrial mutants (data not shown). In addition, their study showed functional transcription of rDNA in the absence of Pol I only for a [rhoo] strain in which an artificial multicopy plasmid carrying an rDNA repeat unit was introduced; the [rhoo] strain carrying an rpa135 deletion and a helper plasmid (pNOY102) was unable to grow on glucose. In contrast, the PSW strains studied here can grow relatively efficiently by transcribing the endogenous rDNA repeats without introduction of any artificial plasmids. Thus, transcription of the endogenous rDNA by Pol II in the [rhoo] strains must have been very inefficient. Furthermore, Pol II transcription of rDNA in [rho°] strains was reported to initiate at the same site (+1) as Pol I, whereas in the PSW strains studied here, rDNA transcription starts at several sites upstream from the Pol I start site. Thus, the relationship between the previously described RNA polymerase switch in [rhoo] strains and the polymerase switch observed in UAF-deficient strains in the current study is not clear.
Comparison with Other Silencing Systems.
In normal yeast cells, Pol II transcription of rDNA is silenced completely. This silencing resembles silencing at the HM loci in that repression of Pol II transcription is complete in wild-type cells, and switching between two alternative metastable states can be revealed only by mutations in components participating in silencing. Thus, as was shown using sir1 mutations (18) or mutations in silencer DNA sequences (19, 20) for silencing at the HM loci, switching between two states of rDNA transcription, non-PSW and PSW, was demonstrated in rrn9 mutant cells (and in other UAF mutants; unpublished observations). It appears that the silenced (non-PSW) state is stabilized by the presence of Pol I-specific transcription factor UAF (Fig. 3B).
Silencing of certain Pol II-transcribed genes inserted into rDNA was recently reported by several workers (21–23). However, the degree of silencing seen in these cases was generally only partial. In addition, attempts to demonstrate switching between on and off transcription states were unsuccessful. It was suggested that Pol II reporter gene expression rapidly alternates between on and off states depending on the Pol I activity of the particular unit where the gene was inserted, leading to the phenotypic equivalent of a stably maintained intermediate expression level (21). In the present system, the non-PSW and PSW states refer to transcription states of the rDNA repeat region as a whole, and reversible switching between the two states was clearly demonstrated. In the above model for silencing of reporter Pol II genes in rDNA, it was suggested that the rDNA chromatin structure responsible for Pol II gene silencing also keeps Pol I activity repressed (21). In contrast, the present work shows that the non-PSW and PSW states are two alternative states; factors that stabilize the non-PSW state (UAF subunits and perhaps other unidentified proteins interacting with UAF), and thus contribute to a complete silencing of Pol II transcription of rDNA, are those required for efficient Pol I transcription. Thus, although silencing of reporter Pol II genes in rDNA might be related to the silencing of Pol II transcription of rDNA described in this paper, the exact relationship between these two phenomena has yet to be elucidated.
In Drosophila some functionally active genes are known to be localized in heterochromatin, including tandemly repeated rDNA transcribed by Pol I (24). It was shown that some heterochromatic genes are functional only in heterochromatin; these heterochromatic genes are silenced when translocated adjacent to euchromatin (1, 25). It was suggested that heterochromatin and euchromatin represent two distinct environments, the heterochromatic environment compatible with the expression of heterochromatic genes and the other compatible with euchromatic gene expression (1, 25). Applying this notion to the present system, one can postulate that the state favoring rDNA transcription by Pol I corresponds to a heterochromatic environment and the state favoring rDNA transcription by Pol II corresponds to a euchromatic environment; the switch between the non-PSW and PSW states may represent a switch between the two different rDNA chromatin states, as discussed below.
Two Possible Alternative States of rDNA.
The absence of functional UAF does not by itself achieve the polymerase switch, but simply makes the non-PSW state metastable, allowing a switch to the PSW state as a random and reversible epigenetic event(s) (Fig. 3B). What then is the nature of the primary events responsible for the polymerase switch? The results of crosses between PSW and non-PSW strains can most simply be explained by a model invoking an alteration of the state of the rDNA template on chromosome XII. Many diploid clones produced tetrads with two distinct haploid segregants, one showing PSW and the other non-PSW (see, e.g., Fig. 4B). These results strongly suggest that the PSW and non-PSW phenotypes are associated with a chromosome, presumably chromosome XII carrying rDNA. The appearance of two distinct types, PSW and non-PSW, of haploid segregants from a single diploid cell may be difficult to explain on the basis of alteration of cytoplasmic materials or nuclear materials such as modification of RNA polymerase(s) or transcription factor(s) or an increase in an extrachromosomal form of rDNA. In fact, we have found that the size of chromosome XII in PSW strains is increased because of a large increase in the number of tandem rDNA repeats as analyzed by pulse field gel electrophoresis (M. Oakes, I. Siddiqi, L. Vu, J. Aris and M. Nomura, unpublished data). [A reversible and heritable change involving a tandem repeat increase can be considered an epigenetic change in a broad sense (see ref. 26).] Thus, the alteration responsible for the PSW phenotype appears to reside in the state of rDNA on chromosome XII, including the length of tandem rDNA repeats and perhaps their localization within the nucleus.
Highly purified UAF contains histones H3 and H4 in addition to Rrn5p, Rrn9p, Rrn10p, and protein p30 (9). UAF binds strongly and specifically to the upstream element of the rDNA promoter carried by protein-free DNA in vitro (8). Thus, actively transcribed rDNA repeats, and perhaps all of the potentially active (but nontranscribed) rDNA repeats, may have UAF bound to the upstream element of the promoter. In fact, the in vivo footprint pattern obtained in a recent analysis of rDNA chromatin in growing yeast cells was consistent with that expected from protection by known specific rDNA-binding proteins, including UAF (27). These considerations support the view that UAF may play a key role in organizing Pol I-specific chromatin structures that are essential for Pol I function and also are responsible for silencing of Pol II transcription. In the absence of the intact UAF structure, the native rDNA chromatin structure may become unstable, allowing rDNA repeat expansion and, with a certain frequency, enabling the rDNA template to be localized at regions suitable for Pol II transcription. We have also observed that deletion of SIR2 in UAF-defective mutants causes an increase, and overproduction of Sir2 protein causes a decrease, in the frequency of a switch from the non-PSW to PSW states (unpublished data). The SIR2 gene is known to play a role in decreasing the rates of mitotic and meiotic recombination within the rDNA locus (28). In addition, an alteration of rDNA chromatin structure in sir2 mutants was recently demonstrated by using micrococcal nuclease and dam methylase (23). Thus, specific rDNA chromatin structures, UAF in particular, appear to play an important role in maintaining a stringent silencing of Pol II transcription of rDNA.
Transcription of rDNA by Pol II in Other Systems.
Transcription from the rDNA promoter mediated by Pol II has been reported in other experimental systems (29–32). These observations may have relevance to the results reported in this paper. The work reported by Smale and Tjian (29) illustrates this point. By using COS monkey cells transfected with a plasmid, they studied expression of the herpes simplex tk gene fused to a human rDNA promoter carried on the plasmid. In the deletion analysis of the upstream region of the promoter, significant transcription by Pol II from two initiation sites (−15 and −20) was observed when deletion extended into the “upstream control element” (33) required for a high level of transcription by Pol I. The upstream control element of mammalian rDNA promoters corresponds to the upstream element of the yeast rDNA promoter where UAF binds to achieve a high level of transcription (8). Although not identified, a factor corresponding to the yeast UAF may exist in mammalian or other metazoan cells, playing a role in organizing a Pol I-specific chromatin structure. Perhaps plasmid DNA carrying the fusion gene without the UCE sequence may fail to form a stable Pol I-specific chromatin structure, allowing transcription of rDNA by Pol II. This scenario is analogous to the RNA polymerase switch observed in UAF-deficient mutants and characterized in this work.
Significance of RNA Polymerase Switch.
We have demonstrated that yeast cells have an inherent ability to synthesize functional rRNA and grow reasonably well without Pol I machinery and in the absence of any artificial helper plasmid. The ease with which the RNA polymerase switch is observed in mutants defective in UAF function suggests a possibility that this second rRNA synthesizing system might serve as a backup system under conditions where mutations or some other environmental conditions impair Pol I machinery, specifically the UAF-mediated Pol I activation system, including the possible rDNA chromatin structure required for transcription by the Pol I machinery. The question of whether this PSW system is in fact used in nature, and if so, under what kind of conditions, remains to be determined.
The presence of the PSW system for rDNA transcription may also be interesting in relation to the evolution of eukaryotic transcription machineries with three nuclear RNA polymerase systems carrying out separate functions. It is known that the archaeal transcriptional machinery consists of a single RNA polymerase, which is similar to eukaryotic RNA polymerases in terms of the number and sequence similarity of the subunits, and two transcription factors, which are homologs of TATA-binding protein and transcription factor IIB, respectively (34, 35). Thus, the archaeal transcription machinery resembles the eukaryotic, rather than the prokaryotic, transcription machinery. This single RNA polymerase system in archaeal organisms transcribes all RNAs, including rRNA, mRNA, and tRNA. The transcription machinery of an ancestor common to both eukaryotic and archaeal organisms might have resembled that of the present-day Archaea, as discussed by Langer et al. (34). It is possible that the emergence of the Pol I system as a specialized transcription machinery devoted to rDNA transcription was not accompanied by an elimination of the previous rDNA transcription system. The latter system, which may have resembled the present-day Pol II system, was perhaps retained but silenced by an epigenetic mechanism, leading to the present-day PSW system. Comparison of rDNA transcription by Pol I with that by the PSW system may be useful in defining unique features of rDNA transcription by Pol I in relation to its regulation, efficiency, and mechanism, as well as rDNA chromatin structures.
Acknowledgments
We thank K. Sutton for technical assistance and Dr. R.A. Young for providing yeast strain Y260. We also thank Drs. S. Arfin, C. L. Greer, and H. Biessmann for critical reading of the manuscript and M. Oakes, J. A. Dodd, and D. Semanko for assistance in its preparation. This work was supported by U.S. Public Health Grant GM-35949.
ABBREVIATIONS
- UAF
upstream activation factor
- Pol I
RNA polymerase I
- Pol II
RNA polymerase II
- PSW
polymerase-switched state
- 5-FOA
5-fluoroorotic acid
References
- 1.Weiler K S, Wakimoto B T. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 2.Elgin S C R. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 3.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 4.Musters W, Knol J, Dekker A F, van Heerikhuizen H, Planta R J. Nucleic Acids Res. 1989;17:9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkens T, Riggs D L, Heck J D, Planta R J, Nomura M. Nucleic Acids Res. 1991;19:5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choe S Y, Schultz M C, Reeder R H. Nucleic Acids Res. 1992;20:279–285. doi: 10.1093/nar/20.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keener J, Josaitis C A, Dodd J A, Nomura M. J Biol Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- 8.Keys D A, Lee B-S, Dodd J A, Nguyen T T, Vu L, Fantino E, Burson L M, Nogi Y, Nomura M. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- 9.Keener J, Dodd J A, Lalo D, Nomura M. Proc Natl Acad Sci USA. 1997;94:13458–13463. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nogi Y, Vu L, Nomura M. Proc Natl Acad Sci USA. 1991;88:7025–7030. doi: 10.1073/pnas.88.16.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nogi Y, Yano R, Nomura M. Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nonet M, Scafe C, Sexton J, Young R. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman F, Fink G R, Hicks J B. Laboratory Course Manual: Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 14.Klootwijk J, de Jonge P, Planta R J. Nucleic Acids Res. 1979;6:27–39. doi: 10.1093/nar/6.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yano R, Nomura M. Mol Cell Biol. 1991;11:754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 17.Conrad-Webb H, Butow R A. Mol Cell Biol. 1995;15:2420–2428. doi: 10.1128/mcb.15.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillus L, Rine J. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 19.Mahoney D J, Marquardt R, Shei G-J, Rose Ab, Broach J R. Genes Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- 20.Sussel L, Vannier D, Shore D. Mol Cell Biol. 1993;13:3919–3928. doi: 10.1128/mcb.13.7.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith J S, Boeke J D. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 22.Bryk M, Banerjee M, Murphy M, Knudsen K E, Garfinkel D J, Curcio M J. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 23.Fritze C E, Verschueren K, Stritch R, Esposito R E. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatti M, Pimpinelli S. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- 25.Wakimoto B T, Hearn M G. Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riggs A D, Mortienssen R A, Russo V E A. In: Epigenetic Mechanisms of Gene Regulation. Russo V E A, Mortienssen R A, Riggs D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 1–4. [Google Scholar]
- 27.Vogelauer M, Cioci F, Camilloni G. J Mol Biol. 1998;275:197–209. doi: 10.1006/jmbi.1997.1451. [DOI] [PubMed] [Google Scholar]
- 28.Gottlieb S, Esposito R E. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 29.Smale S T, Tjian R. Mol Cell Biol. 1985;5:352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopata M A, Cleveland D W, Sollner-Webb B. Proc Natl Acad Sci USA. 1986;83:6677–6681. doi: 10.1073/pnas.83.18.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhar V N, Miller D A, Kulkarni A B, Miller O J. Mol Cell Biol. 1987;7:1289–1292. doi: 10.1128/mcb.7.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doelling J H, Pikaard C S. Nucleic Acids Res. 1996;24:4725–4732. doi: 10.1093/nar/24.23.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haltiner M M, Smale S T, Tjian R. Mol Cell Biol. 1986;6:227–235. doi: 10.1128/mcb.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langer D, Hain J, Thuriaux P, Zillig W. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reeve J N, Sandman K, Daniels C J. Cell. 1997;89:999–1002. doi: 10.1016/s0092-8674(00)80286-x. [DOI] [PubMed] [Google Scholar]
- 36.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]