Abstract
Several classes of the myosin superfamily are distinguished by their “double-headed” structure, where each head is a molecular motor capable of hydrolyzing ATP and interacting with actin to generate force and motion. The functional significance of this dimeric structure, however, has eluded investigators since its discovery in the late 1960s. Using an optical-trap transducer, we have measured the unitary displacement and force produced by double-headed and single-headed smooth- and skeletal-muscle myosins. Single-headed myosin produces approximately half the displacement and force (≈6 nm; 0.7 pN) of double-headed myosin (≈10 nm; 1.4 pN) during a unitary interaction with actin. These data suggest that muscle myosins require both heads to generate maximal force and motion.
Muscle shortening is driven by a cyclical interaction between the contractile proteins myosin and actin. During this cycle the dimeric molecular motor, myosin, transduces chemical energy into mechanical work. Although the contractile system has been investigated extensively, the details of this process and the functional significance of myosin’s “double-headed” structure have remained unclear since the dimeric structure was discovered in the late 1960s (ref. 1; see refs. 2 and 3 for review). Based on the available biochemical and mechanical data (4–8), early models assumed that the two heads of myosin act independently (9, 10). Enzymatic assays clearly showed that the actin-activated ATPase activity per head was the same for proteolytically prepared myosin-head fragments (subfragment-1, S1) and for single- and double-headed heavy meromyosin (HMM; ref. 4). An even more compelling example of independent head action was observed when reconstituted actomyosin threads were used. In these experiments, ensembles of single-headed myosin generated half the force of native myosin (5). More recently, by using an optical trap, single S1 heads were shown to move actin and produce force, suggesting that a dimeric structure is not necessary for the production of work in vitro (11). Additional single-molecule experiments have shown that in synthetic thick filaments, single- and double-headed myosins produce similar unitary step displacements (12).
However, evidence suggesting a functional difference between single- and double-headed myosin species came from measurements of the binding of S1 and HMM to actin in the absence of nucleotide (6, 7). These data showed a smaller association constant for HMM than would be expected if the heads bound independently, implying that the two heads are sterically constrained from simultaneously interacting with actin. More recent kinetic studies comparing S1 and HMM suggest that the two heads may be coordinated in their transition from the weakly bound to the strongly bound state (13).
Because the functional significance of myosin’s two-headed structure remains ambiguous, we have performed a single-molecule mechanical comparison between single- and double-headed smooth- and skeletal-muscle myosins. Using an optical-trap assay capable of measuring the unitary forces (F) and displacements (d) produced by single myosin molecules (14–17), we eliminated potential uncertainties associated with ensemble measurements (i.e., assays requiring large populations of motor molecules). Here, we present direct mechanical evidence suggesting that the two heads of muscle myosin are required to produce maximal force and motion.
METHODS
Protein Preparation.
Single-headed myosin was prepared from chicken pectoralis myosin by hydrophobic interaction chromatography essentially as described by Kalabokis et al. (18). Myosin was digested briefly with papain in 0.2 M ammonium acetate (4, 19). After removal of the soluble S1, undigested myosin, single-headed myosin, and rod were resuspended in 1.4 M ammonium sulfate/20 mM imidazole, pH 6.8/2 mM MgCl2/0.2 mM EDTA/3 mM NaN3/0.5 mM DTT/0.5 mM ATP and applied to a 1 × 30-cm column of Toyopearl ether 650 S (TosoHaas, Montgomeryville, PA) equilibrated in the same buffer. Bound proteins were eluted with a 400- ml gradient of ammonium sulfate from 1.4 M to 1.2 M. The rod fraction does not bind to the column under these conditions, and single-headed myosin elutes first, followed by myosin. Fractions were analyzed by nondenaturing PAGE, and pools containing pure single-headed myosin and double-headed myosin were concentrated by dialysis overnight against 4–5 volumes of saturated ammonium sulfate (see Fig. 1). Single-headed smooth-muscle myosin was prepared from turkey gizzard myosin by a similar procedure, except that it was applied to the column in 1.5 M ammonium sulfate.
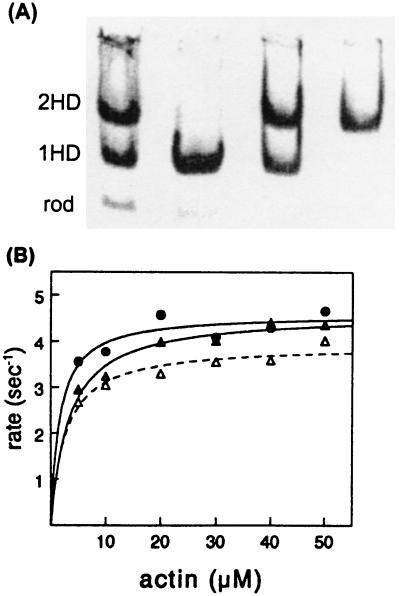
Figure 1.
Characterization of single-headed and double-headed chicken pectoralis-muscle myosin purified by hydrophobic interaction chromatography. (A) Native gel electrophoresis of fractions pooled from a Toyopearl column. Lanes from left to right: applied papain digest, single-headed myosin (1HD) pooled from first peak, mixture of myosin species pooled from fractions between peaks, and double-headed myosin (2HD) pooled from second peak. Most of the rod does not bind to the column. (B) Actin-activated ATPase activity (maximum velocity per head) of 1HD (filled triangles) = 4.6 s−1; of 2HD (open triangles) = 3.9 s−1; and of native myosin (circles) = 4.6 s−1. The Km values were less than 5 μM for all three species.
An S1–biotin construct was prepared by cloning an 87 amino acid sequence segment from the Escherichia coli biotin carboxyl carrier protein (20) after the N-terminal 855 amino acids of the smooth-muscle myosin heavy chain. This construct is biotinylated at a Lys residue located 35 amino acids from the C terminus of the fusion protein (20) during expression in Sf9 cells.
The actin-activated ATPase activities of all myosin species were determined at 25°C in 50 mM KCl/4 mM MgCl2/2.0 mM ATP/10 mM imidazole/1 mM DTT, pH 7.5 (21).
Optical-Trap Experiments.
Details of the optical-trap instrumentation and experimental procedures for the three-bead assay have been published elsewhere (14–17). In brief, flow-cells were loaded with 1 μg/ml single- or double-headed myosin (the S1–biotin construct was loaded into flow-cells after creation of a binding layer with the following sequence of treatments; 0.1 mg/ml biotin-labeled goat anti-mouse IgG, 0.5 mg/ml BSA wash, 0.1 mg/ml streptavidin, and a second 0.5 mg/ml BSA wash). After a 2-min incubation period to allow for myosin to bind to the nitrocellulose-coated surface, BSA (0.5 mg/ml) was added to block the coverslip surface. The chamber was then washed with motility buffer (25 mM imidazole⋅HCl, 4 mM MgCl2, 10 mM DTT, 1 mM EGTA, 25 mM KCl, oxygen scavenger, and 1 μM or 10 μM Na2ATP at pH 7.4) to remove any unbound protein. The final addition to the flow-cell included a dilute concentration of tetramethylrhodamine isothiocyanate phalloidin-labeled F-actin and N-ethylmaleimide (NEM)-myosin-coated polystyrene beads (NH2 beads; 1-μm diameter; Polysciences) in motility buffer. Experiments were initiated by capturing an NEM-myosin-coated bead in each optical trap and securing an actin filament (5–10 μm) to both beads. After pretensioning (≈4 pN) the bead–actin–bead system (16), the filament was brought into proximity of the myosin-coated surface where unitary events were recorded.
We typically measured d at a low single-trap stiffness of ≈0.03 pN/nm (7–8 nm rms thermal motion for the entire pretensioned bead–actin–bead assembly), whereas F was measured under feedback control with an apparent single-trap stiffness of ≈2 pN/nm. The feedback loop included an acoustooptic modulator (AOM), which was used to deflect the laser beam rapidly, countering any force imposed by myosin. The correction signal used to steer the AOM was calibrated to give a measure of force. Filtered displacement traces (low-pass filter cutoff at 2 kHz) and outputs from the force-feedback device were recorded and later digitized at 4 kHz.
Caution is warranted when interpreting d and F measured in this assay. Ideally, the only relevant compliances should exist in the optical trap and in the motor protein itself. However, compliance within the mode of actin-filament attachment to the beads exists and will lead to an underestimate of myosin’s force and motion-generating potential (16, 22, 23). In fact, based on an analysis of our system, 4 pN of pretension on the bead–actin–bead system may lead to as much as a 13% underestimate in our reported displacement amplitudes (see figure 7A of ref. 16). If we also consider the low bandwidth of the feedback device (i.e., ≈140 Hz), then force events in some cases may not represent the power stroke but rather the load-bearing capacity of the molecule as the feedback system pulls back on myosin after the power stroke. Assuming that myosin’s stiffness is the same before and after the power stroke (i.e., no hysteresis in the force–displacement curve), this measurement is still a meaningful parameter that can be used to compare the mechanical characteristics of single- and double-headed myosins.
Mean-Variance (MV) Analysis.
Estimates of d, F, and both displacement and force event durations (ton) were obtained from single-molecule measurements by MV analysis (15), a statistical technique originally developed for ion-channel studies (24). MV analysis involves a simple transform of the raw time-series data into a three-dimensional “histogram,” which we then fit to obtain estimates of unitary parameters. An MV histogram is created by sliding a time window of variable width (20 ms for the data presented here) over the raw data and plotting the mean and variance calculated at each point. The histogram’s third dimension is the number of points (i.e., counts) with a given mean and variance. There are two types of populations that normally exist in an MV histogram (see Fig. 2b): one corresponding to the high-variance baseline data when myosin is not attached to actin and another to the lower-variance “event” data when myosin is bound to actin. This variance reduction is caused by an increase in the effective system stiffness as myosin attaches to actin and allows the event and baseline populations to be separated in two dimensions with higher resolution.
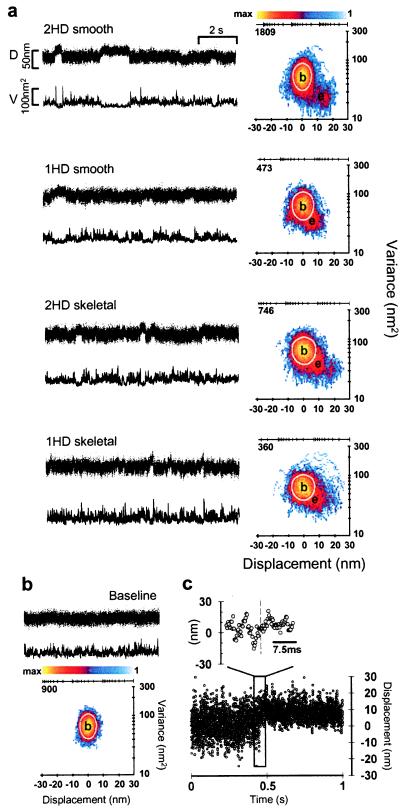
Figure 2.
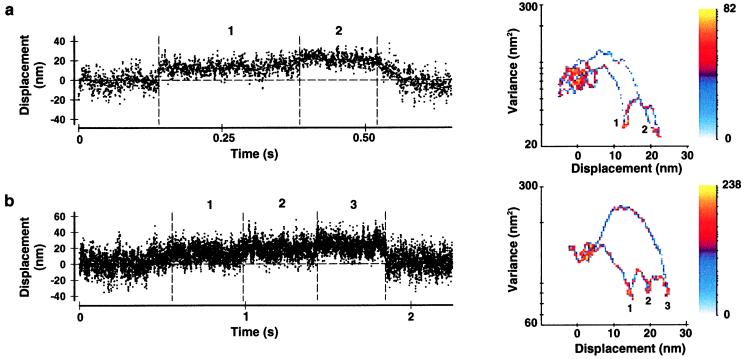
Representative displacement data for single-headed (1HD) and double-headed (2HD) smooth- and skeletal-muscle myosins. (a) Raw time series (Left) and MV histograms (Right) representing 2HD and 1HD smooth- and skeletal-muscle myosin records. Beneath each position record (D) is the running variance (V) calculated for that segment of data by using a 20-ms window. MV histograms in a were calculated with a 20-ms window by transforming the entire record (≈30–60 s in length) from which representative traces were taken (see Methods). Note that the event populations (e) shown in 1HD histograms are roughly half the distance from baseline (b) when compared with 2HD MV transform. (b) Representative baseline record with accompanying MV histogram. Note the lack of an event population in the baseline transform. The color bars in a and b represent the palette used to color code the number of counts (i.e., points with a given mean and variance) in a given bin. In this representation, bins with zero counts are colored white (background). The axes above each histogram in a and b represent the log-scaled distribution of the color bar and are accompanied by the maximum number of counts in each case. The white boundary on each histogram represents the 95% confidence limit of the fit to the baseline population. (c) The expanded view shows the rapid rise of a displacement step (complete in ≤2 ms).
Estimating d and F requires knowledge of the position of each population relative to all others. Therefore, an iterative curve-fitting routine is employed to fit each population with a Gaussian function in the mean dimension and a χ2 function along the variance axis. At limiting myosin surface density, the predominant event population has a positive mean. Although negative event populations can be detected at times, these populations have a small fraction of the volume of the positive populations (15). In this study and previous ones, we have reported only the predominant positive event-population statistics.
To estimate ton, the total counts (i.e., volume) of the event population are recorded at different window widths (15, 24). Volume at a given window width is representative of the number of events with a duration equal to or greater than that width. Accordingly, it is possible to estimate the average event duration by fitting volume vs. window width with the expression V = k·ton·e−W/ton. Here, V is the volume of the population, k is the number of events, ton is average event duration, and W is window width. Raw data analyzed in this study were collected by using myosin from multiple preparations.
RESULTS AND DISCUSSION
Purification and Characterization.
To determine whether myosin’s dimeric structure confers a functional advantage to the molecule, we measured the enzymatic and mechanical characteristics of single- and double-headed smooth- and skeletal-muscle myosins. Single-headed skeletal-muscle myosin was prepared by limited papain digestion of chicken skeletal-muscle myosin (19) and separated from the remaining undigested myosin and rod by hydrophobic interaction chromatography (Fig. 1; see Methods). This chromatographic procedure is far superior in resolving power compared with earlier ion-exchange methods (19). The column does not retain the rod, whereas the single-headed myosin elutes first in a broad peak, followed by the double-headed myosin. Fractions containing pure single-headed myosin or pure double-headed myosin were pooled from the front of the first peak or the back of the second peak, respectively. Native gel patterns of the concentrated pools are shown in Fig. 1. After SDS/PAGE, only two high molecular mass bands are seen for single-headed skeletal myosin; these correspond to the intact myosin heavy chain and the rod heavy chain in equimolar amounts. The light-chain pattern is identical for single- and double-headed skeletal-muscle myosin with little proteolytic cleavage (data not shown). The actin-activated ATPase activities (on a per-head basis) and Km values measured for single-headed and double-headed myosins chromatographed from the same column were very similar (Fig. 1, see legend). These protein preparations were used for the mechanical measurements described below.
Single-headed smooth-muscle myosin also was prepared by limited papain digestion and chromatographed by hydrophobic interaction chromatography, but for this myosin, papain cleaves the N terminus of the regulatory light chain in addition to the heavy chain. This cleavage does not affect the enzymatic activity of the single-headed smooth-muscle myosin, whose activity is independent of phosphorylation (25), but proteolysis of the regulatory light chain prevents phosphorylation of the double-headed myosin isolated from the same column, which remains inactive. Therefore, it was necessary to use native, phosphorylated smooth-muscle myosin as the double-headed species in the mechanical experiments. Both the skeletal- and smooth-muscle single-headed myosins moved actin filaments at rates comparable to those of the native myosins in an in vitro motility assay (data not shown; refs. 8 and 25).
Mechanical Comparison.
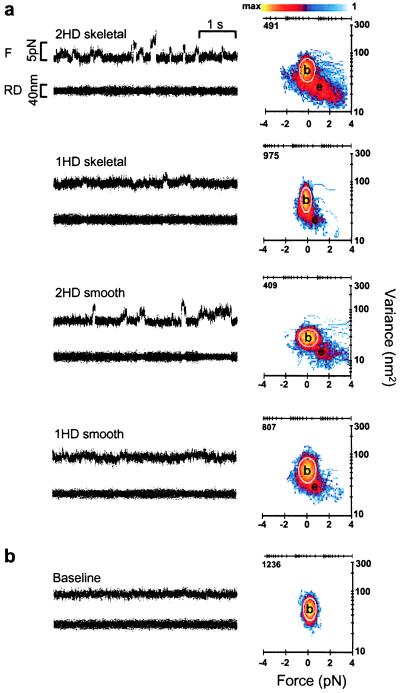
Using the optical-trap assay, we performed a direct mechanical comparison between single- and double-headed myosins at the single-molecule level. Representative d (Fig. 2a) and F (Fig. 3a) records displaying unitary events are shown for single- and double-headed smooth- and skeletal-muscle myosins.
Figure 3.
Representative force data for single-headed (1HD) and double-headed (2HD) skeletal- and smooth-muscle myosins. (a) Raw time series (Left) and MV histograms (Right) representing 2HD and 1HD skeletal- and smooth-muscle myosin records. Beneath each F record is the residual displacement (RD) for that segment of data. MV histograms in a were calculated with a 20-ms window by transforming the entire record (≈30–60 s in length) from which representative traces were taken (see Methods). Note that the event populations (e) shown in 1HD force histograms are roughly half the distance from baseline (b) when compared with 2HD MV transform. (b) Representative baseline record with accompanying MV histogram. Note the lack of an event population in the baseline transform. The color bar in a represents the palette used to color code the number of counts (i.e., points with a given mean and variance) in a given bin. In this representation, bins with zero counts are colored white (background). The axes above each histogram in a and b represent the log-scaled distribution of the color bar and are accompanied by the maximum number of counts in each case. The white boundary on each histogram represents the 95% confidence limit of the fit to the baseline population.
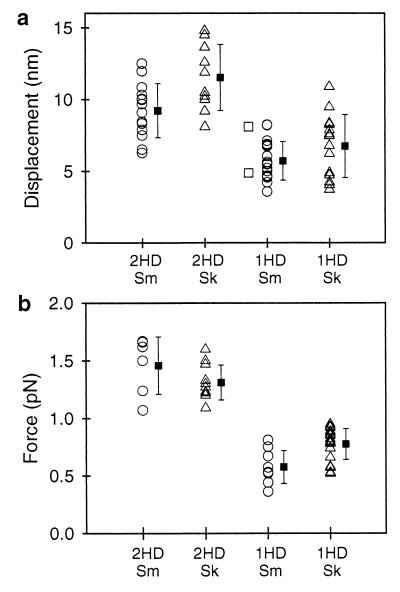
Event amplitudes (d and F) and their ton were determined for multiple records by using MV analysis (refs. 15 and 24; Figs. 2a and 3a). Distributions of d and F produced by single- and double-headed myosins in multiple experiments estimated by MV analysis are plotted in Fig. 4 a and b. The standard deviations associated with each distribution most likely reflect the effects of biological variability, the random orientation of protein bound to the surface, and the slight possibility that more than one myosin molecule contributes to the response. These data indicate that, on average, single-headed smooth- and skeletal-muscle myosins produce half the displacement (62% and 58%, respectively) and half the force (40% and 62%, respectively) compared with double-headed myosins (Fig. 4; Table 1). The displacement and force ton (see Table 1) were the same for single- and double-headed myosins.
Figure 4.
Scatter plots representing the distributions of d (a) and F (b) produced by single-headed (1HD) and double-headed (2HD) smooth-muscle (Sm, open circles) and skeletal-muscle (Sk, open triangles) myosins. The means and standard deviations are represented for each distribution. Displacements produced by a biotin-labeled smooth-muscle S1 construct are shown adjacent to the 1HD smooth data (open squares). Each entry represents the fit from a single MV histogram, which may be comprised of 50–100 unitary events depending on the record duration and event density (see Methods).
Table 1.
Summary of unitary mechanical parameters
| Myosin type | d, nm | ton†, ms | F, pN | ton‡, ms |
|---|---|---|---|---|
| Smooth 2HD | 9.2 ± 0.5 (14) | 167 ± 23 (14) | 1.5 ± 0.1 (6) | 284 ± 54 (5) |
| Smooth 1HD | 5.7 ± 0.3* (18) | 150 ± 19 (14) | 0.6 ± 0.1* (9) | 263 ± 49 (8) |
| Skeletal 2HD | 11.6 ± 0.7 (10) | 48 ± 2 (2) | 1.3 ± 0.1 (11) | 166 ± 25 (7) |
| Skeletal 1HD | 6.8 ± 0.6* (14) | 44 ± 7 (11) | 0.8 ± 0.1* (20) | 149 ± 52 (6) |
Data are presented as the mean ± SEM. The quantities in parentheses represent the number of MV histograms contributing to the mean. 1HD, single-headed; 2HD, double-headed.
P < 0.05 vs. 2HD.
Unloaded ton for displacement data collected at 10 μM ATP.
Loaded ton for force data collected at 10 μM for smooth-muscle myosin and 1 μM ATP for skeletal-muscle myosin.
These data suggest that the inherent displacement generated per myosin head is 5–6 nm (≈6–7 nm after compliance correction; see Methods). In support of this conclusion, an expressed biotiylated smooth-muscle S1 also generated displacements comparable to those for single-headed smooth-muscle myosin (Fig. 4a, open squares). In contrast, double-headed smooth- and skeletal-muscle myosins generated ≈10 nm displacements (≈13 nm after compliance correction), as we had reported (15). Other evidence confirming these results can be found in experiments at higher myosin surface densities (≈2–5 μg/ml), where “staircase” events (i.e., a rapid sequence of multiple steps) were produced by multiple molecules (Fig. 5). In these records, initial attachments were followed by steps that were ≈10 nm apart for double-headed (Fig. 5a) and ≈5 nm apart for single-headed (Fig. 5b) skeletal-muscle myosin. These multiple steps are detected easily both in the time series and the MV transform.
Figure 5.
Displacement time series from experiments in which the myosin surface density was sufficiently high to allow the production of multiple events in rapid succession (i.e., staircases) from more than one molecule. (a) For double-headed (2HD) skeletal-muscle myosin, the distance between steps is ≈10 nm, as indicated by the MV transform displayed (Right). (b) The steps are separated by ≈5 nm for single-headed (1HD) skeletal-muscle myosin. The 2HD and 1HD MV histograms were calculated with an 80-ms and a 320-ms window, respectively. The maximum number of counts is indicated on each histogram. Vertical dashed lines indicate the probable starting and ending points for each step of the staircase.
The single-headed myosin displacements of 5–6 nm reported here are slightly greater than the displacement reported for skeletal S1 (≈3.5 nm; ref. 11) and significantly lower than that recently reported for single-headed myosin (10–15 nm; refs. 12 and 26). The ≈10-nm steps for double-headed myosin, however, are twice the size of those measured for skeletal-muscle HMM (≈4–5 nm; refs. 11, 22, and 23). The lower absolute values for HMM compared with our double-headed myosin data may relate to the method of data analysis. The skeletal HMM estimates were determined by using a running position variance or a positional correlation coefficient between trapped beads to indicate actomyosin interactions (22). For comparison, we used a running variance technique to score events (see Fig. 2, V traces) and to generate composite displacement histograms from single- and double-headed smooth- and skeletal-muscle myosins. In all cases, lower absolute displacement values were obtained when compared with the MV analysis (≈50% lower; data not shown). However, double-headed myosin displacements were still greater than those of single-headed myosins. A detailed consideration of the differences in the method of analysis is beyond the scope of this paper. Strikingly, regardless of the analysis technique, single-headed myosins produced smaller displacements than double-headed myosins.
Potential Artifacts.
Can a double-headed myosin’s ability to produce twice the force and motion of single-headed species be considered an artifact of the single-molecule assay? These experiments were performed at low ATP (1–10 μM) to extend myosin’s attached lifetime and facilitate data analysis. As such, the larger steps observed for double-headed myosins may be a consequence of the low-ATP, rigor-like condition (27). If the first head remains attached in rigor after its power stroke, a discernable plateau in the position data should be evident before attachment of the second head. Within the 2-ms temporal resolution of our detection system (15), there was no evidence for two independent steps (see Fig. 2c). Furthermore, if the first head remained bound in rigor, it would be expected to create a drag force that might hinder the progress of the second head. Because double-headed myosins produce twice the displacement of single-headed myosins (within experimental error), it is not likely that this drag force existed. Even more compelling are recent experiments at saturating ATP conditions (1 mM), where the displacements produced by double-headed smooth-muscle HMM are ≈10 nm (17).
The difference in single- and double-headed myosin displacements also could be attributed to a force-dependent (i.e., nonlinear) compliant element external to the myosin molecule. If this element existed, then the change in position variance on myosin attachment would differ for single- and double-headed myosin; this behavior was not evident in our data (data not shown). The larger steps produced by double-headed myosins, therefore, are not likely an artifact of limiting ATP or stray compliance and must be a functional property of the molecule.
Possible Mechanisms.
We propose two possible mechanisms for the production of larger steps by double-headed myosin. These mechanisms share the need for two heads to generate maximal force and motion but differ in how the two heads are related functionally. In the first case, the two heads attach to actin and perform work in a coordinated manner, where each head contributes half of the total force and displacement. In the second case, the first head merely guides the second head to its optimal interaction so that effectively only one head performs maximal work.
For the coordinated case, the two heads must perform their power strokes in rapid succession, i.e., detachment of the first head is coincident with or immediately followed by the attachment of the second head. Detachment of the first head could be enhanced by intramolecular strain imposed by the second head’s attachment or by translocation of the filament itself. Similar mechanisms have been proposed for kinesin, another dimeric molecular motor (28–30). Indeed, Huxley (31) had proposed originally that muscle shortening accelerated the detachment of crossbridges that were negatively strained. In contrast, under loaded conditions, both myosin heads could remain attached to actin and contribute equally to force, which requires sufficient flexibility in the head–rod junction to allow both heads to attach simultaneously to adjacent actin monomers. Unraveling of the coiled-coil at the head–rod junctions has been shown in both smooth-muscle HMM (21, 32) and striated-muscle myosin (33). Moreover, electron micrographs of F-actin decorated with HMM show that both heads can attach simultaneously to actin in rigor (34).
An alternate explanation is that the first head serves to tether and guide the second head to its optimal interaction with actin. As was recently shown (12), the displacement generated by a single myosin molecule depends on the relative orientation between the myosin head and actin. Thus, the first head may serve to orient the second head, allowing it to generate its maximal displacement (≈10 nm) and force. This model differs from the previous one in that only one head contributes to the molecule’s work production.
Given the current data, it may be possible to distinguish between these two models. The equivalent force and displacement ton values (see Table 1) measured for single- and double-headed species imply that the first head’s contribution to the event duration might be negligible. For example, under loaded conditions where the simultaneous attachment of two heads would be expected to prolong the event duration, these times were similar for both single- and double-headed myosins. Therefore, it is possible that only one head actually performs the work, while the other serves to guide the operative head to its maximally effective orientation. It is important to realize that this conclusion relies heavily on the equivalent ton values observed under loaded conditions. Without any rigorous characterization of the kinetic step or steps that constitute this duration, it may be premature to assume a specific mode by which the two heads interact to generate force and motion.
Conclusions.
These single-molecule studies have uncovered a functional role for muscle myosin’s dimeric structure, whereby two heads are better than one in producing force and motion. The recent finding that expression of single-headed myosin II in Dictyostelium discoideum results in a loss of cytokinesis and cell surface receptor capping further emphasizes the functional significance of a double-headed structure in vivo (35). It should be noted that other dimeric molecular motors exhibit mechanical coordination between heads, perhaps to enhance the efficiency of directed motion. For example, kinesin, which is a processive, microtubule-based cargo carrier (36), has negative interhead cooperativity so that only one head is bound strongly to the microtubule at a time. Because kinesin’s duty cycle (i.e., the fraction of cycle time spent in the attached state) is very high (>90%), negative cooperativity is required to ensure continuous forward motion. In contrast, muscle myosins operate with a much lower duty cycle (<5% under unloaded conditions; ref. 37), an essential feature for high speeds of muscle shortening. Thus, the contractile system in muscle has evolved in which the two heads of myosin interact in a manner that enhances muscle performance by maximizing the displacement and force developed per unit of time per myosin molecule.
Acknowledgments
The authors would like to thank Eric Hayes and Janet Vose for their technical assistance, Don Gaffney for excellent custom software development, and the University of Vermont Muscle Club for numerous helpful discussions. This work was supported by National Institutes of Health Grants HL54568 to K.T. and D.W. and AR17350 to S.L.
ABBREVIATIONS
- HMM
heavy meromyosin
- S1
myosin subfragment 1
- d
unitary displacement
- F
unitary force
- ton
average event duration
- MV
mean-variance
References
- 1.Slayter H S, Lowey S. Proc Natl Acad Sci USA. 1967;58:1611–1618. doi: 10.1073/pnas.58.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooke R. Physiol Rev. 1997;77:671–697. doi: 10.1152/physrev.1997.77.3.671. [DOI] [PubMed] [Google Scholar]
- 3.Cope M J T, Whisstock J, Rayment I, Kendrick-Jones J. Structure. 1996;4:969–987. doi: 10.1016/s0969-2126(96)00103-7. [DOI] [PubMed] [Google Scholar]
- 4.Margossian S S, Lowey S. J Mol Biol. 1973;74:313–330. doi: 10.1016/0022-2836(73)90376-8. [DOI] [PubMed] [Google Scholar]
- 5.Cooke R, Franks K E. J Mol Biol. 1978;120:361–373. doi: 10.1016/0022-2836(78)90424-2. [DOI] [PubMed] [Google Scholar]
- 6.Highsmith S. Biochemistry. 1978;17:22–26. doi: 10.1021/bi00594a004. [DOI] [PubMed] [Google Scholar]
- 7.Margossian S S, Lowey S. Biochemistry. 1978;17:5431–5439. doi: 10.1021/bi00618a017. [DOI] [PubMed] [Google Scholar]
- 8.Harada Y, Noguchi A, Kishino A, Yanagida T. Nature (London) 1987;326:805–808. doi: 10.1038/326805a0. [DOI] [PubMed] [Google Scholar]
- 9.Huxley A F, Simmons R M. Nature (London) 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 10.Eisenberg E, Hill T L, Chen Y. Biophys J. 1980;29:195–227. doi: 10.1016/S0006-3495(80)85126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molloy J E, Burns J E, Kendrick-Jones J, Tregear R T, White D C. Nature (London) 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka H, Ishijima A, Honda M, Saito K, Yanagida T. Biophys J. 1998;75:1886–1894. doi: 10.1016/S0006-3495(98)77629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conibear P B, Geeves M A. Biophys J. 1998;75:926–937. doi: 10.1016/S0006-3495(98)77581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finer J T, Simmons R M, Spudich J A. Nature (London) 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 15.Guilford W H, Dupuis D E, Kennedy G, Wu J, Patlak J B, Warshaw D M. Biophys J. 1997;72:1006–1021. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupuis D E, Guilford W H, Wu J, Warshaw D M. J Muscle Res Cell Motil. 1997;18:17–30. doi: 10.1023/a:1018672631256. [DOI] [PubMed] [Google Scholar]
- 17.Lauzon A-M, Tyska M J, Rovner A S, Freyson Y, Warshaw D M, Trybus K M. J Muscle Res Cell Motil. 1998;19:825–837. doi: 10.1023/a:1005489501357. [DOI] [PubMed] [Google Scholar]
- 18.Kalabokis V N, Vibert P, York M L, Szent-Gyorgyi A G. J Biol Chem. 1996;271:26779–26782. doi: 10.1074/jbc.271.43.26779. [DOI] [PubMed] [Google Scholar]
- 19.Margossian S S, Lowey S. J Mol Biol. 1973;74:301–311. doi: 10.1016/0022-2836(73)90375-6. [DOI] [PubMed] [Google Scholar]
- 20.Cronan J E J. J Biol Chem. 1990;265:10327–10333. [PubMed] [Google Scholar]
- 21.Trybus K M, Freyzon Y, Faust L Z, Sweeney H L. Proc Natl Acad Sci USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta A D, Finer J T, Spudich J A. Proc Natl Acad Sci USA. 1997;94:7927–7931. doi: 10.1073/pnas.94.15.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veigel C, Bartoo M L, White D C S, Sparrow J C, Molloy J E. Biophys J. 1998;75:1424–1438. doi: 10.1016/S0006-3495(98)74061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patlak J B. Biophys J. 1993;65:29–42. doi: 10.1016/S0006-3495(93)81041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cremo C R, Sellers J R, Facemyer K C. J Biol Chem. 1995;270:2171–2175. doi: 10.1074/jbc.270.5.2171. [DOI] [PubMed] [Google Scholar]
- 26.Ishijima A, Kojima H, Funatsu T, Tokunaga M, Higuchi H, Tanaka H, Yanagida T. Cell. 1998;92:161–171. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 27.Hackney D D, Clark P K. Proc Natl Acad Sci USA. 1984;81:5345–5349. doi: 10.1073/pnas.81.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Block S M. J Cell Biol. 1998;140:1281–1284. doi: 10.1083/jcb.140.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hancock W O, Howard J. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young E C, Mahtani H K, Gelles J. Biochemistry. 1998;37:3467–3479. doi: 10.1021/bi972172n. [DOI] [PubMed] [Google Scholar]
- 31.Huxley A F. Prog Biophys. 1957;7:255–318. [PubMed] [Google Scholar]
- 32.Trybus K M. J Biol Chem. 1994;269:20819–20822. [PubMed] [Google Scholar]
- 33.Knight P J. J Mol Biol. 1996;255:269–274. doi: 10.1006/jmbi.1996.0022. [DOI] [PubMed] [Google Scholar]
- 34.Craig R, Szent-Gyorgyi A G, Beese L, Flicker P, Vibert P, Cohen C. J Mol Biol. 1980;140:35–55. doi: 10.1016/0022-2836(80)90355-1. [DOI] [PubMed] [Google Scholar]
- 35.Burns C G, Larochelle D A, Erickson H, Reedy M, De Lozanne A. Proc Natl Acad Sci USA. 1995;92:8244–8248. doi: 10.1073/pnas.92.18.8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard J. Annu Rev Physiol. 1996;58:703–729. doi: 10.1146/annurev.ph.58.030196.003415. [DOI] [PubMed] [Google Scholar]
- 37.Harris D E, Warshaw D M. J Biol Chem. 1993;268:14764–14768. [PubMed] [Google Scholar]