Abstract
MACiE (Mechanism, Annotation and Classification in Enzymes) is a database of enzyme reaction mechanisms, and is publicly available as a web-based data resource. This paper presents the first release of a web-based search tool to explore enzyme reaction mechanisms in MACiE. We also present Version 2 of MACiE, which doubles the dataset available (from Version 1). MACiE can be accessed from http://www.ebi.ac.uk/thornton-srv/databases/MACiE/
INTRODUCTION
Enzymes are proteins that catalyse the repertoire of chemical reactions found in nature, and as such are vitally important molecules. What is so fascinating about these proteins is that they have a wonderful diversity and can carry out highly complex chemical conversions under physiological conditions and retain their stereospecificity and regiospecificity, unlike many organic chemical reactions. They range in size and can have molecular weights of several thousand to several million Daltons, and still they can catalyse reactions on molecules as small as carbon dioxide or nitrogen, or as large as a complete chromosome.
Although enzymes are large molecules, the actual catalysis only takes place in a small cavity, the active site. It is here that a small number of amino acid residues contribute to catalytic function, and where the substrates bind. With the advent of structure determination methods for proteins and by using clever chemical/biochemical experimental design, scientists have been able to propose catalytic mechanisms for many enzymes. Although a great deal of knowledge exists for enzymes, including their structures, gene sequences, mechanisms, metabolic pathways and kinetic data, it tends to be spread between many different databases and throughout the literature. Most web resources relating to enzymes [such as BRENDA (1), KEGG (2), the IUBMB Enzyme Nomenclature website (http://www.chem.qmul.ac.uk/iubmb/enzyme/) (3) and IntEnz (4)] focus on the overall reaction, accompanied in some cases by a textual or graphical description of the mechanism. However, this does not allow for detailed in silico searching of the chemical steps which take place in the reaction. MACiE (5) combines detailed stepwise mechanistic information [including 2-D animations (6)], a wide coverage of both chemical space and the protein structure universe, and the chemical intelligence of the Chemical Markup Language for Reactions (CMLReact) (7). This usefully complements both the mechanistic detail of the Structure–Function Linkage Database (SFLD) for a small number of rather ‘promiscuous’ enzyme superfamilies (8) and the wider coverage with less chemical detail provided by EzCatDB (9), which also contains a limited number of 3D animations. Entries in MACiE are linked, where appropriate, to all of these related data resources.
DATASET AND CONTENT
The dataset for MACiE version 2 was devised to increase the enzyme reaction space coverage of MACiE while trying to keep structural homology to a minimum. Each entry added in the new version was selected so that it fulfils the following criteria:
The EC sub-subclass was not previously in MACiE.
There is a three-dimensional crystal structure of the enzyme deposited in the Protein Data Bank (wwPDB) (10).
There is a mechanism available from the primary literature which explains most of the observed experimental results.
The enzyme is unique at the H level of the CATH code (11), unless the homologue already in MACiE has a significantly different chemical mechanism.
Using the above criteria MACiE was expanded from 100 entries in version 1 to a total of 202 entries, which span 199 EC numbers (version 1 spanned 96 EC numbers) and covers a total of 862 reaction steps. There are almost 4000 EC numbers defined, but the number of different reaction mechanisms needed to bring about all these overall transformations is not clear. For example, the serine protease family of proteins has many different substrates, but the mechanisms are broadly similar. In contrast the β-lactamase enzymes, which have the same EC number, have four completely different mechanisms. Within the EC code, the fourth digit usually defines the substrate specificity, which can be very variable in large enzyme families—but the reaction mechanisms for enzymes with the same first three digits are usually essentially the same. In total there are 224 EC sub-subclasses, with only 181 having known structures (12). Of these MACiE covers 158, i.e. 87%. However, there are probably many more mechanisms that are yet to be defined or discovered.
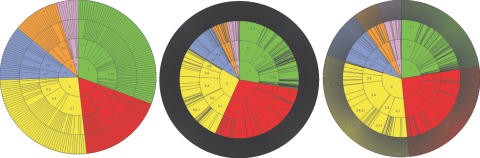
As can be seen from Figure 1, MACiE covers a good proportion of the EC reaction space, with an average relative difference between the size of corresponding EC classes of 4%, with the transferases having the largest difference. When the coverage with respect to EC code present in the PDB is examined, it can be seen that MACiE again represents the coverage of enzymes with known structures very well, with an average relative difference between the corresponding EC classes in MACiE of 5%.
Figure 1.
EC wheels showing the EC coverage of MACiE Version 2 (left), the complete EC space (centre) and the coverage of EC space in the PDB by unique EC serial numbers (right).
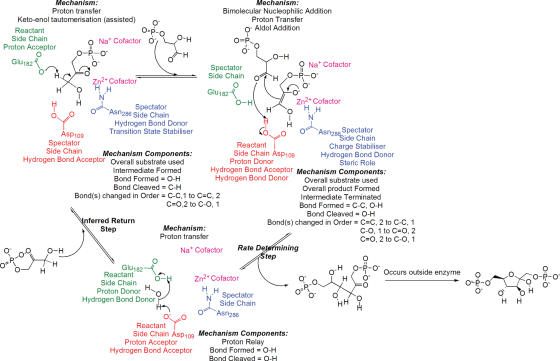
All entries in MACiE contain overall reaction annotation including the information detailed in Table 1. Each elementary reaction or step within an entry is fully annotated as is detailed in Figure 2, this includes comments that have been added by the annotators. An extension of the content from MACiE Version 1 is the addition of inferred return steps. These are explicitly labelled as being inferred in the comment field and are necessary to return the enzyme to a state where it is ready to undergo another round of catalysis.
Table 1.
Overall reaction annotation content
| Catalysis and reaction specific information | Non-catalysis specific information |
|---|---|
| Enzyme name (common IUPAB/JCBN name) | PDB code |
| EC code | Non-catalytic domain CATH code |
| Catalytic residues involved | Non-catalytic UniProt code |
| Cofactors involved | Species name (common and scientific) |
| Reactants and products | Other database identifiers, e.g. EzCatDB, SFLD, etc. |
| Catalytic domain CATH code | Literature references |
| Catalytic UniProt code | |
| Bonds involved, formed, cleaved, changed in order | |
| Reactive centres | |
| Overall reaction comments |
Figure 2.
An example of the annotation found in a MACiE entry. Reaction shown corresponds to fructose-bisphosphate aldolase (entry 52).
There is sometimes more than one proposed mechanism that is consistent with the available experimental data. In MACiE, we have attempted not only to choose the best supported mechanism, but also where possible to annotate enzymes with reasonable alternative mechanisms. Unfortunately, in the current release such annotations are only available as comments on the stage or overall reaction, although future releases of MACiE will include full entries for these alternatives.
Further details of the annotation process and a glossary of terms used can be found on the MACiE website (http://www.ebi.ac.uk/thornton-srv/databases/MACiE/documentation/ and http://www.ebi.ac.uk/thornton-srv/databases/MACiE/glossary.html, respectively).
DATABASE STRUCTURE
The challenge with MACiE has been to capture and usefully represent all the different catalytic steps that occur during the course of an enzymatic reaction. These reactions may consist of any number of steps, and in MACiE we have reactions ranging from 1 step to 16 steps. The representation of these reactions has evolved from a flat file entered in a commercially available chemical database program (ISIS/Base) to the highly structured and powerful CMLReact (7), which is an application of XML (the eXtensible Markup Language). The final step in this evolution has been the conversion of the CMLReact into the relational database format of MySQL.
CMLReact has a heirarchical structure, facilitating its conversion into the relational database format of MySQL. The conversion relies on the CML Schema and requires the MACiE entries to be consistent with the Schema, which adds an internal consistency check into our authoring process. Each CML tag-type becomes an MySQL table; each tag becomes a row in that MySQL table; each attribute of that tag corresponds to a column in the MySQL table. The tree structure of the CML is preserved in the MySQL version; for each row of each table, there are columns specifying which row of which other table corresponds to the row's parent tag in the CML version.
The CML version of MACiE, which is the official archive version, is available from the website as individual entries, and the new website uses the relational version of MACiE to perform the online analysis and searching.
DATABASE FEATURES
The original release of MACiE contained static images and annotation for the overall reaction and each step associated with the mechanism; it also included an animated reaction mechanism for approximately half the reactions then in MACiE. Links to various related resources, such as the RCSB PDB (13), IUBMB nomenclature database, CATH, EzCatDB, PDBSum (14), BRENDA, the Catalytic Site Atlas (15), KEGG and the Enzyme Structures Database, were also included. This new release extends these links to include the Macromolecular Structures Database (MSD) (16), SFLD, UniProt (17), and replaces the IUBMB nomenclature database links with links to IntEnz. The new features in MACiE are detailed in the following sections.
Searching MACiE
There are two levels of search implemented in MACiE. The basic level searches are implemented from the main page (http://www.ebi.ac.uk/thornton-srv/databases/MACiE) and are mainly for accessing the entries from the top level, i.e. for searching entries in MACiE by EC code, enzyme name, etc. The complex searches are all available from the query pages of MACiE (http://www.ebi.ac.uk/thornton-srv/databases/MACiE/queryMACiE.html) and are mainly for searching for specific mechanisms, mechanism components or residues and their functions in the reaction steps, although there are some overall reaction searches implemented as well. Table 2 lists the searches available in MACiE and the Supplementary Data contain a detailed listing of the searches available.
Table 2.
Searches available in MACiE
| Basic | Complex |
|---|---|
| MACiE entry identifier | Species name (overall annotation) |
| Current EC codes | Overall reactants and products |
| Obsolete EC codes | Reaction comments (overall reactions and steps) |
| Catalytic Domain CATH codes | Amino acid residues (up to six residues) |
| All CATH codes | Step mechanisms and/or mechanism components (single and combinations of) |
| PDB code | Chemical changes |
| Enzyme name | Chemical changes with mechanism or mechanism components |
| Catalytic Domain UniProt Codes | Chemical changes with amino acid residues |
| All UniProt Codes | Amino acid residues with mechanism or mechanism components |
| Chemical changes with amino acid residues and mechanisms or mechanism components | |
| Alternative mechanisms |
The following sections describe searching by EC code, PDB code or enzyme name, all of which use heuristics to extend the coverage of MACiE.
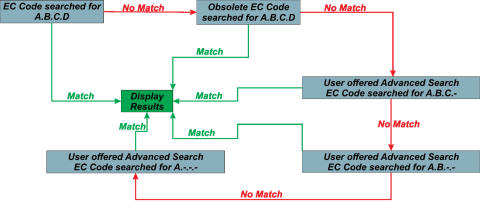
EC code
The EC code search implemented in MACiE is detailed in Figure 3 and can be accessed at any point in the scheme shown. The search for current EC numbers will always walk up the EC code tree until it finds a match, no matter at what level the search is entered. Thus the search will always return a result. As the EC code of enzymes may change over time, a search for obsolete EC codes has also been implemented, although this search will not always return a result. However, it should be noted that the higher up the EC hierarchy search has gone, the less likely it is that the returned mechanism will be a match to the query. The obsolete EC code search works in the same way as the current EC code.
Figure 3.
EC code search heuristics.
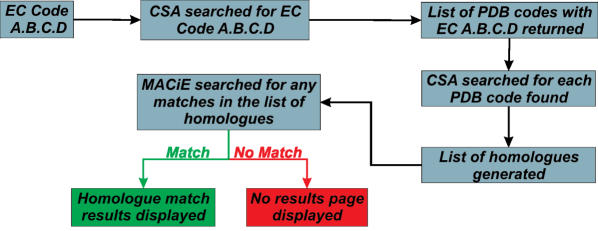
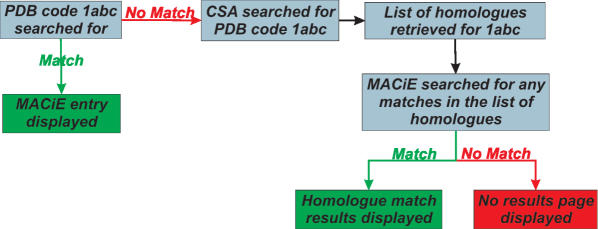
If no matches are found at the serial number level of the EC code, an advanced search option will allow the user to search for a structural homologue of an enzyme with a given EC code, which is shown in Figure 4 and described below. This advanced search option takes the entered EC code and finds the PDB codes of all of the matches to that EC code in the Catalytic Site Atlas (CSA). A homology search is then performed on those PDB codes for a match in MACiE. This homology search is described in more detail in the following section.
Figure 4.
Advanced EC search heuristics.
The CSA is a database of catalytic residues in proteins of known structure. It contains much less mechanistic information than MACiE, but has a considerably wider coverage of protein structures than MACiE does. This wider coverage is partly because the CSA contains not only manually annotated entries, but also contains entries that are automatically annotated based on sequence alignment to the manual entries.
PDB code
There are over 19 000 crystal structures relating to enzymes deposited in the PDB. As MACiE entries require extensive literature searching and analysis, only a small fraction of these PDB entries are covered explicitly, 202 in total. However, we have used the CSA to identify homologues of these enzymes, extending this coverage to 7528 PDB codes.
Figure 5 details the search performed in MACiE, when a protein structure described by a PDB code is entered. Although the entries returned by this search will be homologues, this does not guarantee that the mechanism and the catalytic residue assignments are the same. This is because the homology method (see below) can retrieve very distant relatives. Owing to this limitation, all homologous entries are compared by EC code, and when there is a divergence between the MACiE entry and the homologue at the serial number level, this is clearly indicated to the user. We also list the amino acid residues that are annotated as catalytic in both MACiE and the CSA. Thus it is clear if there is any difference between EC numbers and catalytic residues. If the EC number differs but the catalytic residues between query and homologue are of identical types, it can be inferred that the mechanisms are likely to be the same, but where both differ, the mechanisms are unlikely to be transferable. From the results page we link both to the MACiE entry and the CSA entry.
Figure 5.
PDB search heuristics.
Homology in MACiE
We have been working to bring MACiE and the CSA closer together. This includes using the CSA to determine homologues (those enzymes which are evolutionarily related) of entries in MACiE. The CSA finds homologues using a PSI-BLAST search (with an E-value cut-off of 0.0005 and five iterations) against all sequences currently in the PDB, plus all sequences in a non-redundant subset of UniProt. The UniProt sequences are included purely in order to increase the range of the PSI-BLAST search by bridging gaps between distantly related sequences in the PDB; only sequences occurring in the PDB are retrieved for entry into the CSA. In the CSA, and thus MACiE, homologous entries are only included if the residues which align with the catalytic residues in the parent literature entry are identical in residue type. In other words, there must be no mutations at the catalytic residue positions. There are, however, a few exceptions to this rule:
In order to allow for the many active site mutants in the PDB, one (and only one) catalytic residue per site can be different in type from the equivalent in the parent literature entry. This is only permissible if all residue spacing is identical to that in the parent literature entry, and there are at least two catalytic residues.
Sites with only one catalytic residue are permitted to be mutant provided that the residue number is identical to that in the parent entry.
Fuzzy matching of residues is permitted within the following groups: [V,L,I], [F,W,Y], [S,T], [D,E], [K,R], [D,N], [E,Q], [N,Q]. This fuzzy matching cannot be used in combination with rules (i) or (ii) above.
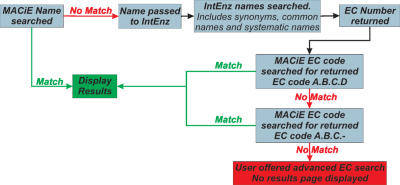
Enzyme name
This is currently implemented as a partial string match, thus entering ‘beta’ will return all the β-lactamases and betaine-aldehyde dehydrogenase. If no results are returned from the partial name search, then the name search heuristics (shown in Figure 6) are implemented.
Figure 6.
Enzyme name search heuristics.
This search utilizes the IntEnz database (4). MACiE searches for a name in IntEnz, either a synonym, alternative name or common name, and returns the EC code of that name. The EC code is then used to search MACiE. If no matches are found to the sub-subclass level of the EC code, the user is offered an advanced EC code search (see Figure 4).
Statistics
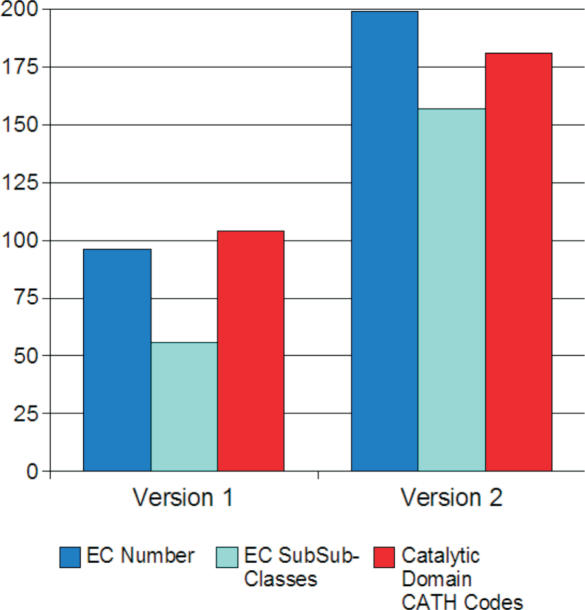
The other major development in MACiE has been the inclusion of database statistics that are all generated on the fly from the SQL tables. A full listing of the statistics available can be found in the Supplementary Data. The growth of MACiE is shown in Figure 7 in terms of EC coverage and CATH coverage.
Figure 7.
Growth of MACiE. This shows the growth in the number of EC codes (blue), EC sub-sub classes (cyan) and catalytic domain CATH codes (red) in MACiE.
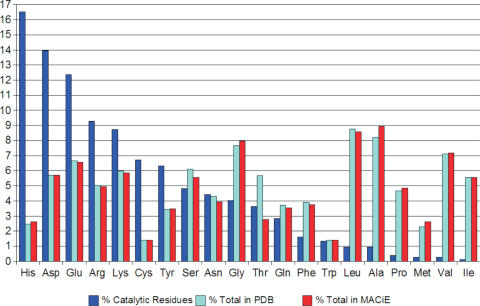
The statistics in MACiE can also be used to examine the function and distribution of amino acid residues (G.L. Holliday, D.E. Almonacid, J.M. Thornton and J.B.O. Mitchell, manuscript in preparation) (see Figure 8), the distribution of mechanism and mechanism components and the bond order changes occurring in each step of the reaction.
Figure 8.
Frequency distribution of amino acid residues. This shows the frequency of catalytic amino acid residues in MACiE (blue), versus the frequency of residues in MACiE (cyan), versus the frequency of residues in the wwPDB (red). The frequency of catalytic amino acid residues in MACiE is calculated by taking the number of residues (of a given type) annotated in MACiE divided by the total number of annotated residues in MACiE, multiplied by 100.
FUTURE DEVELOPMENTS
MACiE is a continually developing resource, and in the future we hope to include 3D data, which will incorporate various statistics and searches related to the analysis of these data. We will also continue to extend the coverage of MACiE to include alternative reaction mechanisms that have been suggested for various enzymes, as well as new mechanisms. Finally, we intend to build a user interface which will allow for chemical diagrams to be drawn and used to search MACiE, an entry process which is more usable and also to implement the classification of enzyme mechanisms that we are developing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We would like to thank the EPSRC (G.L.H. and J.B.O.M.), BBSRC (G.J.B. and J.M.T.—CASE studentship in association with Roche Products Ltd; N.M.O.B. and J.B.O.M.—grant BB/C51320X/1), the Wellcome Trust, EMBL, IBM (G.L.H. and J.M.T.), the Chilean Government's Ministerio de Planificación y Cooperación and the Cambridge Overseas Trust (D.E.A.) for funding and Unilever for supporting the Centre for Molecular Science Informatics. J.W.T. is funded by a European Molecular Biology Laboratory studentship, and is also affiliated with Cambridge University Department of Chemistry. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schomburg I., Chang A., Ebeling C., Gremse M., Heldt C., Huhn G., Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.IUBMB. Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of enzyme-catalysed reactions. 2005.
- 4.Fleischmann A., Darsow M., Degtyarenko K., Fleischmann W., Boyce S., Axelsen K., Bairoch A., Schomburg D., Tipton K.F., Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004;32:D434–D437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holliday G.L., Bartlett G.J., Almonacid D.E., O'Boyle N.M., Murray-Rust P., Thornton J.M., Mitchell J.B.O. MACiE: a database of enzyme reaction mechanisms. Bioinformatics. 2005;21:4315–4316. doi: 10.1093/bioinformatics/bti693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holliday G.L., Mitchell J.B.O., Murray-Rust P. CMLSnap: animated reaction mechanisms. Internet J. Chem. 2004;7 Article 4. [Google Scholar]
- 7.Holliday G.L., Murray-Rust P., Rzepa H.S. Chemical Markup, XML, and the World Wide Web. 6. CMLReact, an XML vocabulary for chemical reactions. J. Chem. Inf. Model. 2006;46:145–157. doi: 10.1021/ci0502698. [DOI] [PubMed] [Google Scholar]
- 8.Pegg S.C.-H., Brown S.D., Ojha S., Seffernick J., Meng E.C., Morris J.H., Chang P.J., Huang C.C., Ferrin T.E., Babbitt P.C. Leveraging enzyme structure–function relationships for functional inference and experimental design: the Structure–Function Linkage Database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 9.Nagano N. EzCatDB: the Enzyme Catalytic-mechanism DataBase. Nucleic Acids Res. 2005;33:D407–D412. doi: 10.1093/nar/gki080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman H.M., Henrick K., Nakamura H. Announcing the worldwide Protein Data Bank. Nature Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 11.Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 12.Martin A.C. PDBSprotEC: a Web-accessible database linking PDB chains to EC numbers via SwissProt. Bioinformatics. 2004;20:986–988. doi: 10.1093/bioinformatics/bth048. [DOI] [PubMed] [Google Scholar]
- 13.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laskowski R.A., Chistyakov V.V., Thornton J.M. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter C.T., Bartlett G.J., Thornton J.M. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golovin A., Oldfield T.J., Tate J.G., Velankar S., Barton G.J., Boutselakis H., Dimitropoulos D., Fillon J., Hussain A., Ionides J.M., et al. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2004;32:D211–D216. doi: 10.1093/nar/gkh078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]