Abstract
Human cytomegalovirus can cause a diverse range of diseases in different immunocompromised hosts. The pathogenic mechanisms underlying these diseases have not been fully elucidated, though the maximal viral load during infection is strongly correlated with the disease. However, concentrating on single viral load measures during infection ignores valuable information contained during the entire replication history up to the onset of disease. We use a statistical model that allows all viral load data sampled during infection to be analysed, and have applied it to four immunocompromised groups exhibiting five distinct cytomegalovirus-related diseases. The results show that for all diseases, peaks in viral load contribute less to disease progression than phases of low virus load with equal amount of viral turnover. The model accurately predicted the time of disease onset for fever, gastrointestinal disease and pneumonitis but not for hepatitis and retinitis, implying that other factors may be involved in the pathology of these diseases.
Keywords: virus replication, models, disease
1. Introduction
Human cytomegalovirus (HCMV) is a highly seroprevalent infection in the human population. In immunocompetent individuals, viral replication is suppressed predominantly, but not exclusively, by CD8+ T-cells (McLaughlin-Taylor et al. 1994; Gillespie et al. 2000; Appay et al. 2002; Bronke et al. 2005a,b; Bunde et al. 2005; Sacre et al. 2005; Sylwester et al. 2005), and the virus has evolved as a broad range of immune manipulation strategies to maintain the homeostatic host–parasite relationship in a clinically asymptomatic form (Tortorella et al. 2000; Lilley et al. 2001; Kaleejta & Shenk 2002; Mocarski 2002; Rehm et al. 2002; Tirabassi & Ploegh 2002; Lilley & Ploegh 2004). In contrast, in the T-cell immunocompromised host (particularly transplant recipients and HIV-infected individuals), high levels of HCMV replication are observed leading to a variety of HCMV end-organ diseases (Emery 2001; Deayton et al. 2004). The anatomical site of HCMV disease varies according to the nature of the patient group, implying that each immunocompromised host may make unique contributions to disease pathogenesis. Thus, bone marrow transplant recipients frequently experience HCMV pneumonitis and gastrointestinal tract disease (Nichols & Boeckh 2000); solid organ recipients experience hepatitis, gastrointestinal tract disease and prolonged fever (Ho 1994), while HIV-infected individuals with CD4 cell counts less than 50 cells μl−1 predominantly experience HCMV retinitis (Dunn & Jabs 1995; Hoover et al. 1996).
The recent application of sensitive methods to quantify viral replication has facilitated an understanding of the viral parameters associated with HCMV disease. These studies have shown that viral load is a dominant risk factor for HCMV disease (Cope et al. 1997; Gor et al. 1998; Spector et al. 1998; Emery et al. 1999a,b; Hassan-Walker et al. 1999; Limaye et al. 2001). In addition, kinetics of replication during the early phases of active replication have been defined and used to estimate the basic reproductive number of HCMV (Emery et al. 2002) and to provide prognostic information in the immunocompromised host (Emery et al. 2000; Schafer et al. 2001). Since the virologic parameters, such as initial viral load, peak viral load, rate of increase in viral load prior to disease onset, are highly correlated it has been difficult, using conventional multivariable models, to ascertain the relative contributions of viral load thresholds and continuous presence of virus to the pathogenesis of different HCMV diseases and to produce estimates of the likely time of onset of disease. In order to address this issue for HCMV pathogenesis, we have utilized a more sophisticated statistical model recently described to investigate the pathogenesis of simian immunodeficiency virus (SIV) infection in rhesus macaques (Macaca mulatta; Regoes et al. 2002). In simple terms, the statistical model allows the relative contributions of peaks in viral load to the pathogenesis of disease to be assessed by computing the total viral turnover prior to disease onset and determining a tolerance threshold for disease, which is related to a peak parameter. Re-iteration of the model with various values of this peak parameter allows the calculation of the optimal value of the peak parameter, which explains a significant proportion of the time to disease observed clinically. In the case of SIV, the model shows that peaks in virus load are not the major pathogenetic factors but that continuous viral presence is a dominant factor (Regoes et al. 2002). In the present report, we show that a similar scenario exists for HCMV pathogenesis in the human host.
2. Material and methods
Sixty-one patients with frequent HCMV load measurements, who suffered from HCMV disease, were analysed. The patients compromised renal transplant (n=12) (Hassan Walker et al. 1999), liver transplant (n=18) (Cope et al. 1997) and bone marrow transplant (n=13) (Gor et al. 1998) recipients and 16 HIV-infected individuals (Bowen et al. 1997). HCMV diseases conformed to the International Workshop definition (Ljungman et al. 2002) and included HCMV retinitis (n=17), gastrointestinal disease (n=7), hepatitis (n=4), pyrexial debilitating disease (n=14) and HCMV pneumonitis (n=14). In addition, a further five cases of presumed HCMV hepatitis were included on the basis of elevated blood alanine aminotransferase (ALT) levels in the absence of any other pathogen. In these cases, peak of ALT was taken as the day of disease. Viral load for HCMV was measured using a quantitative–competitive PCR method on the DNA extracted from the whole blood (level of sensitivity: 200 genomes ml−1 blood) described extensively elsewhere (Fox et al. 1992, 1995). The total number of viral load measurements used in the analysis was 388. In the transplant recipients, viral load monitoring occurred from the day of transplant weekly for the first three months and then routinely on a monthly basis, whereas in AIDS patients routine monitoring occurred when CD4 cell counts fell below 100 cells μl−1 blood every 2–4 weeks. The treating physicians were not aware of the viral load measurements, and the treatment was initiated on clinical grounds, i.e. evidence of HCMV disease.
3. Model used to evaluate contributions of peaks of viral load to disease
We used a previously developed statistical model (Regoes et al. 2002) to regress the time to disease against the viral load. The model takes into account the temporal variation in the virus load. By fitting the model to the HCMV load data we estimate a parameter p, which measures the importance of peaks of virus load for disease progression. For p>1, high peaks became the dominant factor in HCMV disease, whereas if p<1 the high peaks contribute under-proportionately to HCMV disease. A full description of the model and the fitting is given in appendix A.
4. Results
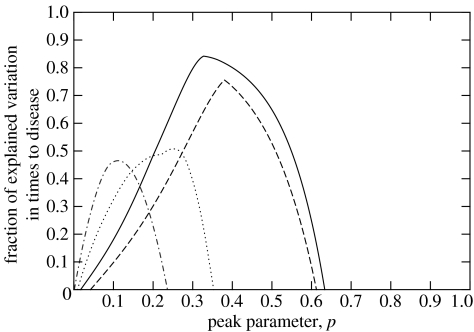
The model outlined earlier was simultaneously fitted to the patients within each HCMV disease category in order to obtain the best estimate for p (pbest). In all types of HCMV diseases under consideration, pbest was less than unity implying that continuous presence of virus was more important than peaks of viral load for determining time to disease. In the case of pyrexial debilitating disease (fever), pneumonitis and gastrointestinal disease, the values of p that explained the greatest amount of the variation in time to disease development were 0.33, 0.25 and 0.38, respectively (figure 1). These values were significantly less than 1 but not significantly different from each other as can be seen from the 95% confidence intervals (CIs) of the fitted peak parameter p listed in table 1. Using the coefficient of fit parameter R2 (equation (A 7)), these values of pbest accounted for 84.2% of the variation in the time to onset of fever, 75.6% of the time to gastrointestinal disease and 50.7% of the time to HCMV pneumonitis.
Figure 1.
Distribution of the peak parameter, p, illustrating the goodness-of-fit, measured by the fraction of explained variation in times to disease for four of the five HCMV diseases under consideration (fever—solid line; pneumonitis—dotted line, gastrointestinal disease—dashed line and hepatitis—dashed-dotted line). All fitted peak parameters are between 0 and 1. The fraction of explained variation in time to disease is significant for all diseases, except for hepatitis (n=4) (see table 1).
Table 1.
Summary of the results obtained for pbest for the HCMV diseases under consideration. (The estimation of the peak parameter was determined by maximization of the variation in times to disease. The 95% confidence intervals were calculated by bootstrapping, and the significance level was determined by an F-test. NA, not applicable.).
| HCMV disease | number of patients | peak parameter (95% CI) | explained variance (%) | significance (p-value) |
|---|---|---|---|---|
| fever | 14 | 0.33 (0.014–0.44) | 84.2 | 3.8×10−6 |
| pneumonitis | 14 | 0.25 (0.12–0.34) | 50.7 | 0.0043 |
| gastrointestinal | 7 | 0.38 (0.058–0.52) | 75.6 | 0.011 |
| hepatitis | 4 | 0.11 (NA) | 46.5 | 0.32 |
| retinitis | 17 | 0.065 (NA) | 2.6 | 0.54 |
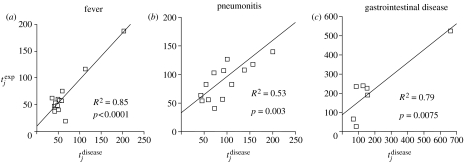
Figure 2 illustrates how well the expected times to disease, , agree with the observed times, , for fever, pneumonitis and gastrointestinal disease. For each of these three diseases, the mean and do not differ significantly (according to a paired t-test), indicating that our model predictions are unbiased. The mean and for pyrexial disease was 65.4±41.7 and 64.2±44 days, respectively, while the comparable mean and for gastrointestinal tract disease was 216±161 and 190±210 days and for pneumonitis was 95.5±40.5 and 98±50 days. For the best fit, the tolerance threshold for disease, τ, was determined to be 854 (ge ml−1) days (95% CI: 59–1958 (ge ml−1) days) for fever; 828 (ge ml−1) days (95% CI: 238–1863 (ge ml−1) days) for pneumonitis and 3041 (ge ml−1) days (95% CI: 159–5884 (ge ml−1) days) for gastrointestinal disease.
Figure 2.
Agreement between the time of disease predicted by the model, , and the observed time to disease, , for the three HCMV diseases where a significant proportion of the variation in time to disease was explained by the model. In the case of gastrointestinal disease, the significance of the regression is dependent on the outlying high data point.
In the case of HCMV hepatitis and retinitis, pbest was also below unity but the model was unable to account for a significant proportion of the variation in time to disease observed clinically. For HCMV hepatitis, pbest was 0.11 accounting for 46.5% of the variation, which did not reach statistical significance. Since this result may reflect the small number of patients with histologically proven HCMV hepatitis, we analysed a further five patients whose sequential HCMV load measures were available and who had elevations in liver function tests (specifically ALT), using the maximum ALT level as the marker for liver-related disease. Interestingly, these analyses indicated that pbest→0, but the model could not explain the variation observed in the time to peak ALT. However, the expected time of peak ALT (45.1±3 days) was an average of approximately 2 days earlier than the actual time to ALT peak (47.3±11.2 days). A similar situation was observed for HCMV retinitis where pbest was 0.065, but the model only accounted for 2.5% of the variation in time to disease. The mean for HCMV retinitis was 124±9 days compared to the mean of 130±43 days (P=0.59), although, as expected from the least-square fit, there was no significant correlation between and (R2=0.04; P=0.43). A summary of these data for all HCMV diseases under consideration is given in table 1.
Since we had estimated the peak parameter p and the tolerance threshold τ from the model for the appearance of fever, gastrointestinal tract disease and pneumonitis, we were interested to determine whether, and at what time, these tolerance thresholds were breached by the HCMV replication in the HIV-infected patient group with HCMV retinitis. The data shown in table 2 reveal that the threshold is reached for each disease significantly prior (between 2 and 3 months) to the appearance of retinitis. These data illustrate the dichotomy between the expected appearance of diseases in the HIV-infected host and the clinical picture where such diseases are either rare or absent prior to the diagnosis of HCMV retinitis.
Table 2.
Calculation of the expected times of appearance of fever, pneumonitis and gastrointestinal disease in AIDS retinitis cases using the previously calculated threshold values, τ. (The mean time for the appearance of HCMV retinitis was 135 (95% CI: 112–158) days after initial detection of virus in the blood (less than 200 ge ml−1). The statistical significance was calculated using the t-test.)
| disease | disease threshold (τ) (ge ml−1) days | expected time of disease in days (95% CI) | difference in texp and tretinitis in days (95% CI) | significance of difference (p-value) |
|---|---|---|---|---|
| fever | 854 | 38.4 (31.5–45.3) | −96.6 (−120.6 to −72.6) | <0.0001 |
| pneumonitis | 829 | 75 (51–100) | −60 (−35 to −84) | <0.0001 |
| gastrointestinal | 3041 | 70.8 (54.5–84.2) | −64.2 (−38.2 to −90.2) | <0.0001 |
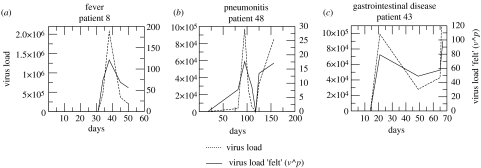
The model suggests with some confidence that for three HCMV diseases, peaks in viral load contribute under-proportionally to HCMV pathogenesis in the immunocompromised host, i.e. total viral turnover is not the best correlate of time to disease. This concept can be illustrated by comparing the actual viral load measured in the blood with the viral load scaled by the exponent p to give an imaginary viral load that is felt by the host during active infection. Three representative examples are shown in figure 3 and illustrate that during periods of low viral loads the host perception is that commensurately more virus is present.
Figure 3.
Comparison of representative longitudinal HCMV load profiles measured in the blood of patients with fever, gastrointestinal disease and pneumonitis with the effective viral load ‘felt’ by the host (viral load scaled by the peak parameter, p). Dashed line, viral load; solid line, virus load ‘felt’ (v^p).
5. Discussion
In the case of HCMV infections in the immunocompromised host, we, and others, have consistently shown using logistic regression models that viral load levels at early times during active infection, at the peak of viraemia and area under the viral load–time curve can provide important prognostic and diagnostic markers for HCMV disease. However, until now, neither the relative contributions of peaks in viral replication in the context of replication rate and turnover of viral infected cells to HCMV pathogenesis have been assessed nor whether the pathogenesis of the distinct HCMV diseases observed in different immunocompromised hosts can be explained by a single pathogenetic mechanism. The results presented here clearly show that for three pathologically distinct HCMV diseases, namely prolonged fever, gastrointestinal tract disease and pneumonitis, peaks in viral load contribute under-proportionally to HCMV disease. Thus, short periods of high viral load contribute less to disease progression than longer periods of lower viral load with an equal amount of viral turnover. The results further imply that it may not be the actual amount of virus present at any given point in time or the amount of viral turnover that dictates whether the hosts suffer from HCMV disease, but that the continuous presence of virus is a major effector mechanism. These results complement our previous analyses of the role of viral load as a risk factor for HCMV diseases (as mentioned earlier) and do not negate a role for continuous periods of high viral load in HCMV pathogenesis; however, they do suggest that the contribution of peaks is smaller than that suggested by the amount of viral turnover during the peak phase. Indeed, as suggested in our previous studies on the viral load at early time-points in active infection (Emery et al. 2000), the current study reinforces the concept that viral load measures at each time-point contain information pertinent to pathogenesis, which can be extracted using the current modelling approach. Using a similar model, the same conclusions have been derived for SIV pathogenesis in experimentally infected rhesus macaques (Regoes et al. 2002). In this paradigm for HIV pathogenesis, the model explained 44% of the variation in time to death from SIV and was more accurate than a viral set-point model.
Two HCMV diseases included in our analyses appear not to segregate with the diseases described earlier. In the case of hepatitis, the model explained 46.5% of variation in time to biopsy proven HCMV disease, but this was not statistically significant and was not improved by the inclusion of an extra five patients with presumed hepatitis. Various explanations, which are not mutually exclusive, can be offered for this observation: firstly, an important, as yet unidentified pathogenic component is missing from the model; secondly, human leukocyte antigen (HLA) matching does not occur between liver transplant donor and recipients, and so the ability of the donor to mount an appropriate immune response against viral infected hepatocytes may be delayed or even absent, thus producing variation in time to clinically apparent hepatitis independent of virologic events and finally, there may be a differential balance between viral lysis and immune-mediated destruction of hepatocytes depending on the viral burden in the infected hepatocytes. Interestingly, HCMV infections have been associated with an increased risk of acute rejection episodes in patients with partial HLA matching but not in patients with a complete HLA match with the donor organ (Ontanon et al. 1998).
In the case of HCMV retinitis, we obtained results similar to the extended dataset of hepatitis patients (pbest for this disease was 0.065 with only 2.6% of the variation being explained by the model). The majority of retinitis is in the HIV-infected patient group who differ from other patients, in that they have a continuously declining immune functionality, which allows HCMV replication to reach a steady state weeks or even months prior to the appearance of retinitis (Spector et al. 1998; Bowen et al. 1997; Emery et al. 1999a,b). It is possible that a specific period of replication is required before the vasculature of the eye can be breached by HCMV and intraocular infection achieved. HIV itself has been shown to affect the ocular vasculature (Dejaco-Ruhswurm et al. 2001; Lim et al. 2001), and it is possible that HIV load itself may be a predisposing factor which accounts for the variation in the time for a patient to experience HCMV retinitis. Nevertheless, despite a small peak parameter value, the model is unable to explain the variation in time to disease implying that other factors, as yet unidentified, are important for the development of retinitis. In this context, the expansion of a fully differentiated, mature effector HCMV specific CD8 T-cell population just prior to HCMV retinitis has been described recently (Bronke et al. 2005a)
An alternative way of considering the HIV-infected patient group is to assess whether, given their HCMV replication history, other HCMV diseases (pneumonitis, gastrointestinal tract disease) would be predicted to occur. Since we know from a number of natural history studies that the first HCMV disease experienced by these patients is retinitis (Dunn & Jabs 1995; Shinkai et al. 1997), we have used the tolerance thresholds calculated for other HCMV diseases to predict the likely time that one would have expected these diseases to occur in the HIV-infected group. The results show that each disease entity, including pneumonitis, would have been expected to occur on average two months prior to the appearance of HCMV retinitis. These results again indicate that host factors, which are important in producing HCMV-related pathology, are absent or severely depleted in the HIV-infected patient group. Interestingly, HCMV DNA is frequently found in the lungs of HIV-patients and yet no overt pathology has been observed (Mann et al. 1997) except in patients with well-preserved CD4 counts, who can presumably mount a cytokine driven T-cell-mediated immune response in the lung (Squire et al. 1992; Barry et al. 2000).
In conclusion, we have shown that peaks of HCMV load contribute much less to the pathogenesis of different HCMV diseases than previously envisaged, and that each viral load value inherently contains important data regarding pathogenesis. These findings have relevance to the deployment of antiviral chemotherapy for HCMV, which should be aimed to maintain levels below the critical tolerance threshold for disease and also identifies certain HCMV diseases whose pathogenesis appear to be distinct.
Appendix A
The total turnover of HCMV, T, over the period prior to disease onset is proportional to the area under the virus load–time curve which is given by the integral of the viral load, v, from the time of HCMV infection to HCMV disease, divided by the duration of a complete replication cycle of the virus, dr:
| A1 |
To ascertain the contribution of peaks in HCMV load to disease, we expand equation (A 1)) for the total viral turnover to the following model, relating virus load in the jth patient, vj, to the time they experienced disease, tjdisease
| A2 |
This model assumes that a universal tolerance threshold (τ) exists that measures the tolerance of all individuals who suffer from HCMV disease irrespective of between patient group variation of tolerance with respect to HCMV infection/disease. Since the right-hand side of equation (A 2) contains the integral over the HCMV load weighted by a factor dependent on viral load, v, it can be rewritten as the integral over the HCMV load weighted by a factor dependent on viral load, v, viz.
| A3 |
In this case, the peak parameter p determines the relative importance of peaks in HCMV load to HCMV disease. For p>1, high peaks became the dominant factor in HCMV disease, whereas if p<1 the high peaks contribute under-proportionately to HCMV disease. Thus, an alternative complementary explanation can be offered; a very large p-value (p≫1) describes a situation where HCMV disease develops when viral load reaches a certain threshold level; when p=1, HCMV disease develops after a certain number of virus particles has been produced; when p is smaller than unity, continuous HCMV burden contributes to HCMV disease disproportionately more than a short and high peak in viral load and when p=0, the total time of the continuous presence of virus is important, irrespective of the viral load.
(a) Fitting the model and experimental data
For a fixed peak parameter p-value, the left-hand side of equation (A 4) is calculated for all cases of HCMV disease as follows:
| A4 |
where tjdisease denotes the time at which the jth patient experienced HCMV disease and vj(t) is the virus load of the jth patient at time t. The geometric mean of the left-hand sides, , yields a universal tolerance threshold, . The value of τ is then used to calculate the expected time of disease for the jth patient according to the following algorithm:
If τ<τj, integrate vj(t)p until the integral equals τ. The time at which this condition is satisfied is the expected time of disease of the jth patient, :
| A5 |
Since τ<τj it follows that , and so this case contains patients who suffer from disease later than expected.
If τ>τj, the viral load progression of the jth patients' is extrapolated assuming that the viral load after the onset of disease is equal to the average of the virus load before disease. Integration of vj(t)p until the integral equals τ yields the expected time of disease of the jth individual (vide supra). In this case, , and so contains patients who experience disease earlier than expected.
We repeat this computation for different values of the peak parameter p between 0 and 10. The best fit is determined by the value of p which minimizes the sum of squares:
where n is the number of patients with the disease under consideration and is calculated for each HCMV disease.
(b) Estimating the quality of the fit
The quality of the model fit can be estimated by the ratio of the explained sum of squares divided by the total sum of squares of the variable regressed (tdisease). The fraction of explained variation R2 is given by:
where n is the number of patients with the disease being assessed. Whether the model explains a significant fraction of the variation in the time to disease can be assessed by an F-test.
Comparison of means between and for each disease under consideration was performed using a paired t-test. P-values equal to 0.05 or less were regarded as significant.
References
- Appay V, et al. Memory CD8+T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 2002;8:379–385. doi: 10.1038/nm0402-379. 10.1038/nm0402-379 [DOI] [PubMed] [Google Scholar]
- Barry S.M, Johnson M.A, Janossy G. Cytopathology or immunopathology? The puzzle of cytomegalovirus pneumonitis revisited. Bone Marrow Transplant. 2000;26:591–597. doi: 10.1038/sj.bmt.1702562. 10.1038/sj.bmt.1702562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen E.F, Sabin C.A, Wilson P, Grifiths P.D, Davey C.C, Johnson M.A, Emery V.C. Cytomegalovirus (CMV) viraemia detected by polymerase chain reaction identifies a group of HIV-positive patients at high risk of CMV disease. AIDS. 1997;11:889–893. doi: 10.1097/00002030-199707000-00008. 10.1097/00002030-199707000-00008 [DOI] [PubMed] [Google Scholar]
- Bronke C, Westerlaken G.H, Miedema F, Tessalaar K, van Baarle D. Progression to CMV end-organ disease in HIV-1 infected individuals despite abundance of highly differentiated CMV-specific CD8+T-cells. Immunol. Lett. 2005a;97:215–224. doi: 10.1016/j.imlet.2004.11.004. 10.1016/j.imlet.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Bronke C, et al. Dynamics of cytomegalovirus (CMV)-specific T-cells in HIV-1 infected individuals progressing to AIDS with CMV end-organ disease. J. Infect. Dis. 2005b;191:873–880. doi: 10.1086/427828. 10.1086/427828 [DOI] [PubMed] [Google Scholar]
- Bunde T, et al. Protection from cytomegalovirus after transplantation is correlated with immediate early 1-specific CD8 T cells. J. Exp. Med. 2005;201:1031–1036. doi: 10.1084/jem.20042384. 10.1084/jem.20042384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope A.V, Sabin C, Burroughs A, Rolles K, Griffiths P.D, Emery V.C. Interrelationships among quantity of human cytomegalovirus (HCMV) DNA in blood, donor–recipient serostatus, and administration of methylprednisolone as risk factors for HCMV disease following liver transplantation. J. Infect. Dis. 1997;176:1484–1490. doi: 10.1086/514145. [DOI] [PubMed] [Google Scholar]
- Deayton J.R, Sabin C.A, Johnson M.A, Emery V.C, Wilson P, Griffiths P.D. Importance of cytomegalovirus viraemia in risk of disease progression and death in HIV-infected patients receiving highly active antiretroviral therapy. Lancet. 2004;363:2116–2121. doi: 10.1016/S0140-6736(04)16500-8. 10.1016/S0140-6736(04)16500-8 [DOI] [PubMed] [Google Scholar]
- Dejaco-Ruhswurm I, Kiss B, Rainer G, Krepler K, Wedrich A, Dallinger S, Rieger A, Schmetterer L. Ocular blood flow in patients infected with human immunodeficiency virus. Am. J. Ophthalmol. 2001;132:720–726. doi: 10.1016/s0002-9394(01)01095-9. 10.1016/S0002-9394(01)01095-9 [DOI] [PubMed] [Google Scholar]
- Dunn J.P, Jabs D.A. Cytomegalovirus retinitis in AIDS: natural history, diagnosis, and treatment. AIDS Clin. Rev. 1995;96:99–129. [PubMed] [Google Scholar]
- Emery V.C. Investigation of CMV disease in immunocompromised patients. J. Clin. Pathol. 2001;54:84–88. doi: 10.1136/jcp.54.2.84. 10.1136/jcp.54.2.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery V.C, Cope A.V, Bowen E.F, Gor D, Griffiths P.D. The dynamics of human cytomegalovirus replication in vivo. J. Exp. Med. 1999a;190:177–182. doi: 10.1084/jem.190.2.177. 10.1084/jem.190.2.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery V.C, Sabin C, Feinberg J.E, Gryzwacz M, Knight S, Griffiths P.D, on behalf of the ACTG 204/GlaxoWellcome 123-014 International CMV Prophylaxis Study Group Quantitative effects of valacyclovir on the replication of cytomegalovirus (CMV) in persons with advanced human immunodeficiency virus disease: baseline CMV load dictates time to disease and survival. J. Infect. Dis. 1999b;180:695–701. doi: 10.1086/314936. 10.1086/314936 [DOI] [PubMed] [Google Scholar]
- Emery V.C, Sabin C.A, Cope A.V, Gor D, Hassan-Walker A.F, Griffiths P.D. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet. 2000;355:2032–2036. doi: 10.1016/S0140-6736(00)02350-3. 10.1016/S0140-6736(00)02350-3 [DOI] [PubMed] [Google Scholar]
- Emery V.C, Hassan-Walker A.F, Burroughs A.K, Griffiths P.D. Human cytomegalovirus (HCMV) replication dynamics in HCMV-naive and -experienced immunocompromised hosts. J. Infect. Dis. 2002;185:1723–1728. doi: 10.1086/340653. 10.1086/340653 [DOI] [PubMed] [Google Scholar]
- Fox J.C, Griffiths P.D, Emery V.C. Quantification of human cytomegalovirus DNA using the polymerase chain reaction. J. Gen. Virol. 1992;73:2405–2408. doi: 10.1099/0022-1317-73-9-2405. [DOI] [PubMed] [Google Scholar]
- Fox J.C, Kidd I.M, Griffiths P.D, Sweny P, Emery V.C. Longitudinal analysis of cytomegalovirus load in renal transplant recipients using a quantitative polymerase chain reaction: correlation with disease. J. Gen. Virol. 1995;76:309–319. doi: 10.1099/0022-1317-76-2-309. [DOI] [PubMed] [Google Scholar]
- Gillespie G.M, et al. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8(+) T lymphocytes in healthy seropositive donors. J. Virol. 2000;74:8140–8150. doi: 10.1128/jvi.74.17.8140-8150.2000. 10.1128/JVI.74.17.8140-8150.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gor D, Sabin C, Prentice H.G, Vyas N, Man S, Griffiths P.D, Emery V.C. Longitudinal fluctuations in cytomegalovirus load in bone marrow transplant patients: relationship between peak virus load, donor/recipient serostatus, acute GVHD and CMV disease. Bone Marrow Transpant. 1998;21:597–605. doi: 10.1038/sj.bmt.1701139. 10.1038/sj.bmt.1701139 [DOI] [PubMed] [Google Scholar]
- Hassan-Walker A.F, Kidd I.M, Sabin C, Sweny P, Griffiths P.D, Emery V.C. Quantity of human cytomegalovirus (CMV) DNAemia as a risk factor for CMV disease in renal allograft recipients: relationship with donor/recipient CMV serostatus, receipt of augmented methylprednisolone and antithymocyte globulin (ATG) J. Med. Virol. 1999;58:182–187. 10.1002/(SICI)1096-9071(199906)58:2%3C182::AID-JMV14%3E3.0.CO;2-Q [PubMed] [Google Scholar]
- Ho M. Advances in understanding cytomegalovirus infection after transplantation. Transplant. Proc. 1994;26:7–11. [PubMed] [Google Scholar]
- Hoover D.R, Peng Y, Saah A, Semba R, Detels R.R, Rinaldo C.R, Jr, Phair J.P. Occurrence of cytomegalovirus retinitis after human immunodeficiency virus immunosuppression. Arch. Ophthalmol. 1996;114:821–827. doi: 10.1001/archopht.1996.01100140035004. [DOI] [PubMed] [Google Scholar]
- Kalejta R.F, Shenk T. Manipulation of the cell cycle by human cytomegalovirus. Front. Biosci. 2002;7:295–306. doi: 10.2741/kalejta. [DOI] [PubMed] [Google Scholar]
- Lilley B.N, Ploegh H.L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. 10.1038/nature02592 [DOI] [PubMed] [Google Scholar]
- Lilley B.N, Ploegh H.L, Tirabassi R.S. Human cytomegalovirus open reading frame TRL11/IRL11 encodes an immunoglobulin G Fc-binding protein. J. Virol. 2001;75:11 218–11 221. doi: 10.1128/JVI.75.22.11218-11221.2001. 10.1128/JVI.75.22.11218-11221.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.C, Cumberland W.G, Minassian S.L, Ransome S.S, Cornish M.J, Terry B.G, Holland G.N. Decreased macular leukocyte velocity in human immunodeficiency virus-infected individuals. Am. J. Ophthalmol. 2001;132:711–719. doi: 10.1016/s0002-9394(01)01201-6. 10.1016/S0002-9394(01)01201-6 [DOI] [PubMed] [Google Scholar]
- Limaye A.P, Huang M.L, Leisenring W, Stensland L, Corey L, Boeckh M. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 2001;183:377–382. doi: 10.1086/318089. 10.1086/318089 [DOI] [PubMed] [Google Scholar]
- Ljungman P, Griffiths P.D, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin. Infect. Dis. 2002;34:1094–1097. doi: 10.1086/339329. 10.1086/339329 [DOI] [PubMed] [Google Scholar]
- Mann M, et al. Lack of clinical utility of bronchoalveolar lavage cultures for cytomegalovirus in HIV infection. Am. J. Respir. Crit. Care Med. 1997;155:1723–1728. doi: 10.1164/ajrccm.155.5.9154883. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Taylor E, Pande H, Forman S.J, Tanamachi B, Li C.R, Zaia J.A, Greenberg P.D, Riddell S.R. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J. Med. Virol. 1994;43:103–110. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- Mocarski E.S. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 2002;10:332–339. doi: 10.1016/s0966-842x(02)02393-4. 10.1016/S0966-842X(02)02393-4 [DOI] [PubMed] [Google Scholar]
- Nichols W.G, Boeckh M. Recent advances in the therapy and prevention of CMV infections. J. Clin. Virol. 2000;16:25–40. doi: 10.1016/s1386-6532(99)00065-7. 10.1016/S1386-6532(99)00065-7 [DOI] [PubMed] [Google Scholar]
- Ontanon J, Muro M, Garcia-Alonso A.M, Minguela A, Torio A, Bermejo J, Pons J.A, Campos M, Alvarez-Lopez M.R. Effect of partial HLA class I match on acute rejection in viral pre-infected human liver allograft recipients. Transplantation. 1998;65:1047–1053. doi: 10.1097/00007890-199804270-00007. 10.1097/00007890-199804270-00007 [DOI] [PubMed] [Google Scholar]
- Regoes R.R, Staprans S.I, Feinberg M.B, Bonhoeffer S. Contribution of peaks of virus load to simian immunodeficiency virus pathogenesis. J. Virol. 2002;76:2573–2578. doi: 10.1128/jvi.76.5.2573-2578.2002. 10.1128/jvi.76.5.2573-2578.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm A, Engelsberg A, Tortorella D, Korner I.J, Lehmann I, Ploegh H.L, Hopken U.E. Human cytomegalovirus gene products US2 and US11 differ in their ability to attack major histocompatibility class I heavy chains in dendritic cells. J. Virol. 2002;76:5043–5050. doi: 10.1128/JVI.76.10.5043-5050.2002. 10.1128/JVI.76.10.5043-5050.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacre K, et al. Repertoire, diversity, and differentiation of specific CD8 T-cells are associated with immune protection against human cytomegalovirus disease. J. Exp. Med. 2005;201:1999–2010. doi: 10.1084/jem.20042408. 10.1084/jem.20042408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer P, Tenschert W, Cremaschi L, Schroter M, Zollner B, Laufs R. Area under the viraemia curve versus absolute viral load: utility for predicting symptomatic cytomegalovirus infections in kidney transplant patients. J. Med. Virol. 2001;65:85–89. 10.1002/jmv.2005 [PubMed] [Google Scholar]
- Shinkai M, Bozzette S.A, Powderly W, Frame P, Spector S.A. Utility of urine and leukocyte cultures and plasma DNA polymerase chain reaction for identification of AIDS patients at risk for developing human cytomegalovirus disease. J. Infect. Dis. 1997;175:302–308. doi: 10.1093/infdis/175.2.302. [DOI] [PubMed] [Google Scholar]
- Spector S.A, Wong R, Hsia K, Pilcher M, Stempien M.J. Plasma cytomegalovirus (CMV) DNA load predicts CMV disease and survival in AIDS patients. J. Clin. Invest. 1998;101:497–502. doi: 10.1172/JCI1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire S.B, Lipman M.C, Bagdades E.K, Mulvenna P.M, Grundy J.E, Griffiths P.D, Johnson M.A. Severe cytomegalovirus pneumonitis in HIV infected patients with higher than average CD4 counts. Thorax. 1992;47:301–304. doi: 10.1136/thx.47.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylwester A.W, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005;202:673–685. doi: 10.1084/jem.20050882. 10.1084/jem.20050882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirabassi R.S, Ploegh H.L. The human cytomegalovirus US8 glycoprotein binds to major histocompatibility complex class I products. J. Virol. 2002;76:6832–6835. doi: 10.1128/JVI.76.13.6832-6835.2002. 10.1128/JVI.76.13.6832-6835.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella D, Gewurz B.E, Furman M.H, Schust D.J, Ploegh H.L. Viral subversion of the immune system. Annu. Rev. Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. 10.1146/annurev.immunol.18.1.861 [DOI] [PubMed] [Google Scholar]