Abstract
Mitochondria originated by permanent enslavement of purple non-sulphur bacteria. These endosymbionts became organelles through the origin of complex protein-import machinery and insertion into their inner membranes of protein carriers for extracting energy for the host. A chicken-and-egg problem exists: selective advantages for evolving import machinery were absent until inner membrane carriers were present, but this very machinery is now required for carrier insertion. I argue here that this problem was probably circumvented by conversion of the symbiont protein-export machinery into protein-import machinery, in three phases. I suggest that the first carrier entered the periplasmic space via pre-existing β-barrel proteins in the bacterial outer membrane that later became Tom40, and inserted into the inner membrane probably helped by a pre-existing inner membrane protein, thereby immediately providing the protoeukaryote host with photosynthesate. This would have created a powerful selective advantage for evolving more efficient carrier import by inserting Tom70 receptors. Massive gene transfer to the nucleus inevitably occurred by mutation pressure. Finally, pressure from harmful, non-selected gene transfer to the nucleus probably caused evolution of the presequence mechanism, and photosynthesis was lost.
Keywords: mitochondria, evolution, protein-targeting, α-proteobacteria, gene transfer, membrane carriers
1. Introduction
Mitochondria evolved from endosymbiotic purple non-sulphur bacteria (α-proteobacteria; John & Whatley 1975, 1977; Andersson et al. 2003), but precisely how has been unclear. The nature of the host and the initial selective advantage of intracellular enslavement have been debated (Martin & Müller 1998; Cavalier-Smith 2002a; Andersson et al. 2003). Less attention has been given to the mechanisms of symbiont-to-organelle conversion (Cavalier-Smith 1983, 1987), which centrally involved novel protein-import mechanisms (Cavalier-Smith & Lee 1985). Origin of the five macromolecular complexes responsible for import (Endo et al. 2003; Wiedemann et al. 2004a; Habib et al. 2005), with about 50 different proteins, is the most fundamental aspect of mitochondrial origins still wanting satisfactory explanation. Here I explain the evolutionary origin of mitochondrial protein-targeting machinery, building on recent discoveries of how components are themselves targeted (Endo et al. 2003; Pfanner et al. 2004; Wiedemann et al. 2004a; Habib et al. 2005) and principles of membrane heredity (Cavalier-Smith 2000, 2004b). The origin of mitochondria involved acquisition of a foreign genome and, more importantly, a foreign membranome (Cavalier-Smith 2004b; the negibacterial envelope: Cavalier-Smith 1983), from which evolved two novel genetic membranes, i.e. membranes arising by growth and division of membranes of the same type (Cavalier-Smith 2004b).
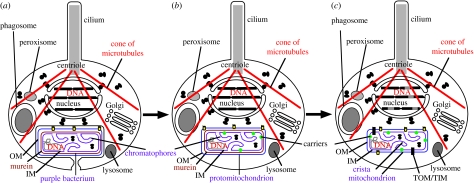
I shall argue that the first enslavement step was uptake of a host carrier protein through the outer membrane (OM) and its insertion into the inner, cytoplasmic membrane (IM) of a photosynthetic purple bacterium that escaped into the host cell's cytoplasm from the food vacuole into which it was initially phagoctyosed (figure 1a,b). Carrier insertion was then made efficient by inserting novel proteins from the host cytosol into the protomitochondrial OM and IM. Finally, massive transfer to the nucleus of symbiont genes so burdened the cell, by adding unneeded or harmful proteins, that an extra mechanism of protein insertion using removable presequences evolved to target their proteins back into mitochondria (figure 1c). This stimulated divergence between host and symbiont ribosomes (Cavalier-Smith 2002a) and coevolutionary divergence of protein-insertion mechanisms involving N-terminal signal peptides for endoplasmic reticulum (ER) and presequences into mitochondria. Improvements in efficiency (see Cavalier-Smith 1987, 2002a) then dramatically reduced protomitochondrial genome size and eliminated its photosynthetic ability, to optimize respiration.
Figure 1.
Origin of mitochondria by permanent internal cell enslavement. (a) Phagocytosis of a photosynthetic purple non-sulphur bacterium (α-proteobacterium) placed it inside the cytoplasm of a protoeukaryote host. Though shown with a cilium, cytoskeleton, endomembrane system, nucleus and peroxisomes, these organelles were probably still actively coevolving in this stem eukaryote, only acquiring their full complement of modern properties during conversion of the purple bacterium into a mitochondrion. (b) The phagosomal membrane failed to fuse with lysosomes, was broken and lost, allowing the purple bacterium to multiply freely in the cytosol of the host that was able to use any photosynthesate leaking from it. Pre-existing outer membrane (OM: blue) proteins, e.g. porins (yellow), allowed host carrier proteins (green: probably arising by gene duplication of a peroxisomal carrier) to enter the bacterial periplasmic space and spontaneously insert into its inner membrane (IM: purple). By extracting photosynthesate for itself, and providing the bacterium with CO2 and minerals, e.g. phosphate, sulphate, the phagotrophic host established a mutualistic endosymbiosis. A pre-existing OM protein evolved into the core protein (Tom40) of the protein translocator of the premitochondrial OM (TOM), allowing numerous other proteins to be inserted to improve small molecule exchanges across its envelope. (c) Following transfer of duplicates of much of the protomitochondrial genome (grey) to the nucleus and integrating them into nuclear DNA, Tim23 IM translocons and OM presequence receptors evolved to retarget many proteins coded by them back into the protomitochondrion, where they would be beneficial, not harmful or wasted. Loss of such genes and others essential for free-living life (Boussau et al. 2004) from the mitochondrial genome permanently enslaved the mitochondrion. Its peptidoglycan murein and genes needed for photosynthesis, but not respiration, were lost during this major streamlining and efficiency increase prior to the last common ancestor (cenancestor) of all eukaryotes.
2. An initially photosynthetic slave
First discussions of the symbiogenetic origin of mitochondria from purple non-sulphur bacteria (John & Whatley 1975, 1977) assumed that the host was anaerobic, and that the symbiont was an aerobically respiring heterotroph, enslaved because of the greater efficiency of ATP production by oxidative phosphorylation that it provided for the first time. However, it is more likely that host and symbiont were both facultative aerobes (Cavalier-Smith 2002a); if the host already had oxidative phosphorylation, the ultimate selective advantage was greater efficiency derived from respiratory and metabolic compartmentation (Cavalier-Smith 2002a). I now argue that the immediate selective advantage for initial enslavement would have been far stronger if the symbiont were not heterotrophic, but photosynthetic instead, with immediate intracellular synergy between a respiring and phagotrophic host (using oxygen and excreting CO2) and a photosynthetic symbiont fixing that CO2 (as in intracellular photosynthetic dinoflagellates in corals). Many photosynthetic microbes exude photosynthesate into the medium: marine phytoplankton excrete 1–40% of total photosynthesate (Smith & Wiebe 1976), cyanobacteria secrete vitamins (Aaronson et al. 1977), and α-proteobacteria secrete glycollate under aerobic conditions (Codd & Turnbull 1975), suggesting that a photosynthetic intracellular ancestor of mitochondria could have established a synergistic symbiosis with host peroxisomes analogous to that in plant photorespiration where glycollate exuded by plastids is recycled by peroxisomes (Cavalier-Smith 1987). Intracellularly cultivating such a strain would immediately benefit the host even prior to the difficult evolution of protein import into it, unlike in the classical heterotrophic symbiont theory. Once mutualistic endosymbiosis was established, mutations giving the consortium a reproductive advantage would be selected. This scenario is plausible, as photosynthetic purple non-sulphur bacteria endosymbiotic in eukaryotic hosts are known (Fenchel & Bernard 1993). A second merit of a photosynthetic symbiont is that their chromatophores (invaginations from the inner membrane that house the dual respiratory/photosynthetic machinery: figure 1a,b) are obvious precursors of mitochondrial cristae, absent in heterotrophic α-proteobacteria like Paracoccus (Cavalier-Smith 2002a). The idea that the symbiont was originally photosynthetic is not new (see Woese 1977 and Searcy 1992), but the reasons for now favouring it are. The 31 most conserved mitochondrial genes group more closely with phototrophic Rhodospirillum rubrum (with tubular chromatophores, the ancestral morphology for cristae given a eukaryote root between unikonts and bikonts: Richards & Cavalier-Smith 2005) than any heterotroph (Esser et al. 2004). I discuss reasons for the loss of photosynthesis at the end of the paper.
3. Carrier evolution
A major difference between a respiring heterotroph like Paracoccus and mitochondria is the presence in the mitochondrial IM of the ATP/ADP carrier protein (AAC) that exchanges ATP and ADP between the cytosol and mitochondrial matrix (John & Whatley 1975). The heterotrophic symbiont theory assumed that enslavement was initiated by inserting AAC into the IM (Cavalier-Smith 1983, 1987). The present photosynthetic symbiont theory is less restrictive, allowing the first enslavement step to be the insertion of any mitochondrial IM carrier able to extract photosynthesate for the host. This would increase the probability of the first step by over an order of magnitude: 18 such mitochondrial carriers have so far been identified (Fiermonte et al. 2004); yeast has 35 genes for this family. The oxaloacetate–sulphate antiporter could extract oxaloacetate from the proteobacterium as carbon and energy source for the host, in exchange for cheap sulphate that the symbiont would require for growth but not be able to obtain for itself. The ATP/phosphate antiporter (Fiermonte et al. 2004) could extract ATP in exchange for phosphate, essential for the symbiont, and get fixed carbon in addition to ATP high-energy phosphate. Either carrier would have benefited the host more than the AAC. Thus the first problem in mitochondrial evolution is the source of these six-helix IM carriers, all belonging to the same family, which lacks close prokaryote relatives (but has more distant ones with 12 helices). The only known family member not located in mitochondria, the ATP importer of peroxisomes (van Roermund et al. 2001), could have generated the first mitochondrial carrier by gene duplication, and subsequent gene duplications would have populated the IM with its present carrier diversity. (A more distant family of six-helix chloroplast envelope IM carriers could not be ancestors of the mitochondrial ones, as chloroplasts originated later (Cavalier-Smith 2000, 2003a); they probably arose by gene duplication of a mitochondrial carrier.)
Intracellular endosymbiosis, probably initiated by primitive phagotrophy (Cavalier-Smith 1987, 2002a), was an essential prerequisite for carrier insertion; extracellular syntrophy, sometimes postulated (Martin & Müller 1998), would not have helped; carrier insertion would probably not be mechanistically possible—if it occurred it would be disadvantageous by extruding proteobacterial metabolites into the environment, not the host. The hypothesis that the host was an anaerobic methanogenic archaebacterium (Martin & Müller 1998), not a facultatively aerobic, phagotrophic protoeukaryote, is untenable, as previously explained (Cavalier-Smith 2002a).
4. Negibacterial origin of TOM40, SAM50 and TIM22
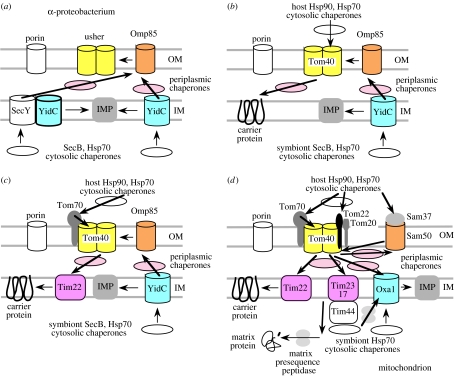
IM carriers enter mitochondria through the OM β-barrel protein pore Tom40, helped by the OM receptor Tom70, periplasmic chaperones and the IM protein-inserter Tim22 complex (Endo et al. 2003; Pfanner et al. 2004; Wiedemann et al. 2004a; figure 2d). Tom40 evolves sufficiently slowly for homologues to be detected in all eukaryotes but not slowly enough for its prokaryote ancestor to be unambiguously identified. I suggest that it evolved from a proteobacterial β-barrel protein like usher (Li et al. 2004), which secretes pilus proteins using periplasmic chaperones; usher and Tom40 are the only known β-barrel proteins with two pores (Li et al. 2004); a relative of both was probably already present in the proteobacterial ancestor of mitochondria (figure 2a). As soon as a new carrier evolved in the host cytosol, it could have entered the periplasm through this pore and interact with pre-existing periplasmic chaperones; in the absence of Tim22 it must either have inserted itself into the IM or, more likely, inserted with the help of pre-existing YidC (figure 2b) or another IM protein (see below). Even if import and insertion were inefficient and even if insertion were random with half the carriers entering the OM, it would supply photosynthesate to the host and initiate enslavement by providing a selective advantage for improving carrier import.
Figure 2.
Origin of the mitochondrial protein-import systems (d) from the ancestral α-proteobacterial protein-export systems (a) The outer membrane (OM) β-barrel proteins (Sam50, Tom40, porins) all evolved directly from symbiont OM proteins and Oxa1/2 evolved from the inner membrane (IM) protein YidC. (b) Enslavement was initiated by the insertion of novel carrier proteins able to enter through Tom40 with the help of pre-existing host and periplasmic chaperone proteins and insert into the IM. The first carrier, possibly descended from the peroxisomal ATP importer, exported ATP or photosynthesate to the host and generated over 30 other carriers by gene duplication. Their import was made efficient by evolution of receptor Tom70, possibly of cyanobacterial origin by lateral gene transfer, inserting by its N-terminal membrane-spanning α-helix, and Tim22 entering via the Tom40 pore (c) Following massive transfer of genes from symbiont to nucleus, the presequence mechanism evolved by gene duplication of Tim22 to Tim23 and Tim17 to generate the inner membrane translocase and the addition of Tom20 and Tom22 to recognize the hydrophobic and positively charged parts, respectively, of the presequences, of Tim44 to transfer the symbiont/matrix chaperone Hsp70 more efficiently onto the emerging preproteins and a matrix peptidase to remove their presequences. Pre-existing periplasmic chaperones diversified and adapted to prevent periplasmic aggregation and improve transfer between membrane–protein complexes, and various less key proteins were added to each to increase stability and improve transfer rates and efficiency. Sec machinery was retained by the excavate jakobid protozoa (Gray et al. 2004), but lost by other mitochondria. Acquisition of presequences by approximately 1000 proteobacterial genes transferred to the nucleus (Esser et al. 2004; greater similarity of many of these to γ- not α-proteobacteria may be an artefact stemming from major divergence during mitochondriogenesis) allowed loss of their symbiont versions and huge mitochondrial genome reduction, raising efficiency by increasing space for matrix enzymes and sparing nutrients and energy previously wasted on multiple copies of their DNA. IMP; inner membrane proteins other than carriers and translocons.
Presence of β-barrel proteins in the OM of mitochondria (e.g. Tom40, Sam50 and porins) and chloroplasts (e.g. Toc75 homologue of Sam50 and their joint negibacterial ancestor Omp85) unequivocally proves that the OM evolved from the bacterial OMs (Cavalier-Smith 1982, 1983), not from the host food vacuole membrane (Schnepf 1964) as many textbooks still suppose, as such membrane proteins are unknown elsewhere in eukaryotes or in their unibacterial relatives: β-barrel membrane proteins are confined to OMs of negibacteria (e.g. proteobacteria, cyanobacteria), chloroplasts, and mitochondria (Schleiff & Soll 2005). I call the mitochondrial ‘intermembrane space’ periplasm (Cavalier-Smith 1983) as it is homologous with bacterial periplasm: I suggest that small Tim mitochondrial periplasmic chaperones evolved from bacterial periplasmic chaperones and that their genes were transferred to the nucleus after Tom40 mechanisms improved by adding Tom5, possibly before Tom70 and Tom20 receptors were added, as they do not use them (Kurz et al. 1999); their mitochondrial gene copies could have been lost once their own import was improved by evolution of Mia40 to help them enter via Tom40 (Chacinska et al. 2004). Similar genic transfer occurred for Sam50, the SAM complex core (Wiedemann et al. 2004a; the TOB complex: Habib et al. 2005) that probably inserts all β-barrel proteins into the mitochondrial OM, including itself (Schleiff & Soll 2005). Comparative sequence and experimental evidence indicates that Sam50 evolved from the negibacterial β-barrel inserter Omp85 (Gentle et al. 2005). This insertion also depends on periplasmic chaperones, which now enter via TOM (Kurz et al. 1999). Although additional proteins were later added from the cytosol to TOM and SAM by insertion mechanisms not requiring passage through Tom40 (Ahting et al. 2005), core proteins of each complex came directly from the symbiont, as predicted (Cavalier-Smith 1987), not the host (Andersson et al. 2003). Most additional proteins (e.g. Tom20, Tom22, Sam37) evolve too fast to determine whether they are of host or symbiont origin. It would be simplest if most came from the host, making the OM chimaeric in origin (Cavalier-Smith 1987).
Tim22 is weakly homologous to LivH, a polytopic cytoplasmic membrane leucine importer of proteobacteria (Rassow et al. 1999), and therefore evolved from a pre-existing inner membrane proteobacterial permease preadapted to import amino acids, not proteins. The even more closely related OM protein OEP16 of chloroplasts probably evolved from Tim22 after it diverged from Tim23/17 (they group on a tree: Rassow et al. 1999). OEP16, unlike bacterial OM proteins, is not a β-barrel, but has four membrane-spanning α-helices (Linke et al. 2004). It probably evolved from Tim22 by gene duplication and insertion into the OM from the cytosol, another example of recruitment of a pre-existing mitochondrial import component for chloroplast import, as postulated (Cavalier-Smith 1982). Possibly LivH, rather than YidC, helped carrier insertion from the outset.
5. Lateral transfer of the TOM70 receptor?
One key protein, Tom70, the receptor for importing carriers and Tim22, probably came neither from host nor symbiont, as I find its only strong Blast hits are to cyanobacteria. As suggested previously (Cavalier-Smith 1987), during the origin of mitochondria the host possibly also harboured symbiotic cyanobacteria, using its waste CO2 and providing oxygen for the mitochondrion. Such intracellular synergy could have allowed this phagotrophic/photosynthetic consortium to out-compete other protoeukaryotic competitors. However, cyanobacteria were not themselves successfully enslaved until somewhat later, by a bikont host after the primary unikont/bikont bifurcation of eukaryotes (Cavalier-Smith 2003a; Richards & Cavalier-Smith 2005). Acquisition of Tom70 from a cyanobacterial symbiont or cyanobacterial food, and its insertion by its N-terminal helical hydrophobic tail, would improve efficiency of carrier import and allow import of other IM proteins with a sufficiently similar structure that it could recognize, notably Tim22—probably initially selected to increase specificity and speed of carrier insertion into the IM (figure 2c). Tim22, strikingly, acts as receptor, pore and energy transducer for insertion—all-in-one (Rehling et al. 2003); adding it alone would give a specific insertion mechanism for multitopic IM proteins. With a basically efficient mechanism for importing carriers (figure 2c), extra proteins could be added to Tom40 to increase stability (Tom6, Tom7) and similarly to Tim22 (e.g. Tim54, which evolves too fast to be sure that it was present in the cenancestor and not an opisthokont invention).
The selective advantage of the first two phases of protein-import evolution is clear: carrier insertion (figure 1b) and efficiency improvement (figure 1c). But what was the selective advantage of evolving the presequence mechanism, with Tom20 and Tom22 receptors, Tim23 IM translocase, novel adaptor (Tim44; Wiedemann et al. 2004a) to transfer translocated proteins to the pre-existing matrix Hsp70 chaperone (together constituting the preprotein translocase-associated motor; Wiedemann et al. 2004a) and matrix peptidase to remove the presequence after import (Wiedemann et al. 2004a) (figure 1c)? This has often been considered the first step in evolution of mitochondrial protein import, but this makes no evolutionary sense as the presequence machinery is inessential for carrier import and too complex to have evolved initially. Even after evolution of efficient carrier import could the cell immediately gain enough benefit by importing any single matrix protein to give sufficient selective impetus for evolving such complex machinery? It would already have everything inside the mitochondrion needed for efficient respiration and metabolism; simple loss by accidental deletion could remove all proteobacterial genes not needed as an enslaved organelle (Boussau et al. 2004) and reduce its genome size about fourfold. But the 1000 or so remaining genes could not be deleted until after they were transferred to the nucleus and the preprotein-import machinery evolved to import their proteins. There would be undoubted eventual future benefit from this by reducing space, material, and energy devoted to DNA transcription and translation in the multiple mitochondria compared with the single nucleus (Cavalier-Smith 1987, 2003b). But natural selection is not prescient. One cannot evolve complex machinery only for a distant conditional future benefit.
6. Gene transfer pressure and protein import
I therefore now propose a novel explanation for the origin of this part of the import machinery: the immediate benefit was to correct the immense phenotypic damage that would inevitably be done by massive transfer of a thousand or more mitochondrial genes to the nucleus. Such transfer has itself never been a problem for understanding symbiogenesis in the sense of being a mechanistic limitation, for even today there is a continual rain of mitochondrial and chloroplast genes into the nucleus (Adams & Palmer 2003; Ricchetti et al. 2004). Once the protomitochondrion was permanently enslaved by evolving efficient carrier import, copies of all its genes would inevitably sooner or later be transferred to the nucleus; as protoeukaryotes had only recently evolved from a posibacterium via a neomuran missing link (Cavalier-Smith 2002b), most of these could probably be transcribed and translated. No selective pressure is needed for such massive transfer. But consider the phenotypic consequences. It is well known to genetic engineers that a significant fraction of randomly chosen foreign genes kill Escherichia coli in high expression plasmids or dramatically lower growth rates. Massive gene transfer to the nucleus would inevitably impose a huge genetic and phenotypic load on the host.
For example, proteobacterial genes for IM proteins would have signal sequences recognizable by host signal recognition particles (SRP) and thus be wastefully or disruptively inserted into host ER after their genes were transcribed intranuclearly. Modifying their hydrophobic signal sequences by making one face positively charged would prevent binding to SRP and insertion into ER and turn them into proto-presequences for mitochondrial import. Only a few bacterial signal sequences are recognizable by TOM (Mukhopadhyay et al. 2005). Addition of Tom22 to the Tom40 complex to recognize the positive charge would enable targeting to the mitochondrial periplasm instead and self-insertion by their presequence into the inner membrane. Multispanning IM proteins like Oxa1 (a mitochondrial relative of proteobacterial YidC, needed for protein insertion from the mitochondrial matrix: figure 2d; whether Oxa1/2 were derived from proteobacterial YidC (Preuss et al. 2005) or a host version of the shorter homologue found in Posibacteria (Tjalsma et al. 2003) is unclear) would be inserted by Tim23; the fact that they transfer directly from TOM to Tim23 without needing periplasmic soluble Tims (Frazier et al. 2003) is consistent with their initial insertion from the cytosol being evolutionarily later than for carriers. I suggest that Tom22 had already been added to Tom40 to increase its stability and/or help carrier import and that it already by chance had a somewhat negatively charged groove on its cytosolic domain preadapted to recognize the positive face of a presequence (mitochondrial presequences have one hydrophobic and one hydrophilic positively charged face (Wiedemann et al. 2004a)). If so, no innovation was necessary in the import machinery that evolved for IM proteins without presequences to allow recognition of presequence-containing IM proteins by Tom22 or their inefficient self-insertion into the IM. Any such protein made in the host cytosol after gene transfer, with a mutation modifying its pre-existing signal sequence by substituting a positively charged for a hydrophobic amino acid, would be correctly retargeted into the IM. The benefit would be greatest for membrane proteins that were most harmful in the ER. After this mechanism became efficient any protein could be retargeted thus, even if the selective advantage was very slight (or indeed absent), by chance mutations of its signal sequence into a presequence.
This phenotypic rescue theory has the same logic as explanations for the origin of RNA editing, spliceosomal introns and the elimination of internal sequences in ciliate macronuclei (Cavalier-Smith 1993a, 2004a; Covello & Gray 1993): harmful mutations generated by extreme mutation pressure can be corrected phenotypically more easily than removing the source of the mutation pressure. In this case, having enslaved the proteobacterium and become totally dependent on it (possibly by then having lost its own respiratory machinery), the host could escape such damaging gene-transfer pressure of many hundreds of gene products only by evolving a generalized protein-import mechanism to return them to their alien donor: the protomitochondrion. This rain of alien genes probably influenced other features of host evolution. It may have forced modifications onto host ribosomes and SRPs to reduce the chances of translating and inserting proteobacterial proteins into the ER or, just as bad, of host ER/secretory proteins into the mitochondrion—a further reason additional to those identified previously (Cavalier-Smith 2002b) why protoeukaryote ribosomes and SRPs diverged so markedly from their archaebacterial sisters. There was probably coevolutionary divergence of the host signal and presequence mechanisms.
Presequence import was probably improved by adding Tom20 to recognize the hydrophobic face of presequences. Being more loosely bound to Tom40 than is Tom22, it could spread around the mitochondrial surface, catching potential importees and bringing them to Tom22/Tom40 for import. Tom20 itself probably evolved from a (host or symbiont) protein with a signal sequence that became its membrane-anchor helix: this helix is recognized by SRP but prevented from ER insertion by a downstream sequence (Kanaji et al. 2000). Efficient import of pre-sequence-bearing IM proteins into the periplasm would prevent harm in the ER, but not directly help mitochondrial function; if inserted into the IM by the presequence they would have the wrong polarity or they might insert instead into the inner side of the OM. However, their partial extrusion into the matrix by Tim22 would allow association with the proteobacterial chaperone Hsp70, which could pull them further, allowing reinsertion into the membrane with the same polarity as when encoded by protomitochondrial genes and thus normal function. Once this began, however crudely, it could have been improved by gene duplication of Tim22 to generate a protoTim23/17 complex (both homologues of Tim22: Meier et al. 2005) and then addition of Tim21 to foster direct binding to TOM, bypassing periplasmic chaperones and Tim44 to increase transfer efficiency to mtHsp70. This would yield efficient generalized machinery for import of any proteobacterial IM protein whose nuclear gene copy mutated to form a presequence and was otherwise compatible with entry without clogging the machinery. Once a protein encoded by a transferred gene was efficiently imported, the original mitochondrial gene copy could be lost. Only two membrane proteins (cytochrome b and Cox1) never achieved this transition (Gray et al. 2004); most did so in the eukaryote cenancestor, but some by chance did so only in certain later diverging lineages. In this way, I suggest, the so-called ‘conservative’ (but in part very innovative) pathway of IM protein import was born.
Inevitably many transferred nuclear genes for matrix proteins would thereafter accidentally acquire presequences and be targeted to the matrix; some proteins are predisposed to have presequences (Lucattini et al. 2004), but that is irrelevant to the initial origin of targeting if that involved only carrier targeting—and the presequence mechanism evolved last. Sooner or later a duplicate of a matrix (or host) signal peptidase would be similarly accidentally retargeted. Any presequence it could recognize would be cleaved, generating a soluble protein; random mutation and selection would bring other matrix presequences into line, rendering all cleavable. As each protein's presequence became efficient, deletion of the mitochondrial version of the gene would immediately be favoured by selection for efficiency, as previously explained (Cavalier-Smith 1987). Gene by gene the mitochondrial genome would diminish, eventually entirely disappearing from the anaerobic hydrogenosomes and mitosomes, which still retain both OM and IM and complex protein-import machinery (Embley et al. 2003). As a consequence of this complicated history, the IM and OM became self-perpetuating, independently of the genomes they once harboured. Contrary to widespread assumptions (Allen et al. 2005), it is unnecessary to postulate special reasons besides difficulty of protein reimport, plus historical accident (Cavalier-Smith 2002b), why most mitochondrial lineages failed to transfer all vital proteins to the nucleus and similarly lose local genomes (de Grey 2005). My argument that presequence-based import of IM proteins preceded that of matrix proteins is consistent with the greater complexity of the latter, additionally requiring Pam16 (Frazier et al. 2004), I suggest added last of all.
7. Broader implications
The molecular determinants of membrane identity that make the mitochondrial OM and IM genetic membranes distinct from others in the eukaryote cell and from their proteobacterial ancestors are the self-insertion abilities, respectively, of the TOM and Tim22 complexes. In each case protein self-complementarity must enable each to recognize and insert itself: the basic principle of membrane heredity (Cavalier-Smith 2000, 2004b), just as nucleotide complementarity is of nucleic acid inheritance. Since these membrane-identity determinants are encoded by DNA, DNA heredity is also essential for membrane heredity. But the converse is equally true for all cells: without membrane heredity mediated by such molecules or the ER Sec61 receptor and Snares in the Golgi and plasma membrane (Cavalier-Smith 2004b), DNA heredity would be impossible for cells. Thus, ever since the last common ancestor of life, DNA heredity and membrane heredity have worked in parallel (Cavalier-Smith 2004b). But it is the origin of the novel membrane heredity of the IM and OM and the insertion mechanisms for IM carriers that are keys to understanding the origin of mitochondria, not mitochondrial genomes (Gray et al. 2004), which do not encode any of these mechanisms. However, as I have tried to show, gene transfer pressure from the protomitochondrial genome probably shaped the third phase of protein-targeting evolution. Much cell megaevolution can only be understood by considering the interplay of genomes and the membranome and of both with the cytoskeleton (Cavalier-Smith 2002a, 2004a). At some stage the proteobacterial peptidoglycan murein was lost; its FtsZ division mechanism for the inner membrane (Kiefel et al. 2004) became supplemented by eukaryotic dynamin for the OM (Nishida et al. 2004).
I have concentrated here on the origin of mitochondria, not their diversification, but must place it in the context of recent evidence that the root of the eukaryote phylogenetic tree lies between unikonts (animals, fungi, Choanozoa, Amoebozoa) and bikonts (plants, chromists, all other protozoa; Stechmann & Cavalier-Smith 2002, 2003; Richards & Cavalier-Smith 2005). This position of the root means that the last common ancestor of all eukaryotes had well-developed aerobic mitochondria. All known groups of anaerobic eukaryotes (protists or fungi) have highly modified mitochondria that have lost oxidative phosphorylation and usually also their genomes, but retain both membranes and their targeting mechanisms as well as synthetic machinery for making iron–sulphur clusters (Embley et al. 2003). Contrary to earlier ideas, these anaerobic lineages all evolved secondarily from aerobic ancestors with oxidative phosphorylation (Embley et al. 2003; van der Giezen et al. 2005). None diverge early on the eukaryote tree (Stechmann & Cavalier-Smith 2002, 2003; Embley et al. 2003; Richards & Cavalier-Smith 2005). There is now no reason to think that there were ever primitively amitochondrial, but otherwise fully developed, eukaryotes. All anaerobic mitochondria arose secondarily from aerobic ones by polyphyletic gene losses (van der Giezen et al. 2005). The subclass of anaerobic mitochondria called hydrogenosomes generates ATP and molecular hydrogen by pyruvate ferredoxin oxidoreductases (PFOR) and hydrogenase (Embley et al. 2003). Both enzymes appear monophyletic within eukaryotes (Embley et al. 2003; van der Giezen et al. 2005), even though hydrogenosomes are found in two unikont and many bikont groups and clearly arose polyphyletically. The simplest way of reconciling both conclusions is if ancestral eukaryotes alternated between aerobic and anaerobic growth. Under aerobic conditions their mitochondria did oxidative phosphorylation. Under anaerobic conditions they generated energy like a hydrogenosome. Comparably versatile protozoa or lower fungi may still exist, and ought to be sought, but all well-studied mitochondria descended from more specialist lineages that lost either PFOR and hydrogenase to become aerobic mitochondria or else lost cytochromes to become either hydrogenosomes or mitosomes. Mitosomes are still more reduced, minute anaerobic mitochondrial derivatives that evolved polyphyletically and are retained only for making iron–sulphur centres (Regoes et al. 2005; van der Giezen & Tovar 2005; van der Giezen et al. 2005), essential for all eukaryotes. Entamoeba mitosomes lost all membrane carriers but one (Chan et al. 2005).
Enslavement of a proteobacterium to make mitochondria probably began as soon as phagocytosis was perfected and peroxisomes originated, at which time the nucleus was itself formed with novel pore complexes created by proteins related to those of the coated vesicle machinery that generated the endomembrane system (Devos et al. 2004) and of intraciliary transport particles of cilia (Jekély & Arendt 2006), which probably co-originated with the nucleus and mitosis (Cavalier-Smith 2002a). However, the last phase of mitochondrial evolution (massive gene transfer to the nucleus) probably post-dated evolution of the nuclear envelope, at least slightly. This is because spliceosomal introns probably evolved from group II introns supplied by the mitochondrial ancestor as an incidental consequence of this gene transfer to the nucleus (Cavalier-Smith 1991). As such introns are spliced very slowly and would be very harmful to the host cell if they evolved in the same compartment as ribosomes, which would probably make proteins truncated by intronic stop sequences (Cavalier-Smith 1991), it is likely that they evolved from self-splicing introns only after the nuclear envelope arose, so the nuclear envelope probably somewhat preceded the major gene transfers.
Finally, why was protomitochondrial photosynthesis lost and respiration retained? The enslaved α-proteobacterium was probably one of many that photosynthesize only under anaerobic conditions. If the host, although a facultative anaerobe, spent at least half its life under aerobic conditions when certain photosynthesis-specific proteins were not expressed, it would be very easy for mutations abolishing photosynthesis to occur during aerobic growth without counterselection until long after they spread considerably. Loss of photosynthesis occurred repeatedly within proteobacteria and among eukaryote algae, so may need no specific explanation. However, as the host was a phagotroph—the first one—it would often have had no shortage of carbon source, for bacterial food can be superabundant. It could have benefited more from increased respiratory efficiency when aerobic than from photosynthesis when anaerobic after evolution of the AAC. Under anaerobic conditions, evolution of PFOR/hydrogenase could enable it to extract some more energy from its prey than by simple glycolysis and have the advantage over photosynthesis of working in the dark. As host and symbiont were probably both facultative aerobes, the optimal environment for the initial enslavement may have been anaerobic/aerobic interfaces or where redox conditions fluctuated unstably between them. As soon as enslavement increased respiration efficiency, its stronger oxygen sink would allow intracellular cultivation also of cyanobacteria; use of their photosynthesate under aerobic conditions could have become more synergistic with mitochondria and peroxisomes than anaerobic photosynthetic contributions by the protomitochondrion. A three way synergy between host, protomitochondrion and symbiotic cyanobacteria could have played a central role in early evolution of both mitochondria and peroxisomes, as suggested previously (Cavalier-Smith 1987); key differences from that earlier proposal are the protomitochondrion being initially photosynthetic, the delay of cyanobacterial-to-plastid conversion (by evolving protein import: Cavalier-Smith 2000) till after the primary eukaryotic divergence into unikonts and bikonts and the autogenous, not symbiogenetic, origin of peroxisomes (Cavalier-Smith 2002a). Thus fluctuating redox levels and fluctuating prey levels could have favoured these changes.
The preceding scenario has obvious implications for the origin of chloroplasts (Cavalier-Smith 1993b, 2000), where similar processes and selective forces were probably at work. A major difference was that enslavement was later, when eukaryote transcriptional and translation machinery had already diverged greatly from that of archaebacteria. This probably made it harder for function of genes transferred into the host nucleus, causing plastids to retain more genes and their ribosomes and protein-insertion machinery to diverge less from their negibacterial ancestors. Only dinoflagellates, which significantly reverted to bacteria-like histone-free nuclear DNA (and evolved novel mechanisms for protein transport across their unusual triple-membrane envelopes) managed to transfer as many plastid genes to the nucleus as did early animals for mitochondria (Cavalier-Smith 2004a). A second major difference is that the pre-existing cyanobacterial OM protein recruited for protein import was Omp85, which became Toc75 (Cavalier-Smith 2000; Schleiff & Soll 2005). As cyanobacterial Omp85 recognizes modern transit sequences (Ertel et al. 2005), Toc75 might originally have been able to recognize them on its own, without help from proteins like Toc139 and Toc34 (Becker et al. 2004); possibly transit sequence recognition evolved more easily and relatively earlier than mitochondrial presequence recognition.
A general feature of the present scenario that space limitations did not permit me to detail is that it well fits the principle that ‘ontogeny recapitulates phylogeny’, i.e. the postulated evolutionary sequence of addition of Tom and Tim subunits generally follows that still observed today, as do their assumed import mechanisms. This scenario therefore solves the chicken and-egg problem of how such complicated machinery evolved, for no major shifts in import mechanism or intermolecular binding properties of the roughly 50 individual proteins comprising the import machinery need have occurred since mitochondria first evolved; molecules added early, e.g. Tim22, do not need receptors putatively added later, e.g. Tom20. Evolution of these key macromolecular complexes is apparently marked by a high degree of conservatism and stasis, despite minor improvements. Specialists will note a few partial exceptions to this that suggest some slight degree of adjustment to ‘early’ mechanisms after later ones were added, but I suggest these simply increased efficiency or rates and were not fundamental. The most important apparent exception to this generalization is Tom40 itself, which is now recognized for import (through other Tom40 pores) by Tom20 and Tom22 (Wiedemann et al. 2004b). Originally it would have been coded by the proteobacterial genome, exported to the periplasm by YidC and into the OM by Omp85 (see figure 2a). Its present requirement for recognition by Tom20 and 22 does not contradict my scenario; it means only that its gene probably remained in the mitochondrion until after Tom20/22 were added to TOM. Such dependence probably could not have evolved if Tom40 came from the host, as sometimes suggested (Andersson et al. 2003). Overall what is striking is that one can formulate a synthesis reflecting known targeting mechanisms and interdependencies, and with selectively and mechanistically plausible intermediate stages. Further work will test the fundamental thesis in more detail and may reveal extra complications.
Note Added In Proof
Perry et al. (2006) suggest that the green plant Tom20 receptor evolved convergently to that of opisthokonts (animals, fungi), by independent origins of presequence binding in different tricopeptide repeat (TPR) paralogues, on the grounds that its transmembrane helix is near the C-terminus not the N-terminus, i.e. its domains are in reverse order compared with opisthokonts. However, the assertion that ‘no genetic mechanisms are known that could generate such a reversal in the order of structural domains’ (Lister & Whelan 2006) is erroneous. Duplications and deletions can easily do so. It is more likely that Tom20 evolved just once, prior to the eukaryotic cenancestor as argued above, and was thus rearranged prior to the green plant cenancestor; a tandem gene triplication followed by four deletions within the cluster could have effected both this rearrangement and the duplication of the single TPR repeat seen in opisthokonts to the double repeat of green plants. That might have been a non-adaptive evolutionary accident. A more interesting possibility is that the symbiogenetic origin of chloroplasts introduced the novel selective advantage of a need for discrimination between mitochondrial presequences and chloroplast transit sequences to prevent chloroplast proteins entering the mitochondrion. Doubling the presequence binding sites per receptor perhaps increased discrimination.
For Tom70, neither a clear evolutionary homologue nor a functional equivalent has been detected in bikonts, though one was recently found in Amoebozoa (Wojtkowska et al. 2005), so Tom70 goes back at least to the ancestral unikont. It is an exaggeration to imply that no sequence-related proteins exist in bikonts (Lister and Whelan 2006; Perry et al. 2006). Toc64 of the plastid OM and mtOM64 of the mitochondrial OM (Chew et al. 2004) have significant sequence similarity to Tom70, but as they have only three, not seven TPR repeats and their functions are unknown it is unclear whether they are orthologous or more distant paralogues.
Sorting out the relationships of the many rapidly evolving divergent eukaryotic TPR paralogues is hard. Functional and Proteomic data are needed for the Tom complex in all bikont supergroups to clarify Tom20 and Tom70 origins.
Acknowledgements
I thank NERC for research grants and NERC and the Canadian Institute for Advanced Research Evolutionary Biology Program for Fellowship support.
References
- Aaronson S, Dhawale S.W, Patni N.J, DeAngelis B, Frank O, Baker H. The cell content and secretion of water-soluble vitamins by several freshwater algae. Arch. Microbiol. 1977;112:57–59. doi: 10.1007/BF00446654. 10.1007/BF00446654 [DOI] [PubMed] [Google Scholar]
- Adams K.L, Palmer J.D. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. 10.1016/S1055-7903(03)00194-5 [DOI] [PubMed] [Google Scholar]
- Ahting U, Waizenegger T, Neupert W, Rapaport D. Signal-anchored proteins follow a unique insertion pathway into the outer membrane of mitochondria. J. Biol. Chem. 2005;280:48–53. doi: 10.1074/jbc.M410905200. [DOI] [PubMed] [Google Scholar]
- Allen J.F, Puthiyaveetil S, Strom J, Allen C.A. Energy transduction anchors genes in organelles. BioEssays. 2005;27:426–435. doi: 10.1002/bies.20194. 10.1002/bies.20194 [DOI] [PubMed] [Google Scholar]
- Andersson S.G, Karlberg O, Canback B, Kurland C.G. On the origin of mitochondria: a genomics perspective. Phil. Trans. R. Soc. B. 2003;358:165–177. doi: 10.1098/rstb.2002.1193. 10.1098/rstb.2002.1193 (See discussion on pages 177–179.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Jelic M, Vojta A, Radunz A, Soll J, Schleiff E. Preprotein recognition by the Toc complex. Embo. J. 2004;23:520–530. doi: 10.1038/sj.emboj.7600089. 10.1038/sj.emboj.7600089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Karlberg E.O, Frank A.C, Legault B.A, Andersson S.G. Computational inference of scenarios for alpha-proteobacterial genome evolution. Proc. Natl Acad. Sci. USA. 2004;101:9722–9727. doi: 10.1073/pnas.0400975101. 10.1073/pnas.0400975101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The origins of plastids. Biol. J. Linn. Soc. 1982;17:289–306. [Google Scholar]
- Cavalier-Smith T. Endosymbiotic origin of the mitochondrial envelope. In: Schwemmler W, Schenk H.E.A, editors. Endocytobiology II. de Gruyter; Berlin: 1983. pp. 265–279. [Google Scholar]
- Cavalier-Smith T. The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann. NY Acad. Sci. 1987;503:55–71. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Intron phylogeny: a new hypothesis. Trends Genet. 1991;7:145–148. [PubMed] [Google Scholar]
- Cavalier-Smith T. Evolution of the eukaryotic genome. In: Broda P, Oliver S.G, Sims P, editors. The eukaryotic genome. Cambridge University Press; Cambridge, UK: 1993a. pp. 333–385. [Google Scholar]
- Cavalier-Smith T. The origin, losses and gains of chloroplasts. In: Lewin R.A, editor. Origin of plastids: symbiogenesis, prochlorophytes and the origins of chloroplasts. Chapman & Hall; New York, NY: 1993b. pp. 291–348. [Google Scholar]
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. 10.1016/S1360-1385(00)01598-3 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of protozoa. Int. J. Syst. Evol. Microbiol. 2002a;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 2002b;52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Protist phylogeny and the high-level classification of protozoa. Eur. J. Protistol. 2003a;39:338–348. 10.1078/0932-4739-00002 [Google Scholar]
- Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote–eukaryote chimaeras (meta-algae) Phil. Trans. R. Soc. B. 2003b;358:109–134. doi: 10.1098/rstb.2002.1194. 10.1098/rstb.2002.1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Chromalveolate diversity and cell megaevolution: interplay of membranes, genomes and cytoskeleton. In: Hirt R.P, Horner D.S, editors. Organelles, genomes and eukaryote phylogeny. CRC Press; London, UK: 2004a. pp. 75–108. [Google Scholar]
- Cavalier-Smith T. The membranome and membrane heredity in development and evolution. In: Hirt R.P, Horner D.S, editors. Organelles, genomes and eukaryote phylogeny. Taylor & Francis; London, UK: 2004b. pp. 335–351. [Google Scholar]
- Cavalier-Smith T, Lee J.J. Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J. Protozool. 1985;32:376–379. [Google Scholar]
- Chacinska A, et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. Embo. J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. 10.1038/sj.emboj.7600389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.W, et al. A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr. Biol. 2005;15:737–742. doi: 10.1016/j.cub.2005.02.068. 10.1016/j.cub.2005.02.068 [DOI] [PubMed] [Google Scholar]
- Chew O, Lister R, Qbadou S, Heazlewood J.L, Soll J, Schleiff E, Millar A.H, Whelan J. A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett. 2004;557:109–114. doi: 10.1016/s0014-5793(03)01457-1. [DOI] [PubMed] [Google Scholar]
- Codd G.A, Turnbull F. Enzymes of glycollate formation and oxidation in two members of the Rhodospirillaceae (purple non-sulphur bacteria) Arch. Microbiol. 1975;104:155–158. doi: 10.1007/BF00447317. 10.1007/BF00447317 [DOI] [PubMed] [Google Scholar]
- Covello P.S, Gray M.W. On the evolution of RNA editing. Trends Genet. 1993;9:265–268. doi: 10.1016/0168-9525(93)90011-6. 10.1016/0168-9525(93)90011-6 [DOI] [PubMed] [Google Scholar]
- de Grey A. Forces maintaining organellar genomes: is any as strong as genetic code disparity or hydrophobicity? BioEssays. 2005;27:436–446. doi: 10.1002/bies.20209. 10.1002/bies.20209 [DOI] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait B.T, Sali A, Rout M.P. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. 10.1371/journal.pbio.0020380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley T.M, van der Giezen M, Horner D.S, Dyal P.L, Foster P. Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Phil. Trans. R. Soc. B. 2003;358:191–201. doi: 10.1098/rstb.2002.1190. 10.1098/rstb.2002.1190 discussion 201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J. Cell Sci. 2003;116:3259–3267. doi: 10.1242/jcs.00667. 10.1242/jcs.00667 [DOI] [PubMed] [Google Scholar]
- Ertel F, Mirus O, Bredemeier R, Moslavac S, Becker T, Schleiff E. The evolutionarily related β-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J. Biol. Chem. 2005;280:28 281–28 289. doi: 10.1074/jbc.M503035200. 10.1074/jbc.M503035200 [DOI] [PubMed] [Google Scholar]
- Esser C, et al. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. 10.1093/molbev/msh160 [DOI] [PubMed] [Google Scholar]
- Fenchel T, Bernard C. A purple protist. Nature. 1993;362:300. doi: 10.1038/362300a0. 10.1038/362300a0 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, De Leonardis F, Todisco S, Palmieri L, Lasorsa F.M, Palmieri F. Identification of the mitochondrial ATP-Mg/Pi transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 2004;279:30 722–30 730. doi: 10.1074/jbc.M400445200. 10.1074/jbc.M400445200 [DOI] [PubMed] [Google Scholar]
- Frazier A.E, Chacinska A, Truscott K.N, Guiard B, Pfanner N, Rehling P. Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell Biol. 2003;23:7818–7828. doi: 10.1128/MCB.23.21.7818-7828.2003. 10.1128/MCB.23.21.7818-7828.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier A.E, et al. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 2004;11:226–233. doi: 10.1038/nsmb735. 10.1038/nsmb735 [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 2005;164:19–25. doi: 10.1083/jcb.200310092. 10.1083/jcb.200310092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.W, Lang B.F, Burger G. Mitochondria of protists. Annu. Rev. Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. 10.1146/annurev.genet.37.110801.142526 [DOI] [PubMed] [Google Scholar]
- Habib S.J, Waizenegger T, Lech M, Neupert W, Rapaport D. Assembly of the TOB complex of mitochondria. J. Biol. Chem. 2005;280:6434–6440. doi: 10.1074/jbc.M411510200. 10.1074/jbc.M411510200 [DOI] [PubMed] [Google Scholar]
- Jekély G, Arendt D. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. BioEssays. 2006;28:191–198. doi: 10.1002/bies.20369. 10.1002/bies.20369 [DOI] [PubMed] [Google Scholar]
- John P, Whatley F.R. Paracoccus denitrificans and the evolutionary origin of mitochondria. Nature. 1975;254:495–498. doi: 10.1038/254495a0. 10.1038/254495a0 [DOI] [PubMed] [Google Scholar]
- John P, Whatley F.R. Paracoccus denitrificans Davis (Micrococcus denitrificans Beijerinck) as a mitochondrion. Adv. Bot. Res. 1977;4:51–115. [Google Scholar]
- Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. 10.1083/jcb.151.2.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefel B.R, Gilson P.R, Beech P.L. Diverse eukaryotes have retained mitochondrial homologues of the bacterial division protein FtsZ. Protist. 2004;155:105–115. doi: 10.1078/1434461000168. 10.1078/1434461000168 [DOI] [PubMed] [Google Scholar]
- Kurz M, Martin H, Rassow J, Pfanner N, Ryan M.T. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell. 1999;10:2461–2474. doi: 10.1091/mbc.10.7.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Qian L, Chen Z, Thibault D, Liu G, Liu T, Thanassi D.G. The outer membrane usher forms a twin-pore secretion complex. J. Mol. Biol. 2004;344:1397–1407. doi: 10.1016/j.jmb.2004.10.008. 10.1016/j.jmb.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Linke D, Frank J, Pope M.S, Soll J, Ilkavets I, Fromme P, Burstein E.A, Reshetnyak Y.K, Emelyanenko V.I. Folding kinetics and structure of OEP16. Biophys. J. 2004;86:1479–1487. doi: 10.1016/S0006-3495(04)74216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Whelan J. Mitochondrial protein import: convergent solutions for receptor structure. Curr. Biol. 2006;16:R197–R199. doi: 10.1016/j.cub.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Lucattini R, Likic V.A, Lithgow T. Bacterial proteins predisposed for targeting to mitochondria. Mol. Biol. Evol. 2004;21:652–658. doi: 10.1093/molbev/msh058. 10.1093/molbev/msh058 [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–44. doi: 10.1038/32096. 10.1038/32096 [DOI] [PubMed] [Google Scholar]
- Meier S, Neupert W, Herrmann J.M. Conserved N-terminal negative charges in the Tim17 subunit of the TIM23 translocase play a critical role in the import of preproteins into mitochondria. J. Biol. Chem. 2005;280:7777–7785. doi: 10.1074/jbc.M412158200. 10.1074/jbc.M412158200 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Ni L, Yang C.S, Weiner H. Bacterial signal peptide recognizes HeLa cell mitochondrial import receptors and functions as a mitochondrial leader sequence. Cell Mol. Life Sci. 2005;62:1890–1899. doi: 10.1007/s00018-005-5178-0. 10.1007/s00018-005-5178-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Misumi O, Yagisawa F, Kuroiwa H, Nagata T, Kuroiwa T. Triple immunofluorescent labeling of FtsZ, dynamin, and EF-Tu reveals a loose association between the inner and outer membrane mitochondrial division machinery in the red alga Cyanidioschyzon merolae. J. Histochem. Cytochem. 2004;52:843–849. doi: 10.1369/jhc.4C6315.2004. 10.1369/jhc.4C6315.2004 [DOI] [PubMed] [Google Scholar]
- Perry A.J, Hulett J.M, Likic V.A, Lithgow T, Gooley P.R. Convergent evolution of receptors for protein import into mitochondria. Curr. Biol. 2006;16:221–229. doi: 10.1016/j.cub.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N, Meisinger C, Lithgow T. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 2004;11:1044–1048. doi: 10.1038/nsmb852. 10.1038/nsmb852 [DOI] [PubMed] [Google Scholar]
- Preuss M, Ott M, Funes S, Luirink J, Herrmann J.M. Evolution of mitochondrial oxa proteins from bacterial YidC: inherited and acquired functions of a conserved protein. J. Biol. Chem. 2005;280:13 004–13 011. doi: 10.1074/jbc.M414093200. 10.1074/jbc.M414093200 [DOI] [PubMed] [Google Scholar]
- Rassow J, Dekker P.J, van Wilpe S, Meijer M, Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. 10.1006/jmbi.1998.2455 [DOI] [PubMed] [Google Scholar]
- Regoes A, Zourmpanou D, Leon-Avila G, van der Giezen M, Tovar J, Hehl A.B. Protein import, replication, and inheritance of a vestigial mitochondrion. J. Biol. Chem. 2005;280:30 557–30 563. doi: 10.1074/jbc.M500787200. 10.1074/jbc.M500787200 [DOI] [PubMed] [Google Scholar]
- Rehling P, et al. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 2003;299:1747–1751. doi: 10.1126/science.1080945. 10.1126/science.1080945 [DOI] [PubMed] [Google Scholar]
- Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2:E273. doi: 10.1371/journal.pbio.0020273. 10.1371/journal.pbio.0020273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T.A, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. 10.1038/nature03949 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Soll J. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. Embo. Rep. 2005;6:1–5. doi: 10.1038/sj.embor.7400563. 10.1038/sj.embor.7400563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf E. Zur Feinstruktur von Geosiphon pyriforme: ein Versuch zur Deutung cytoplasmatischer Membranen und Kompartimente. Arch. Microbiol. 1964;49:112–131. [Google Scholar]
- Searcy D. Origins of mitochondria and chloroplasts from sulfur based symbioses. In: Hartman H, Matsuno K, editors. The origin and evolution of the cell. World Scientific; Singapore: 1992. pp. 47–78. [Google Scholar]
- Smith D.F, Wiebe W.J. Constant release of photosynthate from marine phytoplankton. Appl. Environ. Microbiol. 1976;32:75–79. doi: 10.1128/aem.32.1.75-79.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. 10.1126/science.1071196 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr. Biol. 2003;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. 10.1016/S0960-9822(03)00602-X [DOI] [PubMed] [Google Scholar]
- Tjalsma H, Bron S, van Dijl J.M. Complementary impact of paralogous oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 2003;278:15 622–15 632. doi: 10.1074/jbc.M301205200. 10.1074/jbc.M301205200 [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J. Degenerate mitochondria. Embo. Rep. 2005;6:525–530. doi: 10.1038/sj.embor.7400440. 10.1038/sj.embor.7400440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J, Clark C.G. Mitochondrion-derived organelles in protists and fungi. Int. Rev. Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- van Roermund C.W, Drissen R, van Den Berg M, Ijlst L, Hettema E.H, Tabak H.F, Waterham H.R, Wanders R.J. Identification of a peroxisomal ATP carrier required for medium-chain fatty acid beta-oxidation and normal peroxisome proliferation in Saccharomyces cerevisiae. Mol. Cell Biol. 2001;21:4321–4329. doi: 10.1128/MCB.21.13.4321-4329.2001. 10.1128/MCB.21.13.4321-4329.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Frazier A.E, Pfanner N. The protein import machinery of mitochondria. J. Biol. Chem. 2004a;279:14 473–14 476. doi: 10.1074/jbc.R400003200. 10.1074/jbc.R400003200 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Truscott K.N, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 2004b;279:18 188–18 194. doi: 10.1074/jbc.M400050200. 10.1074/jbc.M400050200 [DOI] [PubMed] [Google Scholar]
- Woese C.R. Endosymbionts and mitochondrial origin. J. Mol. Evol. 1977;10:93–96. doi: 10.1007/BF01751802. 10.1007/BF01751802 [DOI] [PubMed] [Google Scholar]
- Wojtkowska M, Szczech N, Stobienia O, Jarmuszkiewicz W, Budzinska M, Kmita H. An inception report on the TOM complex of the amoeba Acanthamoeba castellanii, a simple model protozoan in mitochondria studies. J. Bioenerg. Biomembr. 2005;37:261–268. doi: 10.1007/s10863-005-6636-y. [DOI] [PubMed] [Google Scholar]