Abstract
The Asian countries chronically infected with avian influenza A H5N1 are ‘global hotspots’ for biodiversity conservation in terms of species diversity, endemism and levels of threat. Since 2003, avian influenza A H5N1 viruses have naturally infected and killed a range of wild bird species, four felid species and a mustelid. Here, we report fatal disseminated H5N1 infection in a globally threatened viverrid, the Owston's civet, in Vietnam, highlighting the risk that avian influenza H5N1 poses to mammalian and avian biodiversity across its expanding geographic range.
Keywords: avian influenza, H5N1, wild species infections, viverrid, Owston's civet, biodiversity conservation
1. Introduction
Wild aquatic birds are the natural reservoir of influenza viruses, and all 16 haemagglutinin (H) and 9 neuramindase (N) subtypes can be found therein. Subtypes H5 and H7 may acquire dramatically enhanced pathogenicity for galliforms (e.g. domestic chickens), but until recently, these highly pathogenic avian influenza (HPAI) viruses have rarely been fatal in aquatic birds such as anseriforms (ducks, geese and swans; Webster et al. 1992).
Surprisingly, the recently emerged H5N1 viruses have acquired pathogenicity for anseriforms and also killed birds in at least 11 of the 27 avian orders (41%), including Charadriiformes (gulls and shorebirds), Ciconiiformes (storks, herons, flamingos and egrets), Columbiformes (pigeons and doves), Falconiformes (eagles, kites and vultures), Gruiformes (cranes), Passeriformes (passerines), Pelecaniformes (pelicans and cormorants), Psittaciformes (parrots and parakeets) and Strigiformes (owls) (OIE 2003; Ellis et al. 2004; FAO 2004; Chen et al. 2005; see table 1 in the electronic supplementary material B). Globally threatened species are listed among these fatal infections.
Currently, circulating forms of H5N1 also pose a risk to a range of mammalian species. H5N1-infected poultry has been a source of transmission to humans and to felids, e.g. tigers (Panthera tigris), leopards (Panthera pardus) (Keawcharoen et al. 2004), and domestic cats Felis catus (Kuiken et al. 2004). In addition, experimental infection of Mustelidae (domestic ferrets, Mustela putorius furo; Govorkova et al. 2005) and mice (Lu et al. 1999) with some strains of H5N1 viruses has been fatal. Experimental inoculation of Suidae (domestic pigs, Sus scrofa) led to infection and mild non-fatal illness without transmission to contact pigs (Choi et al. 2005), while infection of long-tailed macaques (Macaca fascicularis) led to acute respiratory symptoms and fever associated with a necrotizing interstitial pneumonia (Rimmelzwaan et al. 2001). Here, we report the transmission of influenza A H5N1 virus to affect a globally threatened viverrid, the Owston's civet (Chrotogale owstoni; figure 1a).
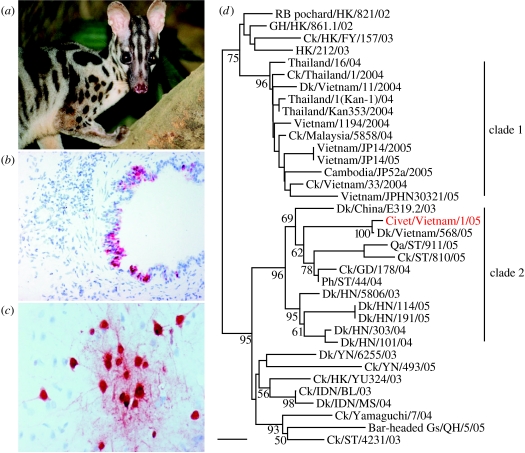
Figure 1.
(a) Owston's civet (Chrotogale owstoni). Immunohistochemistry using a monoclonal antibody against the N protein showed (b) positive red staining of the bronchial epithelium of the lung and (c) positive staining of cerebral neurons of the cerebral cortex. (d) Phylogenetic relationship of the haemagglutinin gene of representative H5N1 influenza A viruses (nucleotide positions 1–1012; scale bar, 0.005 nucleotide changes per site). Methods for genetic analysis have been described previously (Li et al. 2004; Chen et al. 2005). Numbers at branches indicate bootstrap values from 1000 replicates. Clade 1 indicates viruses from the Thailand and Vietnam outbreak in 2004–2005; clade 2 indicates the position of the civet virus with those viruses from southern China. BH gull, brown-headed gull (Larus brunnicephalus); BHG, black-headed gull (Larus ichthyaetus); Ck, chicken; Dk, duck; GD, Guangdong; GH, grey heron (Ardea cinerea); Gs, goose; HK, Hong Kong; HN, Hunan; Pf, peregrine falcon (Falco peregrinus); Ph, pheasant; RB pochard, rosy-billed pochard (Netta peposaca); ST, Shantou; YN, Yunnan.
2. Material and methods
(a) The Owston's Civet Conservation Program (OCP)
The Owston's Civet Conservation Program is located in Cuc Phuong National Park, northern Vietnam, and coordinates an international conservation breeding program for the Owston's civet. In June 2005, the centre held 23 Owston's civets in 15 enclosures arranged into four separate blocks. Ten animals were housed individually, six in pairs, one adult female was held with her two offspring and there was one family group comprising four animals. Cage mesh in 12 of the enclosures would permit small mammals, reptiles and birds to enter, and none of the enclosures were roofed. The animals were fed a diet consisting of fruit purchased from a local market, hard-boiled duck eggs, pork and a range of live animals including earthworms collected from surrounding agricultural land, grasshoppers, stick insects, crabs, fish and frogs.
(b) Virological investigation
Swabs and tissue homogenates were placed in virus transport medium and stored frozen at −70 °C until testing. The swab, or homogenate in virus transport medium, was inoculated into the allantoic cavity of embryonated eggs and into Mardin Darby Canine Kidney cells to isolate influenza viruses and onto FRhK-4 cells to attempt to isolate SARS-like coronaviruses, as previously described (Chan et al. 2002; Peiris et al. 2003). Virus isolates were identified by haemagglutination inhibition tests using subtype-specific reference sera and by reverse transcription polymerase chain reaction (RT–PCR).
RNA was extracted from these clinical specimens using the RNeasy Mini Kit (Qiagen, Chatsworth, CA) and tested by RT–PCR for the influenza A matrix gene, influenza A H5 haemaglutinin and for SARS coronavirus replicase gene (Peiris et al. 2003). Methods for genetic sequencing and phylogenetic analysis have been described previously (Chen et al. 2005).
Serum was collected from 16 other apparently healthy civets held within the centre. The sera were tested by a microneutralization test for neutralizing antibody to H5N1 virus (Choi et al. 2005).
(c) Immunohistochemistry of tissues
Lung and brain tissues were fixed in 10% neutral buffered formalin before processing and embedding in paraffin. For immunohistochemistry, the monoclonal antibody to influenza A NP clone HB65 was used courtesy of T. Kuiken, Erasmus Medical University, Netherlands. Brain from a masked palm civet (Paguma larvata) from Hong Kong that had died of trauma was used as negative control. (See electronic supplementary material A.)
3. Results and discussion
(a) Lethal infection of H5N1 virus in viverrids
On 24 June 2005, one juvenile female Owston's civet showed appetite loss and neurological symptoms, including convulsions, and was dead by the next morning. Two days later (27 June), two animals which had shared an enclosure with the index case displayed similar symptoms followed by hind limb paralysis and both had died by 29 June. Autopsy showed pulmonary oedema with mild parenchymal inflammation without evidence of hyaline membrane formation. Moderately severe meningitis was present, together with cerebral oedema and hypoxic change of the neurons. Foci of necrosis were seen in the liver.
H5N1 virus was isolated from all three animals. H5N1 virus was detected in the lungs, brain, kidney and intestinal tract by virus isolation, by RT–PCR and by detection of viral antigen using immunohistology on organ impression smears or histological sections (figure 1b,c). Within the lung there was staining of bronchial epithelium and pneumocytes with the monoclonal antibody to influenza nucleoprotein, but no significant areas of necrosis were seen in these regions of infection. Influenza virus antigen was detected in the inflammatory exudates in the meninges and electron microscopy demonstrated viral particles consistent with influenza virus. Prominent staining of the neurons of the cerebral cortex and cerebellum was seen but again this was not accompanied by a significant inflammatory cell infiltrate. Thus, the virus appears to have disseminated to involve multiple organs as was previously seen in felids (Keawcharoen et al. 2004) with neurotropism being a prominent feature of the disease (figure 1c).
Phylogenetic analysis of all eight gene segments revealed that this virus was distinct from the dominant H5N1 virus genotype Z associated with poultry outbreaks and transmission to humans in Vietnam, Thailand and Cambodia (Li et al. 2004). Instead, it belongs to the newly described H5N1 genotype G viruses (Chen et al. 2006) which have been previously been isolated from poultry in mainland China and Vietnam (figure 1d). The haemagglutinin gene retains the motif of basic amino acids (QRERRRKR) in the connecting peptide characteristic of HPAI H5N1 viruses. It has not acquired the Lys 627 amino acid substitution in the PB2 protein, which is associated with virulence for mammals (Hatta et al. 2001).
In contrast to previous instances of infection of tigers and leopards, these civets were not fed dead poultry and the source for the infection of the civets remains unknown. However, unexplained poultry deaths were reported in villages neighbouring the National Park. The time course of the infection in the civets is compatible with transmission from the index case to others within the same cage. There was no disease or seroconversion in the other 16 civets held at the centre.
(b) Implications of H5N1 for biodiversity conservation and wildlife health
This discovery of lethal H5N1 infection in Owston's civet extends the range of known mammalian hosts for the virus to include viverrids, a second host family, and Chrotogale, a fourth genus, within the mammalian order Carnivora. This highlights the additional risks that avian influenza A H5N1 viruses may pose to mammalian and avian biodiversity across its expanding geographical range.
Overexploitation for the illegal wildlife trade, combined with habitat loss and fragmentation, continue to pose the major threat to biodiversity in Southeast Asia. Almost all species of Viverridae present in Vietnam, including the Owston's civet, have been over-hunted for human consumption within the illegal wildlife trade (Bell et al. 2004). The ease with which H5N1 virus appears to cross species-barriers suggests that, in addition to poultry exposure, the illegal wildlife trade presents a further potential route for human infection with this virus. This has been vividly demonstrated by the first appearance of H5N1 in Europe and the United Kingdom occurring in illegally smuggled eagles seized in Belgium (Van Borm et al. 2005) and birds imported into the United Kingdom for the pet trade (OIE 2005).
In addition to the two orders containing domestic poultry (Anseriformes and Galliformes), affected avian orders listed in table 1 (see electronic supplementary material B) include those with the largest number of species (Passeriformes, over 5800 spp.) and together comprise 84% of known bird species and 60% of the non-Passeriformes. The wide range of avian orders affected suggests that HPAI H5N1 may be highly pathogenic in many avian species. In terms of risk of exposure to H5N1-infected poultry, one might predict that those species at greatest risk in the wild are those which: (i) live commensally around domestic poultry; (ii) are occasional visitors to areas containing poultry or poultry excreta; or (iii) include affected poultry and wild bird species in their diet. Those mammalian orders likely to be at highest risk of infection through dietary routes are the Carnivora, Primates, Rodentia, Dasyuromorphia (marsupial carnivores) and Didelphimorphia (American opossums). However, species from a range of other orders are occasional consumers of birds, for example, the greater false vampire bats (order Chiroptera) and armadillos (order Edendata) may also scavenge vertebrate carcasses (Macdonald 2001). The well-developed olfactory-based communication systems of mammals, often involving close inspection of urine and faeces, may also increase their susceptibility to infection through the oro-nasal route.
HPAI H5N1-related mortality may have most impact on those taxa already threatened by anthropogenic causes of population decline such as habitat loss, over-hunting and introduced predators. Of 4473 mammalian species evaluated in the 2004 IUCN Red List, 37.7% were listed as globally threatened or near threatened, compared to 20.2% of 9839 species of birds evaluated. When the percentage of assessed taxa listed as threatened or near-threatened (IUCN Red List 2004) is compared among avian orders in which fatal infection has been recorded, the Galliformes emerges as the order with the highest percentage of threatened/near threatened species (37.4%: 107/286) followed by the Psittaciformes (34%: 127/374), Gruiformes (33.0%: 73/221) and Pelecaniformes (33.9%: 22/65), Columbiformes (29%: 97/335), Strigiformes (26.3%: 51/194), Ciconiformes (23.5%: 31/132) and Anseriformes (20.8%: 35/168). High percentages of species in these mammalian orders known, or most likely, to be exposed to HPAI H5N1 are also listed as threatened or near threatened in the IUCN Red List (2004), including 37.2% (96/258) of the Carnivora and 58.8% (161/274) of the Primates. Although HPAI H5N1 has not yet been reported in Australia or North and South America, the marsupial carnivores are also likely to be at risk and high percentages of these are also threatened/near-threatened on both continents.
4. Conclusions
This finding highlights the need for: (i) monitoring and surveillance for H5N1 avian influenza viruses in other wild and domesticated mammal species; and (ii) enhanced biosecurity for endangered species in H5N1-endemic areas, particularly in protected areas, captive breeding facilities and in the wildlife trade. It also underlines the need for interdisciplinary collaboration among virologists, veterinarians, conservation biologists and public health sector workers in addressing emerging infectious diseases which pose significant threats to domestic livestock, human and wildlife health.
Finally, this report illustrates the ease with which this influenza A H5N1 virus can cross species barriers and reinforces the pandemic concern engendered by its progressively increasing geographic range.
Supplementary Material
Non-poultry animal species in which death due to natural infection with currently circulating strains of avian influenza H5N1 has been recorded.
References
- Bell D.J, Roberton S, Hunter P.R. Animal origins of SARS: possible links with the wildlife trade. Phil. Trans. R. Soc. B. 2004;359:1107–1114. doi: 10.1098/rstb.2004.1492. doi:10.1098/rstb.2004.1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H, Maldeis N, Pope W, Yup A, Ozinskas A, Gill J, Seto W.H, Shortridge K.F, Peiris J.S.M. Evaluation of the Directigen FluA+B test for rapid diagnosis of influenza virus type A and B infections. J. Clin. Microbiol. 2002;40:1675–1680. doi: 10.1128/JCM.40.5.1675-1680.2002. doi:10.1128/JCM.40.5.1675-1680.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith G.J.D, Zhang S.Y, Qin K, Wang J, Li K.S, Webster R.G, Peiris J.S.M, Guan Y. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. doi:10.1038/nature03974 [DOI] [PubMed] [Google Scholar]
- Chen H, et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl Acad. Sci. USA. 2006;103:2845–2850. doi: 10.1073/pnas.0511120103. doi:10.1073/pnas.0511120103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.K, et al. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J. Virol. 2005;79:10 821–10 825. doi: 10.1128/JVI.79.16.10821-10825.2005. doi:10.1128/JVI.79.16.10821-10825.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T.M, et al. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathol. 2004;33:492–505. doi: 10.1080/03079450400003601. doi:10.1080/03079450400003601 [DOI] [PubMed] [Google Scholar]
- FAO 2004 Update on the avian influenza situation (as of 15/06/2004)—Issue no. 16. FAOAIDEnews: avian influenza disease emergency. Avian Influenza technical Task Force, FAO, Rome & Bangkok.
- Govorkova E.A, et al. Lethality to ferrets of H5N1 influenza isolated from humans and poultry in 2004. J. Virol. 2005;79:2191–2198. doi: 10.1128/JVI.79.4.2191-2198.2005. doi:10.1128/JVI.79.4.2191-2198.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. doi:10.1126/science.1062882 [DOI] [PubMed] [Google Scholar]
- IUCN Red List 2004 accessed at www.iucn.org on 20th March 2006.
- Keawcharoen J, et al. Avian influenza in tigers and leopards. Emerg. Infect. Dis. 2004;10:2189–2191. doi: 10.3201/eid1012.040759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T, Rimmelzwaan G, Van Amerongen G, Baars M, Fouchier R, Osterhaus A. Avian H5N1 influenza in cats. Science. 2004;306:241. doi: 10.1126/science.1102287. doi:10.1126/science.1102287 [DOI] [PubMed] [Google Scholar]
- Li K.S, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. doi:10.1038/nature02746 [DOI] [PubMed] [Google Scholar]
- Lu X, Tumpey T.M, Morken T, Zaki S.R, Cox N.J, Katz J.M. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 1999;73:5903–5911. doi: 10.1128/jvi.73.7.5903-5911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald D.W, editor. The new encyclopaedia of mammals. Oxford University Press; Oxford, UK: 2001. [Google Scholar]
- OIE 2003 Pathogenic H5N1 avian influenza in waterfowl and wild birds, Final Report, Hong Kong, China 30 July 2003.
- OIE 2005 Disease information, 18 October 2005; no. 43. Web Accessed on 14th December 2005. See http://www.oie.int/eng/info/hebdo/AIS_47.HTM#Sec17
- Peiris J.S.M, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. doi:10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan G.F, Kuiken T, van Amerongen G, Bestebroer T.M, Fouchier R.A, Osterhaus A.D. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J. Virol. 2001;75:6687–6691. doi: 10.1128/JVI.75.14.6687-6691.2001. doi:10.1128/JVI.75.14.6687-6691.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Borm S, Thomas I, Hanquet G, Lambrecht B, Boschmans M, Dupont G, Decaestecker M, Snacken R, van den Berg T. Highly pathogenic H5N1 influenza virus in smuggled eagles, Belgium. Emerg. Infect. Dis. 2005;11:702–705. doi: 10.3201/eid1105.050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G, Bean W.J, Gorman O.T, Chambers T.M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Non-poultry animal species in which death due to natural infection with currently circulating strains of avian influenza H5N1 has been recorded.