Abstract
Recent molecular investigations of marine samples taken from different environments, including tropical, temperate and polar areas, as well as deep thermal vents, have revealed an unexpectedly high diversity of protists, some of them forming deep-branching clades within important lineages, such as the alveolates and heterokonts. Using the same approach on coastal samples, we have identified a novel group of protist small subunit (SSU) rDNA sequences that do not correspond to any phylogenetic group previously identified. Comparison with other sequences obtained from cultures of heterotrophic protists showed that the environmental sequences grouped together with Telonema, a genus known since 1913 but of uncertain taxonomic affinity. Phylogenetic analyses using four genes (SSU, Hsp90, alpha-tubulin and beta-tubulin), and accounting for gamma- and covarion-distributed substitution rates, revealed Telonema as a distinct group of species branching off close to chromist lineages. Consistent with these gene trees, Telonema possesses ultrastructures revealing both the distinctness of the group and the evolutionary affinity to chromist groups. Altogether, the data suggest that Telonema constitutes a new eukaryotic phylum, here defined as Telonemia, possibly representing a key clade for the understanding of the early evolution of bikont protist groups, such as the proposed chromalveolate supergroup.
Keywords: Telonema, Telonemia, environmental sequences, phylogeny, covarion substitution pattern
1. Introduction
Phylogenetic inferences of gene sequences and genome rearrangements have recently resolved some of the most intriguing evolutionary relationships between protist lineages (Baldauf et al. 2000; Stechmann & Cavalier-Smith 2002). By combining gene trees with ultrastructural and biochemical traits, evidence is growing for at least six distinct supergroups of eukaryotes; i.e. the opisthokonts, Amoebozoa (together forming the unikonts), Rhizaria, Excavata, chromalveolates and Plantae, each possibly forming separate kingdoms or subkingdoms (Cavalier-Smith 2003, 2004b; Keeling 2004). In recent years, increasing number of studies have applied clone libraries of DNA sampled from various environments to investigate the protist diversity. A large number of sequences and clades belonging to all the eukaryotic lineages have been detected with this method, but very few sequences seem to constitute entirely unknown supergroups (Berney et al. 2004; Richards & Bass 2005). It is possible that all major groups of living eukaryotes have already been described (Berney et al. 2004). However, the small subunit rRNA (SSU) gene typically used to construct phylogenies of environmental sequences is known to be prone to long-branch attraction, covarion substitutions patterns and other artefacts that may violate the methods applied (Lopez et al. 1999; Philippe 2000; Huelsenbeck & Ronquist 2001; Galtier 2001; Huelsenbeck 2002). In addition, since it is virtually impossible to combine multigene-phylogenies and ultrastructural studies of such uncultured species, it is difficult to rule out the possibility that some sequences may constitute novel high-level taxa (Berney et al. 2004). In our search for hitherto undetermined groups of eukaryotes, we aimed at combining data from clone libraries of the SSU gene, and ultrastructural features and multiple-gene phylogenies of cultured species. Furthermore, we included all undetermined marine sequences from earlier analyses of environmental clone libraries (Berney et al. 2004; Cavalier-Smith 2004a), and performed phylogenetic analyses allowing both gamma- and covarion-distributed substitution patterns (Yang 1996; Huelsenbeck 2002). Here, we present several new eukaryote sequences obtained from the environment and show that they group together with sequences obtained from cultures of Telonema, a genus known since 1913 but of uncertain taxonomic affinity (Griessmann 1913). Based on phylogenies of single- and multi-gene sequences and unique ultrastructural features of cultured Telonema species, we deduce that the Telonema-group constitutes a novel lineage, here defined as phylum Telonemia, which probably is evolutionarily related to the chromist lineages.
2. Material and methods
(a) Cultures
Two cultures of Telonema subtilis were isolated from surface water sampled at the Astan station off Roscoff (English Channel, Brittany, France) on 12 April and 11 July 2000. Water pre-filtered through a 3 μm filter was enriched with K medium (at 1/100 dilution) and grown at 15 °C under 12 h : 12 h light–dark cycles at 150 μmol quanta m−2 s−1. These cultures are deposited in the Roscoff Culture Collection (RCC: www.sb-roscoff.fr/phyto/collect.html). Cultures of Telonema antarcticum were isolated from surface seawater samples taken at the inner Oslofjord winter 1998 and grown in enriched seawater f/2 or IMR/2, transferring weekly to a vigorously growing culture of the cryptophyte Rhodomonas sp. The cultures were held at 17 °C on a 14/10 h light/dark cycles 230 μmol quanta m−2 s−1.
(b) Electron microscopy
Transmission electron microscopy (TEM) of T. subtilis was performed by following standard procedures as described in Eikrem & Moestrup (1998). Scanning electron microscopy (SEM) of T. antarcticum and T. subtilis and TEM of T. antarcticum were conducted according to Klaveness et al. (2005).
(c) Clone libraries and sequencing
Samples were collected from surface waters of the English Channel off Roscoff (Astan station, 12 April and 10 December 2000) and of the Mediterranean Sea off Blanes (Catalan coast, 25 June 2001) and pre-filtered through a 3 μm filter. DNA was collected on 0.2 μm filters and extracted by classical methods as described elsewhere (Romari & Vaulot 2004). The SSU rRNA gene was amplified by PCR using primers described in Moon-van der Staay et al. (2000) and cloned using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA). Positive clones were partially sequenced (550 bp) by Qiagen Genomics Sequencing Services using the internal primer Euk528f (5′-GCG GTA ATT CCA GCT CCA A-3′), and selected clones were fully sequenced by the same company. The T. antarcticum SSU sequence was generated as previously described (Klaveness et al. 2005). Hsp90, alpha- and beta-tubulin sequences from Telonema were amplified with various degenerate PCR primers and subsequently cloned and sequenced as done for the SSU sequences.
(d) Phylogenetic analysis
The evolutionary origin of Telonema was inferred from phylogenetic reconstruction of single- and concatenated sequences obtained from both cultured strains and environmental clones (SSU only). Different phylogenetic methods were used, including Bayesian inferences (Huelsenbeck & Ronquist 2001), maximum likelihood (ML: Guindon & Gascuel 2003), parsimony and distance (Swofford 1998; Felsenstein 2004) methods. All phylogenetic analyses were performed at the freely available University of Oslo Bioportal (http//:www.bioportal.uio.no/).
In the Bayesian analysis of the SSU sequences, variable substitution rates across sites were accounted for by using gamma-shaped distribution of site rate probability (G) and an invariable site parameter (I; together G+I). In addition, since deep branching phylogenetic analyses can be seriously affected by covarion (hereafter only COV) substitution processes, in which homologous sites evolve with different rates across the tree (Lopez et al. 1999a; Galtier 2001b; Huelsenbeck 2002), we conducted Bayesian inferences with MrBayes v. 3.0 and 3.1 (Huelsenbeck & Ronquist 2001), allowing sites to switch between invariable and variable states (Huelsenbeck 2002); i.e. off>on and on>off (G+COV), and finally together with both a constant proportion of invariable site parameter and the covarion model (G+I+COV). To test the significance of covarion rate patterns in the datasets, we did a statistical comparison between all these models in a Bayesian framework by applying the Bayes factor. The Bayes factor is defined as the posterior probability of a hypothesis, given that the prior probabilities of the alternative evolutionary models are equal, and was calculated as the ratio of marginal-likelihood values (i.e. the harmonic mean value) obtained from stationary phases of Markov chain Monte Carlo (MCMC) runs, as suggested by Newton & Raftery (1994). We interpreted the Bayes factor according to the guidelines provided by Kass & Raftery (1995), in which a Bayes factor of 10 is defined as the limit for decisive evidence for favouring the tested model. The general time reversible model was implemented in Bayesian analyses of SSU sequences. Priors for all other model parameters were set to default values. Metropolis coupling was used with three heated (temperature parameter 0.2) and one cold chain. Randomly generated trees were used as a starting point for MCMC chains that carried out for 1–20 000 000 generations. Sampling of trees was done every 100 generations for a total of 10 000–20 000 trees. Burn-in of trees (i.e. sampled before the MCMC chains reached convergence) was set to 3000–15 000 trees based on assessment of the likelihood plots. Consensus of the remaining trees was used to calculate the posterior probabilities of the clades. Two separate runs were performed to confirm the stationarity of the chains; the clade probabilities and likelihood scores of the received trees were very similar between the independent runs, supporting our conclusion that the chains produced a reasonable sample from the posterior distribution within the chosen burn-in period. The SSU tree was also estimated using PAUP* (Swofford 1998), on the basis of ML distances (MLDIST; G+I parameters) and 10 heuristic searches, random addition of sequences and tree-bisection-reconnection branch swapping. Non-parametric bootstrap analyses were done with 100 pseudoreplicates and analysed as for the original datasets, except using one heuristic search for each replicate.
For Hsp90, alpha- and beta-tubulin genes (amino acid sequences), Bayesian inferences were performed with the use of JTT and WAG substitution models and the G, I and COV parameters. Parsimony and distance non-parametric bootstrap support were inferred using Seqboot (Felsenstein 2004), PROTPARS, PROTDIST (JTT model) and NEIGHBOR programs (Phylip package; Felsenstein 2004), applying global rearrangements and five jumbles. Beta-tubulin tree with the inclusion of Kathablepharis and Leucocryptos sequences was reconstructed with an ML method in Phyml (Guindon & Gascuel 2003).
3. Results and discussion
(a) Telonema identified in environmental clone libraries and cultures
Partial sequencing (500 bp starting from position 528) of the SSU clone libraries revealed the presence of many sequences that did not present any significant similarities to sequences available in gene databases (Massana et al. 2004; Romari & Vaulot 2004), indicating a wide and unknown genetic diversity in marine coastal plankton similar to what was recently established for open ocean environments (Lopez-Garcia et al. 2001; Moon-van der Staay et al. 2001). Screening of protist cultures from the English Channel by the same sequencing approach uncovered two cultures containing sequences with high similarity to five of the clone library sequences. Both cultures contained T. subtilis, a small (2–6×4–10 μm) phagotrophic species first described by Griessmann (1913) from Roscoff and Naples in 1913. In parallel, we isolated from the Oslofjord a novel species, T. antarcticum (Klaveness et al. 2005), with an SSU sequence very similar to the environmental and T. subtilis sequences.
(b) Distinct origin of Telonema inferred from multiple sequences analysis applying covarion substitution model
To investigate the evolutionary origin of the Telonema species and the Telonema-like sequences, we obtained full length SSU, Hsp90, alpha-tubulin and beta-tubulin sequences from cultured strains and environmental clones (SSU only).
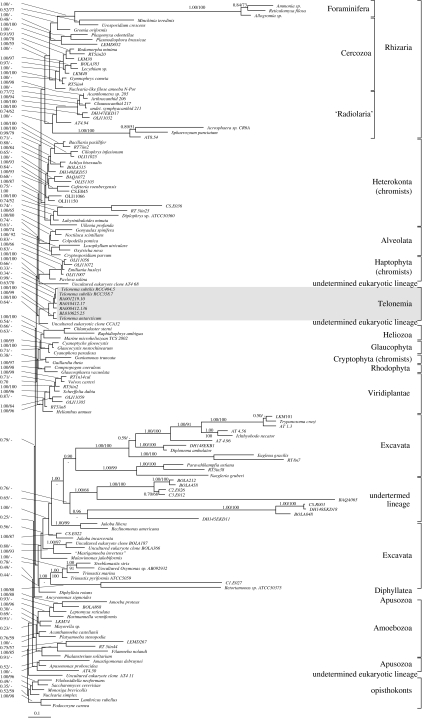
The inferences of the SSU alignment (144 taxa and 1159 characters) clustered seven Telonema and Telonema-like sequences as a monophyletic group with high posterior probability (PP) and bootstrap support (PP=1.0, MLDIST=100%), in which the environmental sequences from Roscoff grouped with T. subtilis, while the sequence obtained from Blanes grouped together with T. antarcticum (figure 1). The Telonema group branched off just below Haptophyta, which was placed at the base of a branch consisting of Heterokonta, alveolates and a large group with Foraminifera, Cercozoa and Radiolaria species (termed Rhizaria; Cavalier-Smith 2002). The other environmental sequences included in the analysis, which have been pointed out as potential undetermined lineages (Berney et al. 2004; Cavalier-Smith 2004a), were almost all grouped within characterized lineages; only three sequences appeared to have uncertain origin; one sequence (Clone CCA32) was weakly supported (PP=0.64) as a sister to Telonema. Some of the environmental sequences placed within the excavates, here described as undetermined lineage, may belong to alveolates, but possibly misplaced because of long-branch attractions (for details, see figure 1; Berney et al. 2004; Cavalier-Smith 2004a). Nearly all other species were grouped together in accordance with previously defined lineages and supergroups, including the opisthokonts, Amoebozoa, Excavata, Rhizaria, Alveolata and Heliozoa. In contrast, the Heterokonta, Haptophyta and Cryptophyta were each well-supported groupings, with the two latter as supported sister groups in protein trees (see also Harper et al. 2005), but were not clustered as the chromalveolate supergroup (Yoon et al. 2002; Harper & Keeling 2003). Likewise, the red algae, plants and glaucophytes were not grouped as a monophyletic Plantae (Rodríguez-Ezpeleta et al. 2005), but belonged to the same branch together with some of the chromists and Heliozoa. Both the latter inconsistencies with the proposed supergroup definitions are widely seen in SSU trees (Van de Peer et al. 2000; Berney et al. 2004). Interestingly, two species with previously unclear origin, Diphylleia rotans and ‘Mastigamoeba invertens’, were weakly placed together with excavates (PP=0.78).
Figure 1.
SSU phylogeny of the Telonema-clade. Tree inferred with Bayesian inference by applying a covarion (G+I+COV) model of evolution. The numbers at the nodes represent Bayesian posterior probability and bootstrap values from distance analyses (G+I) above 50%. The undetermined lineage has previously been identified as phylotypes of uncertain taxonomic affiliation, and its grouping with the excavates may result from long-branch artefacts (Berney et al. 2004; Cavalier-Smith 2004a). For accession numbers of sequences used, see table S1 in electronic supplementary material.
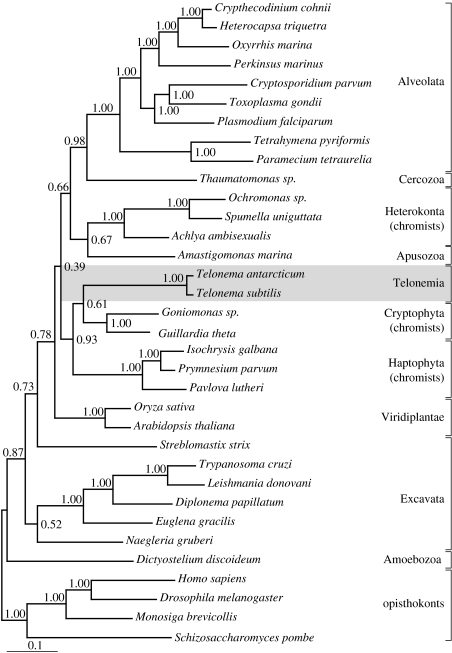
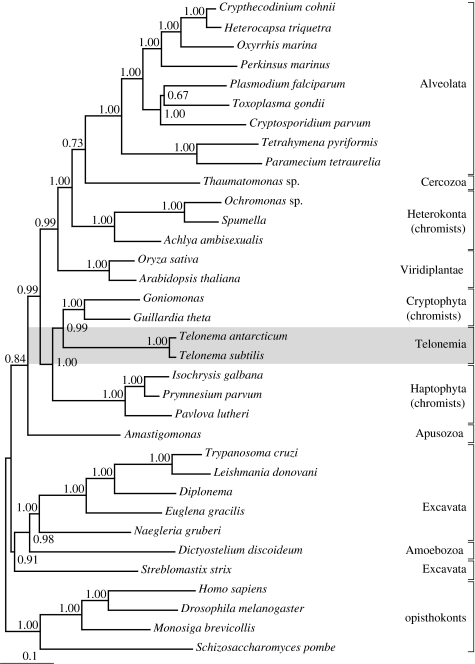
Consistent with the SSU tree, the analyses of Hsp90 (34 taxa and 476 amino acid characters; figure 2) and Hsp90+SSU (figure 3) placed Telonema as a distinct lineage. Both Hsp90 and Hsp90+SSU trees grouped the Telonema clade together with the haptophytes and cryptophytes as a monophyletic lineage (PP=0.93 and PP=1.00, respectively). The other groups were largely grouped as in the SSU tree, but comparison between the two protein trees show different placement of the plants, Amastigomonas marina (apusozoan) and Streblomastix strix (excavates).
Figure 2.
Hsp90 phylogeny of Telonema. Tree reconstructed using Bayesian inference and G+I model. Telonema is placed in a clade composed of cryptophytes and haptophytes. (Accession numbers shown in table S1, electronic supplementary material.)
Figure 3.
Concatenated Hsp90+SSU tree reconstructed by Bayesian inference applying a mixed G+I and G+I+COV evolutional model for each gene partition, respectively.
Analysis of single-gene alpha-tubulin (40 taxa and 355 characters) and beta-tubulin (39 taxa and 383 characters) sequences resulted in mutually incongruent tree topologies (results not shown). The tubulin sequences did not increase the resolution of the phylogeny of the Telonema-group or other well-established groups when concatenated with the Hsp90 and SSU sequences, but the Telonema species were still placed together with the haptophyte and cryptophyte sequences in combined Hsp90+tubulin tree (figure S1; electronic supplementary material). The topological differences in the SSU, Hsp90 and concatenated Hsp90+SSU trees, moving Telonema a few internal branches in the trees, clearly indicate that Telonema is a deep, distinct group of its own, but is also indicating possible evolutionary relationship to the haptophytes and cryptophytes.
Since Telonema was placed close to cryptophytes in the protein trees, additional analyses of SSU and beta-tubulin were performed with the inclusion of sequences from Kathablepharis and Leucocryptos, recently suggested to constitute a sister group to cryptophytes (Cavalier-Smith 2004b; Okamoto & Inouye 2005). The inferred trees confirmed recent analysis by Okamoto & Inouye (2005) in supporting the sister grouping of the kathablepharids and cryptophytes SSU (MLDIST tree), and the unresolved placement of the kathablepharids in beta-tubulin tree (ML tree). However, the inclusion of the kathablepharid sequences did not change the placement of Telonema significantly. Telonema was not attracted to the kathablepharids or cryptophytes (results not shown).
(c) Covarion substitution patterns in SSU and tubulin sequences
For almost all genes, one of the covarion models fitted the data better than the G+I model with at least a Bayes factor of 40.56, which is several magnitudes higher than 10 usually regarded as decisive evidence for the model (Newton & Raftery 1994; Kass & Raftery 1995). Only the Hsp90 data received insignificant Bayes factor differences. Importantly, applying the G+I+COV and G+COV models in analysis of tubulin-containing data clearly increased the posterior probability for the monophyly of excavates in the concatenated protein tree (figure S1; electronic supplementary material). These changes suggest that the covarion-like processes in the evolution of tubulin genes may violate the underlying model of the phylogenetic inferences if not accounted for, possibly causing inconsistencies in previous multi-sequence trees (see, for instance, Hampl et al. 2005; Simpson et al. 2006). However, using the different substitution rate models on the Hsp90 sequences resulted in very similar posterior probability values, indicating that the phylogeny of this gene is not very sensitive to model choice. Since the G+COV clearly fitted best to the alpha- and beta-tubulin sequences, we applied this substitution model in the analysis of the concatenated Hsp90+alpha-tubulin+beta-tubulin sequences. It should be noted that contrary to expected results from testing nested models in a ML framework, estimation of evolutionary models using Bayesian inferences does not always receive highest harmonic mean likelihood score for the most general model (for insights and discussion, see Holder & Lewis 2003; Nylander et al. 2004).
(d) Ultrastructural features of Telonema indicate relationship to chromists and alveolates
Members of the genus Telonema are biflagellated and have a proboscis-like structure located at the flagellar pole (figure 4a–c). The ultrastucture of T. subtilis and T. antarcticum revealed characters that provide clues for the evolutionary position of the Telonema clade, including mitochondria with tubular cristae (tubulocristae) and a highly complex cytoskeleton composed of layers of microtubuli and microfilaments (figure 4d,f). Additional traits defining the phylogeny of the group were exclusively identified in T. antarcticum; these are characteristic peripheral vacuoles located just beneath the cell membrane (figure 4b,d,f) and tripartite tubular hairs on the long flagellum (figure 4e). Chloroplasts were not observed in any of the investigated species.
Figure 4.
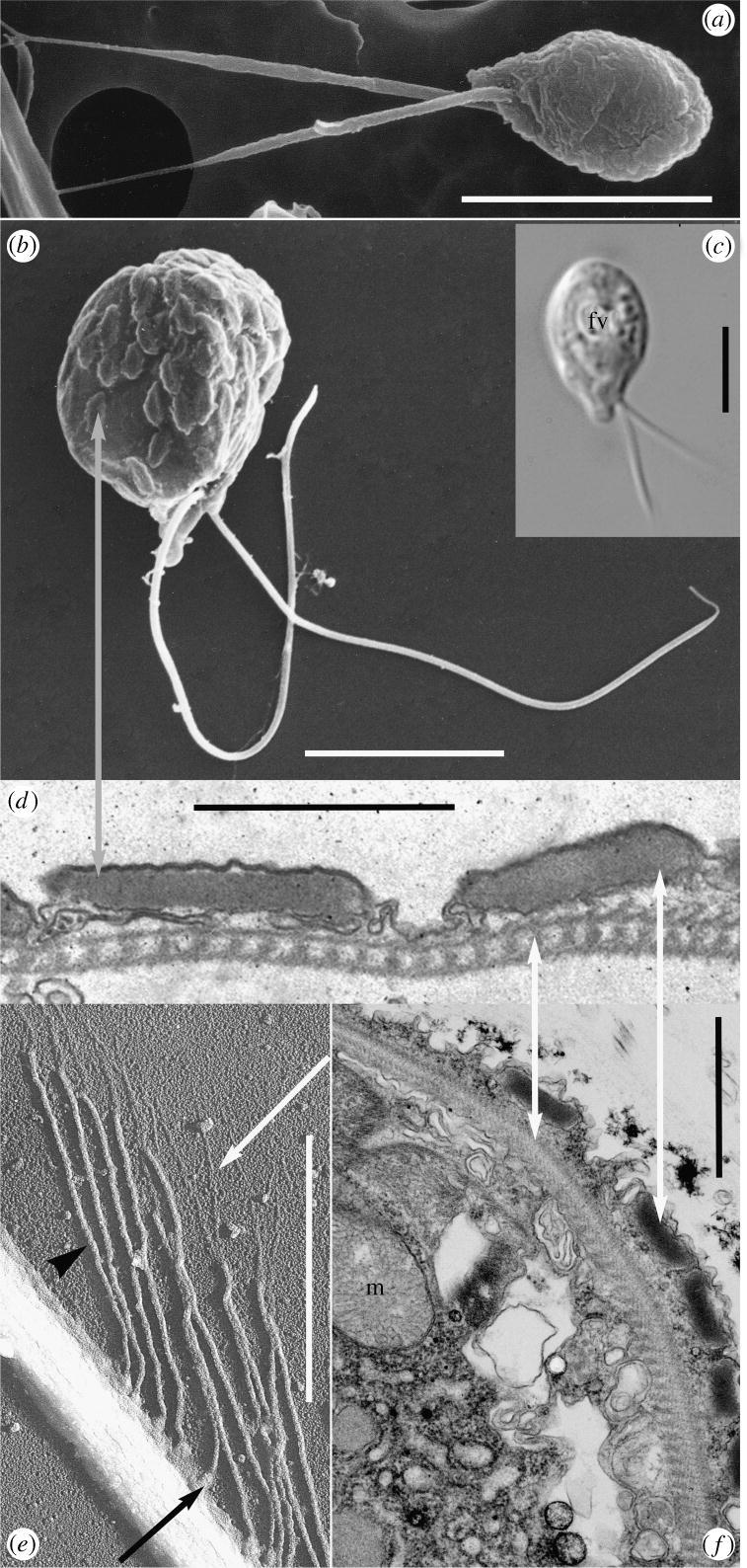
Morphology and ultrastructure of Telonema. (a–c) Whole cell: (a) Telonema subtilis scanning electron micrograph from natural sample (Gulf of Naples); (b) cultured T. antarcticum from the Oslofjord, showing cortical alveoli (grey arrow); (c) light micrograph of cultured cell (RCC 404 from Roscoff), fv, food vacuole. (d–f) Sub-cellular components of T. antarcticum: (d) section through peripheral vacuoles and cytoskeleton (white arrows); (e) detail of flagellum with flagellar tubular tripartite hairs as revealed by shadow cast whole-mount (see white arrow: distal filament; arrowhead: shaft; black arrow: base); (f) longitudinal section of embedded T. antarcticum, showing the cortical alveoli-like peripheral vacuoles, complex cytoskeleton (white arrows), m, mitochondrion with tubular crista. (a–c) Scale bar, 5 μm; (d–f) scale bar, 1 μm.
Cortical alveoli, similar to the peripheral vacuoles identified in T. antarcticum, are one of the main character defining the alveolates, but membranous structures situated just beneath the plasmalemma have also been described in the heterokont classes Raphidophyceae and Dictyochophyceae (Hara et al. 1985; Ishida et al. 2000), as well as among the glaucophytes (Plantae; Cavalier-Smith 1999, 2002). In addition, haptophytes appear to contain similar structures (as their subsurface cisterna act in the process of directed deposition of Golgi-derived scales to the cell surface; Brown & Romanovicz 1976). It is unclear whether these peripheral vacuoles of Telonema are homologous structures already present in a primitive form before the radiation of the alveolates, chromists and Plantae (i.e. the corticates), but they could have evolved independently as an adaptation for strengthening the cell and for pelagic life forms (Cavalier-Smith 2003). On the basis of this single character, Telonema could be related to any of these groups, but both the inferred sequence phylogeny and the presence of tubulocristate mitochondria suggest that Telonema is more closely related to alveolates and some of the chromists than to the glaucophytes. A detailed comparison of kathablepharids and Telonema reveals substantial ultrastructural differences, suggesting distant relationship (table S2 in electronic supplementary material; Lee & Kugrens 1991; Vørs 1992; Kugrens & Clay 2003; Klaveness et al. 2005).
In contrast to many other groups of protists, the heterokonts swim with a forward directed flagellum that pulls the cell owing to a number of tubular hairs attached to the surface of the flagellum (Andersen 2004). Typically, the heterokonts are supposed to have tubular flagellar hairs composed of a hollow base and tubular shaft as well as non-tubular distal fibres, the so-called tripartite hairs (Inouye 1993). Since the tripartite hairs are synthesized in a composite process involving synthesis of distinct components in the endoplasmatic reticulum (ER) and Golgi apparatus, as well as precise assembly and transport to the surface of the flagella, it seems unlikely that such hairs would have evolved several times independently. Tripartite hairs have until now been regarded as a synapomorphic trait defining the heterokonts (for review and references, see Andersen 2004).
(e) Combining ultrastructure and molecular phylogenies: is Telonema a deep-branching chromalveolate?
On the basis of the complex biosynthesis and marginal distribution of tripartite hairs, Telonema should be related to the heterokonts. However, phylogenetic analyses of all gene sequences do not support this hypothesis. Instead, the protein trees place Telonema closer to other chromist groups, the cryptophytes and haptophytes. Interestingly, the cryptophytes also possess similar tubular flagellar hairs (that usually are bipartite), produced in the ER (Hibberd 1971; Inouye 1993). The chromists have recently been placed with the alveolates in the chromalveolates (Cavalier-Smith 2003; Keeling 2004), together constituting a large and highly diverse group of ecologically important heterotrophic and photosynthetic species, such as coccolithophorids, diatoms, brown seaweeds (together, the chromists), ciliates, apicomplexans (harbouring malaria-causing agents) and dinoflagellates (together, the alveolates). The chromists and alveolates are almost never shown as a single clade in nucleus-encoded gene phylogenies (Keeling 2004; Bachvaroff et al. 2005), but phylogenies of concatenated chloroplast sequences and the identification of rare gene replacements unique for these groups have brought together support for the monophyly of both the chromists and chromalveolates (see Cavalier-Smith 2003; Harper & Keeling 2003; Andersen 2004; Keeling 2004; Patron et al. 2004). If the chromists are monophyletic (Cavalier-Smith 2003; Keeling 2004), the most parsimonious explanation for the evolution of these hairs would be an inheritance from a common ancestor of chromists and Telonema (Cavalier-Smith 2002). However, the discrete cytoskeleton identified in Telonema has not been found in any of the chromist lineages, but resembles the cortical structures in alveolates and some excavates (Huttenlauch & Stick 2003).
4. Conclusions
(a) Telonemia: a new phylum related to the kingdom Chromista
In summary, on the basis of ultrastructure (i.e. tubular mitochondrial crista, peripheral vacuoles, tubular tripartite hairs and the complex cytoskeleton), we conclude that Telonema is distinct from all established protist phyla, and constitutes a deep eukaryotic lineage, here defined as a separate phylum, Telonemia, phylum novum (international code for zoological nomenclature, ICZN). The molecular trees confirm the distinctiveness of Telonemia, but also show affinity to some of the chromists. Thus, it is possible that Telonemia branched off deeply from one of the chromist lineages (before the cytoskeleton was reduced in the majority of known chromists), or possibly diverged before the separation of chromists and alveolates (if chromalveolates are monophyletic; see concatenated Hsp90+SSU trees, figure 3)—and thus may be one of the earliest known groups with affinity to the chromalveolates. Telonema has previously been classified as an order Telonemida in the phylum Opalozoa, kingdom Protozoa (Cavalier-Smith 1993), but the presence of tripartite hairs fits better to the definition of kingdom Chromista (Cavalier-Smith 2004b). If Telonemia should be classified within Chromista, we believe that an erection of a new subkingdom would be the most natural, given the ultrastructural differences between Telonema and the current subkingdoms Cryptista (cryptophytes and kathablepharids) or Chromobiota (haptophytes and heterokonts). More sequences from additional phylogenetic markers will be needed to test the evolutionary origin of Telonemia and the proper taxonomic placement within the protist kingdoms.
The present work clearly highlights the importance of establishing cultures in parallel to the acquisition of sequences direct from the environment when aiming to address the evolution of eukaryotes. Finally, our results also emphasize the notion that the so-called ‘uncultivated’ species (Pace 1997) may very well correspond to existing taxonomic entities (Berney et al. 2004; Cavalier-Smith 2004a)—for example, established from field observations by classical protistologists—but for which no cultures have been available or, if available, were not examined in detail until now. The (re)discovery of the Telonema-clade is a glaring example.
(b) Formal description of a new phylum, the Telonemia, phylum novum (ICZN)
Phagotrophic and biflagellate protists of pyriform shape with flagella emerging on opposite sides of a short protruding antapical rostrum. A food vacuole may be located anteriorly at the stout end of cell, in front of centro-lateral large nucleus. Food uptake in depression in antero-ventral region. Characteristic band of vesicles is located laterally. Cell with tubulocristate mitochondria and with characteristic subcortical lamina of microtubuli supporting layers of microfilaments oriented slightly offset from a right angle to each other.
Acknowledgments
We thank J. G. Ormerod for the critical reading of the manuscript and Emilita R. Nerli for technical assistance. We also thank Johan Nylander and Thomas Cavalier-Smith for discussion, Cédric Berney for SSU alignment and anonymous referees for helpful suggestions. This work was supported in part by grants from the Norwegian Research Council (to K.S.J. and D.K.) and the programs PICODIV (EVK3-CT-1999-00021), PicManche, CNRS-Aventis fund, CNRS PROOF BIOSOPE, CPER Souchothèque de Bretagne and CRB (to D.V.).
Supplementary Material
References
- Andersen R.A. Biology and systematics of heterokont and haptophyte algae. Am. J. Bot. 2004;91:1508–1522. doi: 10.3732/ajb.91.10.1508. [DOI] [PubMed] [Google Scholar]
- Bachvaroff T.R, Sanchez Puerta M.V, Delwiche C.F. Chlorophyll c containing plastid relationships based on analyses of a multi-gene dataset with all four chromalveolate lineages. Mol. Biol. Evol. 2005;22:1772–1782. doi: 10.1093/molbev/msi172. doi:10.1093/molbev/msi172 [DOI] [PubMed] [Google Scholar]
- Baldauf S.L, Roger A.J, Wenk-Siefert I, Doolittle W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. doi:10.1126/science.290.5493.972 [DOI] [PubMed] [Google Scholar]
- Berney C, Fahrni J, Pawlowski J. How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2004;2:13. doi: 10.1186/1741-7007-2-13. doi:10.1186/1741-7007-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.M, Romanovicz D.K. Biogenesis and structure of Golgi-derived cellulosic scales in Pleurochrysis. I. Role of the endomembrane system in scale assembly and exocytosis. Appl. Polym. Symp. 1976;28:537–585. [Google Scholar]
- Cavalier-Smith T. The protozoan phylum Opalozoa. J. Eukaryot. Microbiol. 1993;40:609–615. [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Protist phylogeny and the high-level classification of Protozoa. Eur. J. Protistol. 2003;39:338–348. doi:10.1078/0932-4739-00002 [Google Scholar]
- Cavalier-Smith T. Only six kingdoms of life. Proc. R. Soc. B. 2004a;271:1251–1262. doi: 10.1098/rspb.2004.2705. doi:10.1098/rspb.2004.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Chromalveolate diversity and cell megaevolution: interplay of membranes, genomes and cytoskeleton. In: Hirt R.P, Horner D.S, editors. Organelles, genomes and eukaryote phylogeny: an evolutionary synthesis in the age of genomics. The Systematics Association Special Volume Series. vol. 68. CRC Press; London: 2004b. pp. 75–108. [Google Scholar]
- Eikrem W, Moestrup O. Structural analysis of the flagellar apparatus and the scaly periplast in Chrysochromulina scutellum sp. nov. (Prymnesiophyceae, Haptophyta) from the Skagerrak and the Baltic. Phycologia. 1998;37:132–153. [Google Scholar]
- Felsenstein, J. 2004 PHYLIP (phylogeny inference package) version 3.6: distributed by the author. Seattle, WA: Department of Genome Sciences, University of Washington.
- Galtier N. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol. Biol. Evol. 2001;18:866–873. doi: 10.1093/oxfordjournals.molbev.a003868. [DOI] [PubMed] [Google Scholar]
- Griessmann K. Über marine Flagellaten. Arch. Protistenk. 1913;32:1–78. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hampl V, Horner D.S, Dyal P, Kulda J, Flegr J, Foster P.G, Embley T.M. Inference of the phylogenetic position of oxymonads based on nine genes: support for Metamonada and Excavata. Mol. Biol. Evol. 2005;22:2508–2518. doi: 10.1093/molbev/msi245. doi:10.1093/molbev/msi245 [DOI] [PubMed] [Google Scholar]
- Hara Y, Inouye I, Chihara M. Morphology and ultrastructure of Olisthodiscus luteus (Raphidophyceae) with special reference to the taxonomy. Bot. Mag. Tokyo. 1985;98:251–262. [Google Scholar]
- Harper J.T, Keeling P.J. Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol. Biol. Evol. 2003;20:1730–1735. doi: 10.1093/molbev/msg195. doi:10.1093/molbev/msg195 [DOI] [PubMed] [Google Scholar]
- Harper J.T, Waanders E, Keeling P.J. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int. J. Syst. Evol. Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. doi:10.1099/ijs.0.63216-0 [DOI] [PubMed] [Google Scholar]
- Hibberd D.J. Observations of the ultrastructure of the flagella and periplast in the Cryptophyceae. Br. Phycol. J. 1971;6:61–72. [Google Scholar]
- Holder M, Lewis P.O. Phylogeny estimation: traditional and Bayesian approaches. Nat. Rev. Genet. 2003;4:275–284. doi: 10.1038/nrg1044. doi:10.1038/nrg1044 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P. Testing a covariotide model of DNA substitution. Mol. Biol. Evol. 2002;19:698–707. doi: 10.1093/oxfordjournals.molbev.a004128. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Huttenlauch I, Stick R. Occurrence of articulins and epiplasmins in protists. J. Eukaryot. Microbiol. 2003;50:15–18. doi: 10.1111/j.1550-7408.2003.tb00101.x. doi:10.1111/j.1550-7408.2003.tb00101.x [DOI] [PubMed] [Google Scholar]
- Inouye I. Flagella and flagellar apparatuses of algae. In: Berner T, editor. Ultrastructure of microalgae. CRC Press; London: 1993. pp. 99–133. [Google Scholar]
- Ishida K, Cavalier-Smith T, Green B.R. Endomembrane structure and the chloroplast protein targeting pathway in Heterosigma akashiwo (Raphidophyceae, Chromista) J. Phycol. 2000;36:1135–1144. doi:10.1046/j.1529-8817.2000.00071.x [Google Scholar]
- Kass R.E, Raftery A.E. Bayes factors. J. Am. Stat. Assoc. 1995;90:773–795. [Google Scholar]
- Keeling P.J. Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 2004;91:1481–1493. doi: 10.3732/ajb.91.10.1481. [DOI] [PubMed] [Google Scholar]
- Klaveness D, Shalchian-Tabrizi K, Thomsen H.A, Eikrem W, Jakobsen K.S. Telonema antarcticum sp. nov., a common marine phagotrophic flagellate. Int. J. Syst. Evol. Microbiol. 2005;55:2595–2604. doi: 10.1099/ijs.0.63652-0. doi:10.1099/ijs.0.63652-0 [DOI] [PubMed] [Google Scholar]
- Kugrens P, Clay B.L. Cryptomonads. In: Wehr J.D, Sheath R.G, editors. Freshwater algae of North America. Ecology and classification. Academic Press; San Diego, CA: 2003. pp. 715–755. [Google Scholar]
- Lee R.E, Kugrens P. Katablepharis ovalis, a colorless flagellate with interesting cytological characteristics. J. Phycol. 1991;27:505–513. doi:10.1111/j.0022-3646.1991.00505.x [Google Scholar]
- Lopez P, Forterre P, Philippe H. The root of the tree of life in the light of the covarion model. J. Mol. Evol. 1999;49:496–508. doi: 10.1007/pl00006572. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Rodriguez-Valera F, Pedros-Alio C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Massana R, Balague V, Guillou L, Pedros-Alio C. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol. Ecol. 2004;50:231–243. doi: 10.1016/j.femsec.2004.07.001. doi:10.1016/j.femsec.2004.07.001 [DOI] [PubMed] [Google Scholar]
- Moon-van der Staay S.Y, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- Moon-van der Staay S.Y, van der Staay G.W.M, Guillou L, Vaulot D, Claustre H, Medlin L.K. Abundance and diversity of prymnesiophytes in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol. Oceanogr. 2000;45:98–109. [Google Scholar]
- Newton M.A, Raftery A.E. Approximate Bayesian inference by the weighted likelihood bootstrap (with discussion) J. R. Stat. Soc. 1994;56:3–48. [Google Scholar]
- Nylander J.A, Ronquist F, Huelsenbeck J.P, Nieves-Aldrey J.L. Bayesian phylogenetic analysis of combined data. Syst. Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. doi:10.1080/10635150490264699 [DOI] [PubMed] [Google Scholar]
- Okamoto N, Inouye I. The katablepharids are a distant sister group of the Cryptophyta: a proposal for Katablepharidophyta divisio nova/Kathablepharida phylum novum based on SSU rDNA and beta-tubulin phylogeny. Protist. 2005;156:163–179. doi: 10.1016/j.protis.2004.12.003. doi:10.1016/j.protis.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Pace N.R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. doi:10.1126/science.276.5313.734 [DOI] [PubMed] [Google Scholar]
- Patron N.J, Rogers M.B, Keeling P.J. Gene replacement of fructose-1,6-bisphosphate aldolase supports the hypothesis of a single photosynthetic ancestor of chromalveolates. Eukaryot. Cell. 2004;3:1169–1175. doi: 10.1128/EC.3.5.1169-1175.2004. doi:10.1128/EC.3.5.1169-1175.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H. Opinion: long branch attraction and protist phylogeny. Protist. 2000;151:307–316. doi: 10.1078/S1434-4610(04)70029-2. doi:10.1078/S1434-4610(04)70029-2 [DOI] [PubMed] [Google Scholar]
- Richards T.A, Bass D. Molecular screening of free-living microbial eukaryotes: diversity and distribution using a meta-analysis. Curr. Opin. Microbiol. 2005;8:240–252. doi: 10.1016/j.mib.2005.04.010. doi:10.1016/j.mib.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey S.C, Roure B, Burger G, Löffelhardt W, Bohnert H.J, Philippe H, Lang B.F. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. doi:10.1016/j.cub.2005.06.040 [DOI] [PubMed] [Google Scholar]
- Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 2004;49:784–798. [Google Scholar]
- Simpson A.G.B, Inagaki Y, Roger A.J. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of ‘primitive’ eukaryotes. Mol. Biol. Evol. 2006;23:615–625. doi: 10.1093/molbev/msj068. doi:10.1093/molbev/msj068 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. doi:10.1126/science.1071196 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 1998. PAUP: phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- Van de Peer Y, Baldauf S.L, Doolittle W.F, Meyer A. An updated and comprehensive rRNA phylogeny of (crown) eukaryotes based on rate-calibrated evolutionary distances. J. Mol. Evol. 2000;51:565–576. doi: 10.1007/s002390010120. [DOI] [PubMed] [Google Scholar]
- Vørs N. Ultrastructure and autecology of the marine, heterotrophic flagellate Leucocryptos marina (Braarud) Butcher 1967 (Katablepharidaceae/Kathablepharidae), with a discussion of the genera Leucocryptos and Katablepharis/Kathablepharis. Eur. J. Protistol. 1992;28:369–389. doi: 10.1016/S0932-4739(11)80001-5. [DOI] [PubMed] [Google Scholar]
- Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. doi:10.1016/0169-5347(96)10041-0 [DOI] [PubMed] [Google Scholar]
- Yoon H.S, Hackett J.D, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc. Natl Acad. Sci. USA. 2002;99:15 507–15 512. doi: 10.1073/pnas.242379899. doi:10.1073/pnas.242379899 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.