Abstract
Phylogenetic analyses imply that multiple engrailed-family gene duplications occurred during hexapod evolution, a view supported by previous reports of only a single engrailed-family gene in members of the grasshopper genus Schistocerca and in the beetle Tribolium castaneum. Here, we report the cloning of a second engrailed-family gene from Schistocerca gregaria and present evidence for two engrailed-family genes from four additional hexapod species. We also report the existence of a second engrailed-family gene in the Tribolium genome. We suggest that the engrailed and invected genes of Drosophila melanogaster have existed as a conserved gene cassette throughout holometabolous insect evolution. In total 11 phylogenetically diverse hexapod orders are now known to contain species that possess two engrailed-family paralogues, with in each case only one paralogue encoding the RS-motif, a characteristic feature of holometabolous insect invected proteins. We propose that the homeoboxes of hexapod engrailed-family paralogues are evolving in a concerted fashion, resulting in gene trees that overestimate the frequency of gene duplication. We present new phylogenetic analyses using non-homeodomain amino acid sequence that support this view. The S. gregaria engrailed-family paralogues provide strong evidence that concerted evolution might in part be explained by recurrent gene conversion. Finally, we hypothesize that the RS-motif is part of a serine-rich domain targeted for phosphorylation.
Keywords: engrailed, invected, Hexapoda, gene duplication, concerted evolution, gene conversion
1. Introduction
Genes of the engrailed-family encode homeodomain containing transcription factors. Drosophila melanogaster (hereafter Drosophila) engrailed is expressed in a reiterated pattern in the posterior of developing segments and is required for segment border formation and the establishment of positional information within segments (Kornberg 1981). In addition, Drosophila engrailed is known to play important roles in wing development (Hidalgo 1994), hindgut formation (Takashima et al. 2002) and neurogenesis (Siegler & Jia 1999).
Widely conserved patterns of expression support an ancestral role for engrailed-family genes in arthropod segmentation (Damen 2002) and neurogenesis (Duman-Scheel & Patel 1999). The extent to which these roles are conserved beyond arthropods is currently unclear and debated (Seaver 2003). However, engrailed-family genes have acquired new roles and diverged and diversified in function, over the course of metazoan evolution. For example, engrailed-family genes are not involved in the generation of metamerism in vertebrates (Seaver 2003), and in some insects, engrailed-family genes have been recruited to help control the development of wing colour patterns (Brunetti et al. 2001).
There is no doubt that engrailed-family genes have duplicated on numerous occasions during metazoan evolution (Gibert 2002). For example, basally branching deuterostomes, such as sea urchins and the cephalochordate Amphioxus, possess a single engrailed-family gene (Dolecki & Humphreys 1988), while vertebrate deuterostomes, possess two (Logan et al. 1992) or more (Ekker et al. 1992; Force et al. 1999) engrailed-family genes. The engrailed-family paralogues of vertebrates are located on different chromosomes and most likely arose via whole genome duplication during chordate evolution (Gibert 2002).
Many arthropods are also known to possess at least two engrailed-family genes. Drosophila has a second engrailed-family gene named invected (Gustavson et al. 1996). The Drosophila engrailed and invected paralogues arose from a duplication event independent to that which occurred during chordate evolution. The genes have similar intron/exon structure, are tightly linked, co-regulated and positioned tail-to-tail (Gustavson et al. 1996).
Two previous studies have attempted to address when this particular engrailed-family gene duplication event occurred. Marie & Bacon (2000) reported the cloning of two engrailed-family genes from the cockroach Periplaneta americana, while Peterson et al. (1998) found at least two engrailed-family paralogues in the milkweed bug Oncopeltus fasciatus and the firebrat Thermobia domestica. The cloning of engrailed-family paralogues from species representing more basally branching insect clades raised the possibility that the duplication which gave rise to engrailed and invected predated the radiation of insects.
Three lines of evidence argued against this scenario however. First, only a single engrailed-family gene has previously been reported for the beetle Tribolium castaneum (Brown et al. 1994), and grasshoppers of the genus Schistocerca (Patel et al. 1989; Dearden & Akam 2001). Second, the topology of engrailed-family gene trees supports the occurrence of multiple duplications during insect evolution since engrailed-family genes from the same insect order typically group together (Peterson et al. 1998; Marie & Bacon 2000). Finally, the engrailed and invected genes of Drosophila and the silk moth Bombyx mori possess a conserved homeobox intron (Hui et al. 1992), that is not found in the engrailed-family paralogues of other insects, including another holometabolous insect, the honeybee Apis mellifera (Walldorf et al. 1989). The probability that homeobox introns were independently inserted at the same location into two existing engrailed-family paralogues seemed remote. The more parsimonious explanation seemed to be that an ancestral gene containing the homeobox intron was duplicated not long before the divergence of the Lepidoptera and Diptera.
Here, we report the cloning of a second engrailed-family gene from the grasshopper Schistocerca gregaria and the existence of a second engrailed-family gene in the recently published T. castaneum genome. We present evidence for two engrailed-family genes from three additional insect species; the ladybird beetle Harmonia axyridis, the Indian stick insect Carausius morosus and the mayfly Ephemera vulgata. We have also cloned partial sequences for two engrailed-family genes from the springtail Folsomia candida.
Recently published genomic data and our S. gregaria engrailed-family gene data constitute good evidence that the concerted evolution of insect engrailed-family paralogues has resulted in phylogenetic analyses that exaggerate the frequency of gene duplication, a possibility first proposed for hexapod engrailed-family genes by Peterson et al. (1998). We argue that the current data are entirely consistent with the hypothesis that the engrailed-family gene duplication that gave rise to Drosophila engrailed and invected predated the radiation of hexapods, although conclusive proof of this must await genomic data from non-holometabolous hexapods. We discuss the significance of these data for the evolution of engrailed-family gene function in hexapods and for the use of gene trees in the study of gene family evolution.
2. Material and methods
(a) Sample type, preparation and RNA extraction
Total RNA was prepared using the QIAGEN RNeasy mini kit. Individual adult A. mellifera were homogenized by flash freezing in liquid nitrogen and grinding using a mortar and pestle. Embryonic tissue from S. gregaria (30, 30% embryos per column) and C. morosus (six late embryos per column), individual larvae of T. castaneum and H. axyridis and individual E. vulgata adults were homogenized mechanically using a rotor–stator homogenizer. H. axyridis larvae were starved for 5 days prior to RNA extraction to avoid contamination from their gut. S. gregaria genomic DNA was prepared using the QIAGEN Genomic-tip 20/G kit.
(b) Cloning
Details of primers and PCR conditions are provided in the electronic supplementary material. An S. gregaria cDNA library—prepared from mixed stage embryos (20–25%) polyA(+) RNA in Lambda Zap II (Stratagene; Dearden & Akam 2001)—was screened using the methods of Mason & Vulliamy (1995).
(c) Whole mount in situ hybridization
Whole mount in situ hybridization was carried out as per Dearden et al. (2000). To produce gene specific probes, Sgen-1 and Sgen-2 cDNA library clones were restricted towards the 5′ end of the 3′UTR, so that in each case only sequence complementary to the 3′UTR was transcribed (see electronic supplementary material). Probes were digested to aid penetration by incubation in an equal volume of 120 mM Na2CO3, 80 mM NaHCO3, pH10.2 at 60 °C for 40 min.
3. Results
(a) Sequence nomenclature: engrailed-family genes and their conserved domains
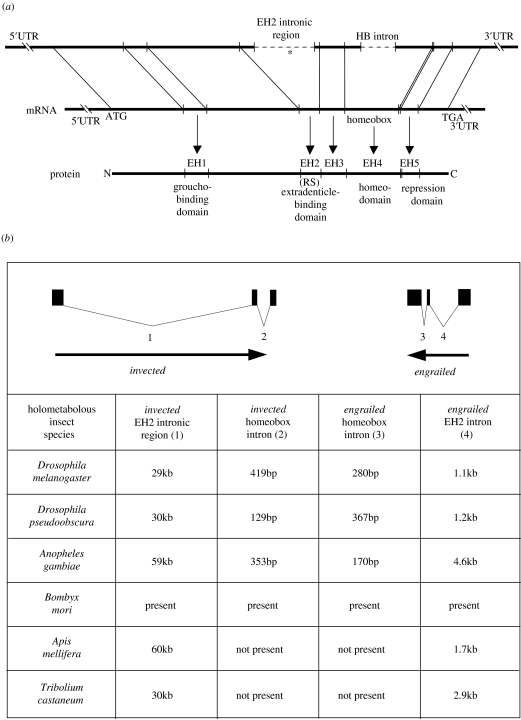
On the basis of comparisons between vertebrate engrailed-family proteins, Logan et al. (1992) identified four engrailed-family homology regions (EH1, EH2, EH3 and EH5), in addition to the homeodomain (EH4; figure 1a). These five regions are common to all metazoan engrailed-family proteins. We use this nomenclature when discussing hexapod engrailed-family proteins, but note that different names have been used by some authors (Marie & Bacon 2000). A widely conserved feature of metazoan engrailed-family genes is an out of frame (+1) intron that is positioned within a codon encoding a highly conserved glycine residue at the C-terminal end of EH2 (figure 1a). We refer to this as the ‘EH2 intron’.
Figure 1.
(a) Diagram showing the relative positions of the engrailed-family homology regions (EH1–EH5). A conserved intron (dashed line) is located within sequence encoding EH2. In a subset of hexapod engrailed-family genes a hexanucleotide microexon (asterisk) is present in this intronic region that introduces an arginine–serine dipeptide (RS) to EH2. A homeobox intron (dashed line) is found only in the engrailed and invected genes of dipteran and lepidopteran species. (b) The genomic arrangement of holometabolous insect engrailed and invected paralogues is conserved. They are linked and orientated tail-to-tail. The relative lengths of homologous introns are also conserved across holometabolous insect species. The EH2 intronic region (1) of invected genes is in the order of tens of kilobases long, and an order of magnitude longer than the EH2 intron (4) of engrailed genes. The homeobox introns (2 and 3) of dipteran and lepidopteran species are only a few hundred bases long. Data for Bombyx mori come from Wu et al. (1999) in which precise sequence lengths were not reported. Some engrailed-family genes contain additional introns not shown in (a) or (b).
Functions have been attributed to EH1, EH2 and EH4 (figure 1a). EH1 binds the co-repressor groucho, which is recruited by engrailed-family proteins to actively repress target genes through modifications in chromatin (Tolkunova et al. 1998). The N-terminal end of EH2 mediates binding to the cofactor Extradenticle (Peltenburg & Murre 1996). Extradenticle binds DNA cooperatively with engrailed-family proteins (Kobayashi et al. 2003). Engrailed-family proteins bind to consensus target sites within regulatory DNA via EH4, more commonly referred to as the homeodomain.
The functions of the other conserved regions are less clear. EH5 is important for target gene repression, but what this domain interacts with is unknown (Han & Manley 1993). EH3 is not highly conserved in arthropod engrailed-family proteins and in the invected proteins of some holometabolous insects has increased in length (figure 2, electronic supplementary material). However, since this region links the extradenticle binding domain and homeodomain, it is thought to be important for cooperative binding (Peltenburg & Murre 1996).
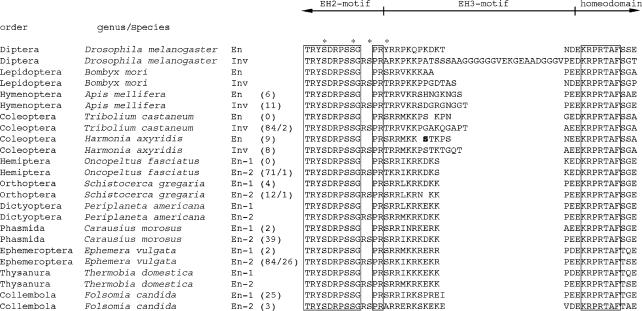
Figure 2.
An alignment of hexapod engrailed-family protein sequence obtained in this study, and for comparison data from Hui et al. (1992), Peterson et al. (1998) and Marie & Bacon (2000). Encompassed EH2 (C-terminal end), EH3 and EH4 (N-terminal end of homeodomain). The number of RT-PCR clones obtained for each sequence is shown in brackets after the genus/species name. In cases where there are two numbers, the second indicates the number of clones recovered that contain a splice variant lacking the hexanucleotide encoding the RS-motif. Residues that are 100% conserved across hexapod species are boxed. A putative polymorphic residue in Harmonia axyridis engrailed is shown in bold; a proline may in some cases replace this serine. The Oncopeltus fasciatus clones were obtained from an RT-PCR experiment using a gene specific primer targeted to sequence encoding EH1. An asterisk is positioned above serine/threonine residues that we hypothesize, in some contexts, might be targets for phosphorylation (see §4).
(b) The conservation of engrailed and invected during holometabolous insect evolution
The genome sequence of a number of holometabolous insect species has recently become available, including several species from the genus Drosophila, another dipteran Anopheles gambiae, the coleopteran T. castaneum and the hymenopteran A. mellifera. In addition, a BAC library has been constructed for the lepidopteran B. mori (Wu et al. 1999). Inspection of the genomic data reveals that all these insect species—which represent four phylogenetically diverse holometabolous insect orders—possess two engrailed-family genes. The intron/exon structure and genomic orientation of the paralogues are remarkably similar (figure 1b). In each case the paralogues are linked and positioned tail-to-tail such that their 3′ exon(s) lie close together. Large EH2 introns in both paralogues mean that the 5′ exon(s) of the genes lie relatively far apart. Together, these data strongly suggest that the engrailed and invected genes of Drosophila existed in the common ancestor of holometabolous insects and that their genomic organization has been conserved during holometabolous insect evolution. Note that the EH2 intron of invected is consistently around an order of magnitude longer than the EH2 intron of engrailed.
A significant inter-species difference is the existence of a conserved homeobox intron in the engrailed and invected genes of dipteran and lepidopteran species. This intron is absent from engrailed and invected in the hymenopteran and coleopteran species. The significance of this will be addressed in §4.
Prior to the Tribolium genome sequence becoming available we cloned fragments of engrailed and invected from another beetle, the ladybird H. axyridis (figure 2, electronic supplementary material). All invected clones shared the same sequence (Accession no. DQ323902), but two distinct populations of clones were recovered for the engrailed paralogue (Accession nos. DQ323900 & DQ323901). Within each of the two populations of clones there is no nucleotide polymorphism. However, the two populations differ at 12 nucleotide positions, including one that translates into a difference at the amino acid level (figure 2, electronic supplementary material). Harmonia axyridis total RNA was isolated from an individual larva and so these results are consistent with the existence of two distinct alleles of engrailed in this individual, perhaps reflecting a high level of nucleotide polymorphism in this polymorphic species of ladybird beetle. However, it cannot be completely ruled out that engrailed has duplicated recently and that there are in fact three engrailed-family genes in this species.
(c) Four non-holometabolous hexapod species each contain two distinct engrailed-family paralogues
We carried out RT-PCR using degenerate primers on total RNA extracted from four non-holometabolous hexapod species, including two hemimetabolous insects, the grasshopper S. gregaria and the stick insect C. morosus, the mayfly E. vulgata (a representative of a basal clade among the winged insects) and the springtail F. candida. In this study, we tentatively consider springtails (Collembola) a hexapod group that branch basally to the Insecta, but we are aware that this traditional view is not supported by some recent molecular phylogenies constructed using mitochondrial DNA sequence data (Cook et al. 2005).
Two distinct engrailed-family sequences were recovered for each of the non-holometabolous species examined (figure 2, electronic supplementary material). In the case of E. vulgata (Accession nos. DQ323896 & DQ323897) and F. candida (Accession nos. DQ323898 & DQ323899) there is a significant number of nucleotide and amino acid differences between sequences from the same species, which is consistent with the existence of at least two engrailed-family paralogues. In the case of the hemimetabolous insects, C. morosus (Accession nos. DQ323894 & DQ323895) and S. gregaria, sequences from the same species were much more similar both at the nucleotide and amino acid levels. However, a high degree of sequence similarity in and around the homeobox may be a general feature of hemimetabolous engrailed-family paralogues (Peterson et al. 1998; Marie & Bacon 2000; see below for S. gregaria).
(d) The arginine-serine dipeptide motif (RS-motif)
The arginine–serine dipeptide motif (or ‘RS-motif’) is specific to a subset of arthropod engrailed-family proteins. The function of the RS-motif is unknown (but see §4). It is positioned towards the C-terminal end of EH2, which is a region encoded by sequence in our RT-PCR clones (figure 2, electronic supplementary material). In the invected genes of holometabolous insects the RS-motif is encoded by a hexanucleotide microexon positioned somewhere within the 20 kb+ EH2 intron or to be more precise the 20 kb+ EH2 ‘intronic region’ since there are in fact introns either side of a microexon (Hui et al. 1992). However, this microexon does not exist in the EH2 intron of holometabolous engrailed genes (Hui et al. 1992). Indeed, using RT-PCR and with reference to the published genome sequence, we have confirmed that the RS-motif is encoded by a microexon in invected (previously named E60 by Walldorf et al. (1989)), but not engrailed (previously named E30 by Walldorf et al. (1989)), in the honeybee A. mellifera (figure 2, electronic supplementary material).
The RS-motif is not restricted to holometabolous insect invected proteins however. For each of the four non-holometabolous hexapod species examined we found that one, and only one, of the two engrailed-family sequences we isolated encodes the RS-motif (figure 2, electronic supplementary material). The RS-motif is also only encoded by one of the paralogues cloned from P. americana by Marie & Bacon (2000) and from O. fasciatus and T. domestica, respectively, by Peterson et al. (1998) (figure 2, electronic supplementary material).
We also found evidence that the RS-motif is encoded by a microexon in non-holometabolous hexapod species. For S. gregaria, O. fasciatus (from experiments described below) and E. vulgata we recovered splice variants of the same gene that lack the hexanucleotide sequence encoding the RS-motif. Clones containing this splice variant were recovered at a particularly high frequency (approx. 25%) from the mayfly E. vulgata, suggesting that in this species it might be expressed at high levels (figure 2, electronic supplementary material).
(e) Two Schistocerca gregaria engrailed-family paralogues: evidence for gene conversion
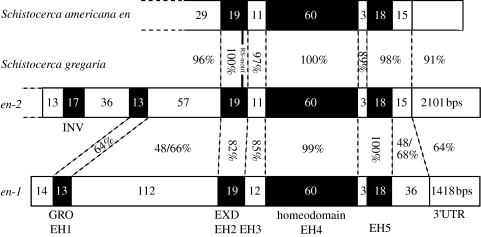
To confirm that there are at least two engrailed-family genes in S. gregaria we screened an embryonic cDNA library using the RT-PCR clones as probes. We recovered two distinct engrailed-family genes (figure 3). One clone contained the entire coding sequence of a gene that does not encode an RS-motif (Sgen-1, Accession no. DQ323891). Another contained what is probably the entire coding sequence of a gene that encodes the RS-motif (Sgen-2, Accession no. DQ323892). The lack of an upstream stop codon and the absence of a convincing Kozak match, means that we cannot rule out the possibility this gene extends further in the 5′ direction. However, sequence encoding all of the engrailed-homology regions (EH1–EH5) and 5′ sequence encoding an invected-specific domain (see below), are present, suggesting that the clone contains most, if not all, of the coding sequence. Sgen-2 is almost identical in sequence, and clearly orthologous, to the engrailed-family gene fragment cloned from Schistocerca americana by Patel et al. (1989) (figure 3).
Figure 3.
Bars represent Schistocerca engrailed-family genes/proteins. The number of amino acids each region encodes is shown within the bars. The location of the RS-motif in S. americana engrailed & Schistocerca gregaria engrailed-2 is indicated by a vertical bar. The percentages between the bars indicate levels of nucleotide identity. The microexon encoding the RS-motif was not included in comparisons. The region between EH1 and EH2, the region C-terminal to EH5 and the 3′UTR are difficult, if not impossible, to align between Sgen-1 and Sgen-2. The percentage identity values shown are arbitrary. Varying gap penalties results in distinct alignments and different percentage identity values. The values here derive from alignments constructed using minimum gap penalties and so maximize levels of sequence identity. For the region between EH1 and EH2 and the region C-terminal to EH5, two percentage identity values are shown, derived from alignments using amino acid and nucleotide sequence, respectively. EXD, extradenticle-binding domain; GRO, groucho-binding domain; INV, invected-specific domain.
By designing gene specific primers flanking the homeobox and carrying out PCR on S. gregaria genomic DNA we confirmed that neither Sgen-1 nor Sgen-2 contain the homeobox intron characteristic of dipteran and lepidopteran engrailed and invected genes (data not shown).
A striking feature of the S. gregaria engrailed-family paralogues is an extremely high level of nucleotide sequence identity in and around the homeobox (figure 3). There is only one nucleotide difference (out of 180 nucleotides) between the homeoboxes of Sgen-1 and Sgen-2. This explains why the existence of the two genes was not evident from clones containing just homeobox sequence (Dearden & Akam 2001). A high level of sequence identity is not a feature of more 5′ or 3′ regions (figure 3). The sequence outside of the engrailed-homology regions is so divergent between the two genes/proteins—both in terms of sequence and length—that it cannot be aligned easily, if at all. Comparing Sgen-1 and Sgen-2, the region between the sequence encoding EH1 and EH2 and the region 3′ to sequence encoding EH5, reveals levels of nucleotide divergence similar to that which exists between the 3′UTRs (figure 3). This is not consistent with a very recent duplication, as might be concluded from homeobox sequence. Sequence outside the engrailed-homology regions has clearly been evolving independently for a significant period of time.
(f) Embryonic expression of Sgen-1 and Sgen-2
To provide a first estimate of whether the function of these two genes has diverged, we examined the embryonic expression of Sgen-1 and Sgen-2 using whole mount in situ hybridization and probes designed to target the 3′UTR of each gene, which have diverged significantly (figure 3 and electronic supplementary material).
The expression of Sgen-1 largely co-localizes with Sgen-2 during S. gregaria segmentation, as has been noted for the engrailed-family paralogues of two other insect species (Peterson et al. 1998; Marie & Bacon 2000). However, as a consequence of carrying out in situ hybridization for both genes on embryos from the same egg pod—which develop more or less in synchrony—we noticed that early antennal and mandibular expression of Sgen-2 appears slightly ahead of Sgen-1 (see electronic supplementary material).
(g) The invected-specific domain
Comparison of the Drosophila and B. mori engrailed and invected genes led to the identification of one invected-specific and two engrailed-specific domains (Hui et al. 1992). Neither of the proposed engrailed-specific domains has proved to be widely conserved across the arthropods, but the invected-specific domain is clearly an ancestral feature of insect engrailed-family proteins. A conserved tetrapeptide (LSVG) is found at the far N-terminal end of all known holometabolous invected proteins. This tetrapeptide is also present at the N-terminal end of the engrailed-family proteins encoded by Paen-2 (Marie & Bacon 2000) and Sgen-2 (this study), but not Paen-1 and Sgen-1. Therefore, hemimetabolous insects are similar to holometabolous insects, in that the engrailed-family paralogue that encodes the RS-motif also encodes the invected-specific domain.
The N-terminal ends of all other non-holometabolous insect engrailed-family proteins are unknown since the most 5′ coding sequence is not included in RT-PCR clones. We designed highly degenerate primers against the sequence encoding the LSVG tetrapeptide and used them in RT-PCR experiments on O. fasciatus total RNA. Interestingly, we managed to amplify additional 5′ sequence—that included sequence encoding EH1—from O. fasciatus total RNA (Accession no. DQ323893 and see electronic supplementary material), but only for the engrailed-family paralogue that encodes the RS-motif (named O.f.en-r2 by Peterson et al. (1998)). These data are consistent with this gene also encoding the LSVG tetrapeptide, a characteristic of invected proteins.
(h) Phylogenetic analyses using N-terminal and C-terminal sequence exhibit distinct topologies
Figure 4 shows two gene trees constructed using Bayesian tree estimation (MrBayes v. 3.1) and amino acid sequence encoded 3′ and 5′ to the EH2 intron (for methods see the electronic supplementary material). Only engrailed-family genes for which the complete coding sequence is available are included in these analyses.
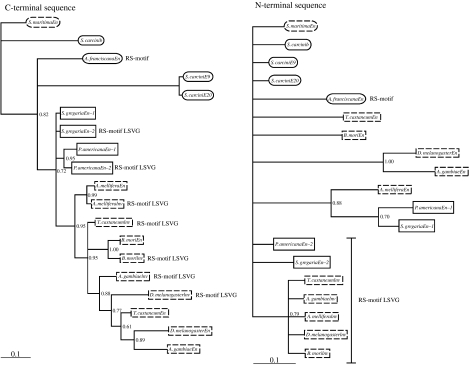
Figure 4.
Phylogenetic trees constructed using C-terminal and N-terminal amino acid sequence from arthropod engrailed-family proteins (for methods see the electronic supplementary material). Boxes with dashed lines, holometabolous insect proteins; Solid boxes, hemimetabolous insect proteins; Solid ellipses, crustacean proteins; Ellipses with dashed lines; myriapod protein. The proteins containing the RS-motif and invected-specific tetrapeptide (LSVG) are labelled. Note that the RS-motif in Artemia fransciscana engrailed is not encoded by a microexon. S. carcini: Sacculina carcini (barnacle/Crustacea, Maxillopoda). S. maritima: Strigamia maritima (centipede/Myriapoda, Chilopoda, Geophilomorpha). Refer to figure 2 (electronic supplementary material) for the full genus/species names of the hexapod species included in this analysis.
The phylogenetic tree constructed using amino acid sequence encoded by the 5′ exon(s) has a topology consistent with the evolutionary history of holometabolous engrailed-family genes inferred from genomic data. The invected proteins of holometabolous insects group together to the exclusion of the engrailed proteins. In the tree constructed using C-terminal amino acid sequence (which includes the homeodomain), engrailed and invected proteins from the same holometabolous species on the whole group together.
Unfortunately, there are relatively few and only short, regions of conservation at the 5′ end of engrailed-family genes and so the relationship between holometabolous and hemimetabolous engrailed-family genes is not resolved by trees constructed using N-terminal amino acid sequence. It is interesting however, that in the tree constructed using C-terminal sequence (figure 4) the proteins encoded by the two Periplaneta paralogues group together, while in the tree constructed using N-terminal sequence (figure 4) the Schistocerca and Periplaneta proteins that lacked the RS-motif group together to the exclusion of the Schistocerca and Periplaneta proteins that contain the RS-motif.
4. Discussion
(a) The origins of Drosophila engrailed and invected
There are now 10 phylogenetically diverse insect orders that contain species known to possess at least two engrailed-family genes. In each case, only one of the two genes encodes the RS-motif, a conserved feature of holometabolous invected proteins. This is also the case in the collembolan F. candida, which is traditionally considered a hexapod species. Where 5′ sequence is available, it is also known that the gene encoding the RS-motif always encodes a second invected-specific domain (the tetrapeptide LSVG). The most parsimonious explanation for these data is that the duplication giving rise to the Drosophila engrailed and invected genes predates the hexapod radiation. If phylogenetic analyses based on mitochondrial DNA sequence are correct, and hexapods and crustaceans are mutually paraphyletic (Cook et al. 2005), the duplication might even predate the divergence of insects and some crustaceans. Most of the crustacean species that have been examined are known to possess multiple engrailed-family genes (Abzhanov & Kaufman 2000; Gibert 2002). At the very least, genomic data and phylogenetic trees constructed using N-terminal amino acid sequence support the duplication predating the radiation of holometabolous insects. The absence of conserved engrailed-specific motifs however, means that it cannot be ruled out that an ancestral hexapod engrailed-family gene encoding the RS-motif and LSVG tetrapeptide was duplicated independently in the lineage leading to holometabolous insects and lineages leading to other, non-holometabolous, hexapod species. The convergent loss from one paralogue of sequence encoding the two invected-specific motifs could plausibly have followed each independent duplication event. Definitive proof that the two paralogues identified in non-holometabolous hexapod species are the orthologues of the engrailed and invected genes of holometabolous hexapods must therefore await genomic data from non-holometabolous hexapod species.
(b) Concerted evolution of the homeoboxes of hexapod engrailed-family genes
Genomic data strongly suggest that engrailed and invected have existed throughout holometabolous insect evolution as a conserved gene cassette. Holometabolous engrailed and invected are consistently found to be closely linked and arranged tail-to-tail. However, phylogenetic trees constructed using C-terminal amino acid sequence, which includes the homeodomain, do not support the proposal that the duplication giving rise to engrailed and invected predated the radiation of holometabolous insects. We believe that misleading phylogenetic analyses are most easily explained by the concerted evolution of sequence in and around the engrailed and invected homeoboxes during holometabolous insect evolution.
One mechanism by which genes can evolve in a concerted fashion is gene conversion. Gene conversion has been defined as ‘the non-reciprocal transfer of information from one DNA duplex to another’ (Szostak et al. 1983). Almost 100% nucleotide identity between the homeoboxes of Sgen-1 and Sgen-2, but significant divergence between the genes elsewhere, represents strong evidence that a homeobox-specific gene conversion event has occurred recently in this hemimetabolous lineage. Indeed, the homeoboxes of engrailed-family genes from the same hemimetabolous species are always more similar to each other at the nucleotide level than they are to the homeoboxes of engrailed-family genes from other hemimetabolous species (data not shown). This is reflected in the grouping of Paen-1 and Paen-2 in a phylogenetic tree constructed using C-terminal (homeodomain) amino acid sequence, but the grouping of Paen-1 and Sgen-1 in a tree constructed using N-terminal sequence. Gene conversion might therefore be a recurrent feature of hemimetabolous engrailed-family gene evolution. Homeobox sequences have clearly diverged between engrailed-family genes from different insect orders. This argues against the nucleotide identity between genes from the same species resulting from selection for nucleotide conservation.
(c) A model for hexapod engrailed-family gene evolution
The absence of genomic data means that the relative position and orientation of hemimetabolous insect engrailed-family paralogues remains unknown. If the duplication that gave rise to Drosophila engrailed and invected did predate the hexapod radiation, then hemimetabolous paralogues, such as Sgen-1 and Sgen-2 are also likely to be linked and arranged tail-to-tail. We propose that the genomic arrangement of hexapod engrailed-family paralogues has predisposed them to recurrent gene conversion between their homeoboxes, which in turn has resulted in phylogenetic trees that overestimate the frequency of gene duplication. Sequence homogenization can also result from unequal crossing over. However, unequal crossing over cannot occur between genes arranged in opposite orientations (Leigh Brown & Ish-Horowicz 1981), and even if the engrailed-family paralogues of non-holometabolous hexapods are arranged in tandem, it is difficult to envisage how this mechanism would result in sequence homogenization specifically restricted to the homeobox and surrounding sequence.
There are precedents for gene conversion occurring between sequences arranged as palindromes. Gene conversion between inverted genes was first demonstrated by Leigh Brown & Ish-Horowicz (1981) in a study on the hsp70 genes of Drosophila species. They proposed that occasional looping of the chromosome brought the linked and inverted hsp70 genes close together and in the same orientation, allowing gene conversion to occur by mechanisms similar to those described in fungi. This type of gene conversion has also been used to explain the concerted evolution of the α-amylase (Hickey et al. 1991; Shibata & Yamazaki 1995) and trypsin (Wang et al. 1999) genes in Drosophila, the hsp82 (Benedict et al. 1996) genes in the mosquito and testis-specific genes positioned within large palindromes on the Y chromosomes of primates (Rozen et al. 2003).
Gene conversion tracts have previously been shown to encompass the entire coding sequence of inverted and linked genes (Shibata & Yamazaki 1995; Benedict et al. 1996). However, gene conversion in Drosophila has been shown to act in a patchwork manner (Curtis & Bender 1991)—specific initiation sites are not used as in some fungi—and gene conversion tracts have been identified within the Drosophila rosy gene that average only 352 bp in length (Hilliker et al. 1994). It is therefore possible that gene conversion tracts are restricted to the homeobox and surrounding sequence of hexapod engrailed-family genes. But why might this be the case? The occurrence of gene conversion depends on high levels of sequence identity (Walsh 1987). The majority of the engrailed-homology regions, including the homeodomain are encoded by sequence at the 3′ end of engrailed-family genes. The 5′ regions of engrailed-family genes—including the sequence encoding EH1—are much less conserved at the nucleotide level. A lack of stabilizing selection in these regions might have meant that following duplication they diverged in sequence at a rate enabling them to escape recurrent gene conversion and its homogenizing effects (Walsh 1987). Gene conversion is also known to occur at a higher frequency between sequences positioned close together (Leigh Brown & Ish-Horowicz 1981; Liao 1999). Due to their tail-to-tail orientation and very large EH2 introns, the 3′ exons of engrailed and invected are much more closely linked than the 5′ exons.
Gene conversion also offers a good explanation for how conserved homeobox introns could come to be fixed in both the engrailed and invected genes of dipteran and lepidopteran species. Gene conversion via a double strand break repair mechanism (Szostak et al. 1983) could have resulted in an intron being transferred from one paralogue to the other.
(d) The role of the engrailed and invected gene cassette in hexapod segmentation
In D. melanogaster engrailed can compensate for the loss of invected function during embryonic development, but invected cannot completely compensate for the loss of engrailed function. Gustavson et al. (1996) proposed that in the rapidly developing Drosophila embryo there is simply not enough time to transcribe and translate the long invected transcript, making transcription of the much shorter engrailed transcript vital for the control of segmentation. This certainly seems to be the case for the segmentation gap gene duplicates knirps and knirps-related. The knirps-related gene has a 19 kb intron (similar in size to the 29 kb EH2 intron in Drosophila invected), whereas the knirps gene only has a 1 kb intron (similar in size to the 1.1 kb EH2 in Drosophila engrailed). A small form of the knirps-related gene, that lacks the 19 kb intron can rescue knirps mutants, but the endogenous gene cannot (Rothe et al. 1992). Also, mutations that lengthen mitotic cycles in the early embryo suppress knirps mutants, presumably because under these conditions there is enough time for the knirps-related gene to be transcribed (Ruden & Jackle 1995).
This study is consistent with others (Peterson et al. 1998; Marie & Bacon 2000) in showing that insect engrailed-family paralogues are largely co-expressed during segmentation. We suspect that in insects exhibiting more primitive, and slower, modes of embryonic development, engrailed-family paralogues may be equally important and act redundantly, in the control of segmentation. The gene duplicates were probably fixed due to divergence in their roles in other aspects of development. Differences in expression and/or function of insect engrailed-family paralogues have been reported for neurogenesis (Siegler & Jia 1999; Marie & Blagburn 2003), hindgut formation (Gustavson et al. 1996) and wing development (Simmonds et al. 1995).
(e) The RS-motif and surrounding sequence: a serine-rich domain targeted for phosphorylation?
The RS-motif is encoded by a microexon in holometabolous insect invected genes. The cloning from primitive insects, such as the mayfly E. vulgata, of splice variants lacking the hexanucleotide encoding the RS-motif suggests that the RS-motif was encoded by a microexon in the hexapod common ancestor. The amino acid residues surrounding the RS-motif are highly conserved. In fact, the amino acid residues in EH2 are 100% conserved across all known hexapod engrailed-family proteins. The RS-motif and surrounding sequence clearly has a conserved and important function.
We hypothesize that this region of hexapod engrailed-family proteins is a serine-rich domain targeted for phosphorylation. A Ca2+-dependent group of protein kinases, exemplified by phosphorylase kinase, require an arginine residue two amino acids on the carboxy-terminal side of their target serine or threonine for function (Krebs & Beavo 1979). Insertion of an RS-motif into the highly conserved domain encoded by EH2 creates two such consensus sites, in addition to one or two that already exist in this region (figure 2, electronic supplementary material).
It is already known that engrailed-family genes encode phosphoproteins. Protein kinase CK2 has been shown to phosphorylate a serine-rich domain lying N-terminal to EH2 in both vertebrate engrailed-2 (Maizel et al. 2002) and Drosophila engrailed (Bourbon et al. 1995). Protein kinase A is known to phosphorylate a serine residue within the homeodomain of vertebrate engrailed-2 (Hjerrild et al. 2004). However, proteins that contain the RS-motif, such as Drosophila invected, have not yet been examined.
It is unclear how phosphorylation of the EH2 domain might affect protein function. However, phosphorylation of vertebrate engrailed-2 by protein kinase CK2 enhances nuclear localization and prevents secretion from COS-7 cells (Maizel et al. 2002). In contrast, the phosphorylation of vertebrate engrailed-2 by protein kinase A decreases its DNA binding affinity (Hjerrild et al. 2004).
(f) Gene conversion and the use of phylogenetics in the study of gene family evolution
Gene trees constructed using C-terminal sequence paint a misleading picture of holometabolous insect engrailed-family gene evolution. What are the wider ramifications of this for the use of phylogenetics in the study of gene family evolution? Fortunately, it appears that gene conversion is largely restricted to sequences that are linked (Liao 1999). Genes that arise via whole genome duplication, interchromosomal segmental duplication and many cases of retrotransposition are likely to evolve independently, free from the homogenizing effects of gene conversion. There is some evidence however that this is not universally the case (Sugino & Innan 2005).
Gene conversion is much more likely to be a factor affecting the evolution of genes that arise through tandem duplication, although these may also be homogenized by unequal crossing over. This study suggests that it is important to determine whether gene conversion has taken place before assuming gene tree topology gives an accurate picture of evolutionary history. However, this may prove problematic, since even if tandemly duplicated genes are no longer undergoing gene conversion, they may once have done so. The latest computer models for gene conversion between tandemly duplicated genes predict that following duplication there will be a period of time—variable in length—before the cycle of divergence through random mutation and homogenization through gene conversion is broken (Teshima & Innan 2004). An accurate understanding of the evolutionary history of some gene families may therefore ultimately rely on the availability of genome sequence from a wider range of organisms.
Acknowledgments
We thank Mike Majerus for providing us with ladybird larvae and we are grateful to Francis Ratnieks for honeybee tissue. Chuck Cook and Kristen Panfilio kindly provided us with springtail and milkweed bug RNA preparations. We also thank Wicken Fen Nature Reserve, Cambridgeshire, UK for permitting us to catch mayflies. A.D.P. was supported by a grant from the Wellcome Trust.
Supplementary Material
Online data includes: Primer sequences and PCR conditions; a description of how 5' Oncopeltus fasciatus engrailed-2 sequence was amplified; sequence and expression data for Sgen-1 and Sgen-2; full coding sequence for Tribolium castaneum engrailed and invected; engrailed-family nucleotide and amino acid alignment files.
References
- Abzhanov A, Kaufman T.C. Evolution of distinct expression patterns for engrailed paralogues in higher crustaceans (Malacostraca) Dev. Genes Evol. 2000;210:493–506. doi: 10.1007/s004270000090. doi:10.1007/s004270000090 [DOI] [PubMed] [Google Scholar]
- Benedict M.Q, Levine B.J, Ke Z.X, Cockburn A.F, Seawright J.A. Precise limitation of concerted evolution to ORFs in mosquito Hsp82 genes. Insect Mol. Biol. 1996;5:73–79. doi: 10.1111/j.1365-2583.1996.tb00042.x. [DOI] [PubMed] [Google Scholar]
- Bourbon H.M, Martin-Blanco E, Rosen D, Kornberg T.B. Phosphorylation of the Drosophila engrailed protein at a site outside its homeodomain enhances DNA binding. J. Biol. Chem. 1995;270:11 130–11 139. doi: 10.1074/jbc.270.19.11130. doi:10.1074/jbc.270.19.11130 [DOI] [PubMed] [Google Scholar]
- Brown S.J, Patel N.H, Denell R.E. Embryonic expression of the single Tribolium engrailed homolog. Dev. Genet. 1994;15:7–18. doi: 10.1002/dvg.1020150103. doi:10.1002/dvg.1020150103 [DOI] [PubMed] [Google Scholar]
- Brunetti C.R, Selegue J.E, Monteiro A, French V, Brakefield P.M, Carroll S.B. The generation and diversification of butterfly eyespot color patterns. Curr. Biol. 2001;11:1578–1585. doi: 10.1016/s0960-9822(01)00502-4. doi:10.1016/S0960-9822(01)00502-4 [DOI] [PubMed] [Google Scholar]
- Cook C.E, Yue Q, Akam M. Mitochondrial genomes suggest that hexapods and crustaceans are mutually paraphyletic. Proc. R. Soc. B. 2005;272:1295–1304. doi: 10.1098/rspb.2004.3042. doi:10.1098/rspb.2004.3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Bender W. Gene conversion in Drosophila and the effects of the meiotic mutants mei-9 and mei-218. Genetics. 1991;127:739–746. doi: 10.1093/genetics/127.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen W.G. Parasegmental organization of the spider embryo implies that the parasegment is an evolutionary conserved entity in arthropod embryogenesis. Development. 2002;129:1239–1250. doi: 10.1242/dev.129.5.1239. [DOI] [PubMed] [Google Scholar]
- Dearden P.K, Akam M. Early embryo patterning in the grasshopper, Schistocerca gregaria: wingless, decapentaplegic and caudal expression. Development. 2001;128:3435–3444. doi: 10.1242/dev.128.18.3435. [DOI] [PubMed] [Google Scholar]
- Dearden P, Grbic M, Falciani F, Akam M. Maternal expression and early zygotic regulation of the Hox3/zen gene in the grasshopper Schistocerca gregaria. Evol. Dev. 2000;2:261–270. doi: 10.1046/j.1525-142x.2000.00065.x. doi:10.1046/j.1525-142x.2000.00065.x [DOI] [PubMed] [Google Scholar]
- Dolecki G.J, Humphreys T. An engrailed class homeo box gene in sea urchins. Gene. 1988;64:21–31. doi: 10.1016/0378-1119(88)90477-5. doi:10.1016/0378-1119(88)90477-5 [DOI] [PubMed] [Google Scholar]
- Duman-Scheel M, Patel N.H. Analysis of molecular marker expression reveals neuronal homology in distantly related arthropods. Development. 1999;126:2327–2334. doi: 10.1242/dev.126.11.2327. [DOI] [PubMed] [Google Scholar]
- Ekker M, Wegner J, Akimenko M.A, Westerfield M. Coordinate embryonic expression of three zebrafish engrailed genes. Development. 1992;116:1001–1010. doi: 10.1242/dev.116.4.1001. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett F.B, Amores A, Yan Y.L, Postlethwaite J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert J.M. The evolution of engrailed genes after duplication and speciation events. Dev. Genes Evol. 2002;212:307–318. doi: 10.1007/s00427-002-0243-2. doi:10.1007/s00427-002-0243-2 [DOI] [PubMed] [Google Scholar]
- Gustavson E, Goldsborough A.S, Ali Z, Kornberg T.B. The Drosophila engrailed and invected genes: partners in regulation, expression and function. Genetics. 1996;142:893–906. doi: 10.1093/genetics/142.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Manley J.L. Functional domains of the Drosophila engrailed protein. Embo. J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey D.A, Bally-Cuif L, Abukashawa S, Payant V, Benkel B.F. Concerted evolution of duplicated protein-coding genes in Drosophila. Proc. Natl Acad. Sci. USA. 1991;88:1611–1615. doi: 10.1073/pnas.88.5.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo A. Three distinct roles for the engrailed gene in Drosophila wing development. Curr. Biol. 1994;4:1087–1098. doi: 10.1016/s0960-9822(00)00247-5. doi:10.1016/S0960-9822(00)00247-5 [DOI] [PubMed] [Google Scholar]
- Hilliker A.J, Harauz G, Reaume A.G, Gray M, Clark S.H, Chovnick A. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics. 1994;137:1019–1026. doi: 10.1093/genetics/137.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerrild M, Stensballe A, Jensen O.N, Gammeltoft S, Rasmussen T.E. Protein kinase A phosphorylates serine 267 in the homeodomain of engrailed-2 leading to decreased DNA binding. FEBS Lett. 2004;568:55–59. doi: 10.1016/j.febslet.2004.05.009. doi:10.1016/j.febslet.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Hui C.C, Matsuno K, Ueno K, Suzuki Y. Molecular characterization and silk gland expression of Bombyx engrailed and invected genes. Proc. Natl Acad. Sci. USA. 1992;89:167–171. doi: 10.1073/pnas.89.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Fujioka M, Tolkunova E.N, Deka D, Abu-Shaar M, Mann R.S, Jaynes J.B. Engrailed cooperates with extradenticle and homothorax to repress target genes in Drosophila. Development. 2003;130:741–751. doi: 10.1242/dev.00289. doi:10.1242/dev.00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T. Engrailed: a gene controlling compartment and segment formation in Drosophila. Proc. Natl Acad. Sci. USA. 1981;78:1095–1099. doi: 10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs E.G, Beavo J.A. Phosphorylation-dephosphorylation of enzymes. Annu. Rev. Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. doi:10.1146/annurev.bi.48.070179.004423 [DOI] [PubMed] [Google Scholar]
- Leigh Brown A.J, Ish-Horowicz D. Evolution of the 87A and 87C heat-shock loci in Drosophila. Nature. 1981;290:677–682. doi: 10.1038/290677a0. doi:10.1038/290677a0 [DOI] [PubMed] [Google Scholar]
- Liao D. Concerted evolution: molecular mechanism and biological implications. Am. J. Hum. Genet. 1999;64:24–30. doi: 10.1086/302221. doi:10.1086/302221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C, Hanks M.C, Noble-Topham S, Nallainathan D, Provart N.J, Joyner A.L. Cloning and sequence comparison of the mouse, human, and chicken engrailed genes reveal potential functional domains and regulatory regions. Dev. Genet. 1992;13:345–358. doi: 10.1002/dvg.1020130505. doi:10.1002/dvg.1020130505 [DOI] [PubMed] [Google Scholar]
- Maizel A, Tassetto M, Filhol O, Cochet C, Prochiantz A, Joliot A. Engrailed homeoprotein secretion is a regulated process. Development. 2002;129:3545–3553. doi: 10.1242/dev.129.15.3545. [DOI] [PubMed] [Google Scholar]
- Marie B, Bacon J.P. Two engrailed-related genes in the cockroach: cloning, phylogenetic analysis, expression and isolation of splice variants. Dev. Genes Evol. 2000;210:436–448. doi: 10.1007/s004270000082. doi:10.1007/s004270000082 [DOI] [PubMed] [Google Scholar]
- Marie B, Blagburn J.M. Differential roles of engrailed paralogs in determining sensory axon guidance and synaptic target recognition. J. Neurosci. 2003;23:7854–7862. doi: 10.1523/JNEUROSCI.23-21-07854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.J, Vulliamy T.J. Screening recombinant DNA libraries by hybridisation and amplification. In: Hames B.D, Higgins S.J, editors. Gene probes 2: a practical approach. Oxford University Press; Oxford, UK: 1995. pp. 31–75. [Google Scholar]
- Patel N.H, Kornberg T.B, Goodman C.S. Expression of engrailed during segmentation in grasshopper and crayfish. Development. 1989;107:201–212. doi: 10.1242/dev.107.2.201. [DOI] [PubMed] [Google Scholar]
- Peltenburg L.T, Murre C. Engrailed and hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. Embo. J. 1996;15:3385–3393. [PMC free article] [PubMed] [Google Scholar]
- Peterson M.D, Popadic A, Kaufman T.C. The expression of two engrailed-related genes in an apterygote insect and a phylogenetic analysis of insect engrailed-related genes. Dev. Genes Evol. 1998;208:547–557. doi: 10.1007/s004270050214. doi:10.1007/s004270050214 [DOI] [PubMed] [Google Scholar]
- Rothe M, Pehl M, Taubert H, Jackle H. Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature. 1992;359:156–159. doi: 10.1038/359156a0. doi:10.1038/359156a0 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek J.D, Minx P.J, Cordum H.S, Waterston R.H, Wilson R.K, Page D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. doi:10.1038/nature01723 [DOI] [PubMed] [Google Scholar]
- Ruden D.M, Jackle H. Mitotic delay dependent survival identifies components of cell cycle control in the Drosophila blastoderm. Development. 1995;121:63–73. doi: 10.1242/dev.121.1.63. [DOI] [PubMed] [Google Scholar]
- Seaver E.C. Segmentation: mono- or polyphyletic? Int. J. Dev. Biol. 2003;47:583–595. [PubMed] [Google Scholar]
- Shibata H, Yamazaki T. Molecular evolution of the duplicated Amy locus in the Drosophila melanogaster species subgroup: concerted evolution only in the coding region and an excess of nonsynonymous substitutions in speciation. Genetics. 1995;141:223–236. doi: 10.1093/genetics/141.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegler M.V, Jia X.X. Engrailed negatively regulates the expression of cell adhesion molecules connectin and neuroglian in embryonic Drosophila nervous system. Neuron. 1999;22:265–276. doi: 10.1016/s0896-6273(00)81088-0. doi:10.1016/S0896-6273(00)81088-0 [DOI] [PubMed] [Google Scholar]
- Simmonds A.J, Brook W.J, Cohen S.M, Bell J.B. Distinguishable functions for engrailed and invected in anterior–posterior patterning in the Drosophila wing. Nature. 1995;376:424–427. doi: 10.1038/376424a0. doi:10.1038/376424a0 [DOI] [PubMed] [Google Scholar]
- Sugino R.P, Innan H. Estimating the time to the whole-genome duplication and the duration of concerted evolution via gene conversion in yeast. Genetics. 2005;171:63–69. doi: 10.1534/genetics.105.043869. doi:10.1534/genetics.105.043869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J.W, Orr-Weaver T.L, Rothstein R.J, Stahl F.W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. doi:10.1016/0092-8674(83)90331-8 [DOI] [PubMed] [Google Scholar]
- Takashima S, Yoshimori H, Yamasaki N, Matsuno K, Murakami R. Cell-fate choice and boundary formation by combined action of Notch and engrailed in the Drosophila hindgut. Dev. Genes Evol. 2002;212:534–541. doi: 10.1007/s00427-002-0262-z. doi:10.1007/s00427-002-0262-z [DOI] [PubMed] [Google Scholar]
- Teshima K.M, Innan H. The effect of gene conversion on the divergence between duplicated genes. Genetics. 2004;166:1553–1560. doi: 10.1534/genetics.166.3.1553. doi:10.1534/genetics.166.3.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkunova E.N, Fujioka M, Kobayashi M, Deka D, Jaynes J.B. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol. Cell Biol. 1998;18:2804–2814. doi: 10.1128/mcb.18.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walldorf U, Fleig R, Gehring W.J. Comparison of homeobox-containing genes of the honeybee and Drosophila. Proc. Natl Acad. Sci. USA. 1989;86:9971–9975. doi: 10.1073/pnas.86.24.9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J.B. Sequence-dependent gene conversion: can duplicated genes diverge fast enough to escape conversion? Genetics. 1987;117:543–557. doi: 10.1093/genetics/117.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Magoulas C, Hickey D. Concerted evolution within a trypsin gene cluster in Drosophila. Mol. Biol. Evol. 1999;16:1117–1124. doi: 10.1093/oxfordjournals.molbev.a026202. [DOI] [PubMed] [Google Scholar]
- Wu C, Asakawa S, Shimizu N, Kawasaki S, Yasukochi Y. Construction and characterization of bacterial artificial chromosome libraries from the silkworm, Bombyx mori. Mol. Gen. Genet. 1999;261:698–706. doi: 10.1007/s004380050013. doi:10.1007/s004380050013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online data includes: Primer sequences and PCR conditions; a description of how 5' Oncopeltus fasciatus engrailed-2 sequence was amplified; sequence and expression data for Sgen-1 and Sgen-2; full coding sequence for Tribolium castaneum engrailed and invected; engrailed-family nucleotide and amino acid alignment files.