Abstract
The FKBP12–rapamycin-associated protein (FRAP; also called RAFT1/mTOR) regulates translation initiation and entry into the cell cycle. Depriving cells of amino acids or treating them with the small molecule rapamycin inhibits FRAP and results in rapid dephosphorylation and inactivation of the translational regulators 4E-BP1(eukaryotic initiation factor 4E-binding protein 1) and p70s6k (the 70-kDa S6 kinase). Data published recently have led to the view that FRAP acts as a traditional mitogen-activated kinase, directly phosphorylating 4E-BP1 and p70s6k in response to mitogenic stimuli. We present evidence that FRAP controls 4E-BP1 and p70s6k phosphorylation indirectly by restraining a phosphatase. A calyculin A-sensitive phosphatase is required for the rapamycin- or amino acid deprivation-induced dephosphorylation of p70s6k, and treatment of Jurkat I cells with rapamycin increases the activity of the protein phosphatase 2A (PP2A) toward 4E-BP1. PP2A is shown to associate with p70s6k but not with a mutated p70s6k that is resistant to rapamycin- and amino acid deprivation-mediated dephosphorylation. FRAP also is shown to phosphorylate PP2A in vitro, consistent with a model in which phosphorylation of PP2A by FRAP prevents the dephosphorylation of 4E-BP1 and p70s6k, whereas amino acid deprivation or rapamycin treatment inhibits FRAP’s ability to restrain the phosphatase.
FKBP12–rapamycin-associated protein (FRAP; also called RAFT1/mTOR) regulates translation initiation by altering the phosphorylation state of the translational regulators 4E-BP1(eukaryotic initiation factor 4E-binding protein 1) and p70s6k (the 70-kDa S6 kinase) (1). Multiple phosphorylation sites in a variety of sequence contexts in 4E-BP1 and p70s6k are rapidly dephosphorylated in many mammalian cell lines after FRAP inhibition by amino acid deprivation (2) or rapamycin treatment (3, 4). Although it is possible that these growth-arresting treatments act by inhibiting the various kinases that phosphorylate 4E-BP1 and p70s6k, it also is possible that rapamycin treatment (resulting in formation of the FKBP12–rapamycin complex) and amino acid deprivation activate a serine/threonine protein phosphatase that dephosphorylates these regulatory sites. Such a mechanism would be analogous to the ability of FKBP12–FK506 to activate the phosphatase activity of PP2B toward para-nitrophenylphosphate (5). It seems unlikely that the p70s6k kinases are directly inhibited by FKBP12–rapamycin because a p70s6k mutant truncated at the N and C termini is readily phosphorylated in vivo in response to mitogens, even in the presence of rapamycin (6–8). Furthermore, PDK1, a serine/threonine kinase that phosphorylates p70s6k at the rapamycin-sensitive site Thr-229, has been shown to be insensitive to rapamycin (9). Many attempts have been made to detect direct phosphorylation of 4E-BP1 and p70s6k by FRAP in vitro. In the case of 4E-BP1, inefficient phosphorylation has been observed (10), but the possibility of contamination from other kinases has not been excluded. Phosphorylation of p70s6k by FRAP has also been reported (11), but only when using an inactive, misfolded fragment of p70s6k that is unresponsive to FRAP regulation in vivo (7, 8). The in vivo relevance of these inefficient in vitro phosphorylations remains uncertain. Here, we examine the possibility that FRAP regulates 4E-BP1 and p70s6k phosphorylation via the serine/threonine protein phosphatase (PP2A).
MATERIALS AND METHODS
Kinase Assays.
Jurkat cells expressing the simian virus 40 large tumor antigen (TAg) (7 × 106) were pelleted and resuspended in cell culture media containing amino acids (RPMI medium 1640 supplemented with 10% FBS, GIBCO/BRL) or lacking amino acids (Dulbecco’s PBS supplemented with 10% dialyzed FBS, GIBCO/BRL). Resuspended cells were treated with 20 nM calyculin A (Alexis Biochemicals, San Diego) or 25 nM rapamycin for 45 min before lysis in 500 μl buffer A (20 mM NaH2PO4 pH 7.2/1 mM Na3VO4/5 mM NaF, 25 mM 2-glycerophosphate/2 mM EGTA/2 mM EDTA/0.5% Triton X-100/1 mM DTT/1 μg/ml leupeptin/1 μg/ml pepstatin/0.2 mM PMSF). Cell lysates were incubated with α-p70s6k polyclonal antibody (Santa Cruz Biotechnology) followed by precipitation with protein A/G PLUS-agarose (Santa Cruz Biotechnology). Kinase reactions were then performed as described (3).
32P Labeling of Glutathione S-Transferase (GST)–4E-BP1.
GST–4E-BP1 was expressed in Escherichia coli from the plasmid pGEX-2T (a gift from Nahum Sonenberg, McGill University, Toronto) and purified by glutathione Sepharose chromatography (Amersham Pharmacia) following procedures recommended by the manufacturer. Purified GST–4E-BP1 was eluted with free glutathione and dialyzed against PBS containing protease inhibitors and DTT as above. Dialyzed GST–4E-BP1 was incubated with 10 units mitogen-activated protein kinase (New England Biolabs) in the presence of 0.1 mM ATP with 0.1 mCi (1 Ci = 37 GBq) [γ-32P]ATP for 15 min at 37°C. The 32P-labeled GST–4E-BP1 was then repurified on glutathione Sepharose and eluted with free glutathione.
Phosphatase Assays.
For assaying cell lysates, 2 × 107 TAg Jurkat cells were treated with or without 25 nM rapamycin for 25 min before lysis with buffer B (50 mM Tris⋅HCl, pH 8.0/0.5 M NaCl/1% Nonidet P-40/protease inhibitors as above). Lysates were clarified by centrifugation. A 120-μl volume of lysate was mixed with 80 μl of 32P-labeled GST–4E-BP1 substrate and incubated at 25°C with shaking. Aliquots of 20 μl were removed at the times indicated and were boiled in SDS loading buffer before separation by using SDS/PAGE. The amount of 32P-labeled GST–4E-BP1 remaining at each time point was quantitated by bioimaging analysis (Fujix Bas 1000). For assaying PP2A immune complexes, 5 × 107 cells were treated for 25 min and lysed as above. Clarified cell lysates were incubated with an α-PP2A polyclonal antibody (Upstate Biotechnology, Lake Placid, NY) followed by precipitation with 15 μl of protein A/G PLUS-agarose (Santa Cruz Biotechnology). Immune complexes were washed three times with buffer B and resuspended in 120 μl of buffer C (50 mM Tris⋅HCl, pH 7.0/10% glycerol/2 mg/ml BSA/2 mM MnCl2/14 mM 2-mercaptoethanol/protease inhibitors as above). Resuspended beads were mixed with 80 μl of 32P-labeled GST–4E-BP1 substrate and incubated at 33°C with shaking. Aliquots were removed and quantitated as above.
Western Blotting.
For the anti-PP2A blot, 3 × 107 TAg Jurkat cells were treated with or without rapamycin for 25 min as above and lysed in 0.5 ml of buffer A with 150 mM NaCl followed by p70s6k immunoprecipitation as above. Lysates and immune complexes were separated by using SDS/PAGE and transferred to Immobilon-P (Millipore). PP2A was visualized with an anti-PP2A mAb (Upstate Biotechnology, Lake Placid, NY) and a horseradish peroxidase-conjugated anti-mouse secondary antibody followed by enhanced chemiluminescence (Amersham Pharmacia). For the anti-hemagglutinin (HA) blot, 2 × 107 TAg Jurkat cells were transfected with 20 μg of the plasmid pKH3 containing HA-tagged, full-length p70s6k or p70s6k truncated at the N and C termini as described (8) (gift from John Blenis, Harvard University Medical School). Transfected cells were grown for 24 h before lysis in buffer A with 150 mM NaCl. Clarified lysates were incubated with an anti-PP2A mAb (Upstate Biotechnology, Lake Placid, NY,) and PP2A was precipitated with protein A agarose (GIBCO/BRL). Immune complexes were washed three times with buffer A plus 150 mM NaCl. Cell lysates and immune complexes were separated by using SDS/PAGE and then were transferred to Immobilon-P (Millipore). HA-tagged proteins were visualized as described above by using 12CA5 ascites as the primary antibody. For the 4E-BP1 blot, NIH 3T3 cells were serum starved for 36 h before treatment with 30 nM rapamycin or 30 nM calyculin A for 60 min or 30 nM rapamycin for 30 min followed by addition of 30 nM calyculin A for a further 30 min. Heat-stable proteins were extracted and separated by using SDS/PAGE before detection with an anti-4E-BP1 polyclonal antibody.
PP2A Phosphorylation.
Sf9 cells (1.5 × 108) were infected for 50 h with baculoviruses for expression of full-length, FLAG-tagged, wild-type or kinase-dead (D2338A) FRAP. Cells were lysed in 8 ml of buffer A and immunoprecipitated with M2 resin (Eastman Kodak). Immune complexes were washed four times with buffer A and once with buffer D (25 mM Hepes, pH 7.7/50 mM KCl/10 mM MgCl2/20% glycerol/0.1% Nonidet P-40/1 mM DTT). Immune complexes were split into three aliquots and mixed with 25 μl of buffer D plus 250 nM calyculin A, 0.1 mM cold dATP, 15 μCi [γ-32P]ATP, and GST–4E-BP1 or PP2A as substrates. GST–4E-BP1 was purified as described above. PP2A was obtained from Upstate Biotechnology (Lake Placid, NY). Approximately 0.1 μg of total protein was added in each case. Kinase reactions were carried out for 15 min at 30°C before quenching with SDS loading buffer and separation of proteins with SDS/PAGE. Proteins were transferred to Immobilon-P (Millipore) and visualized by autoradiography.
RESULTS AND DISCUSSION
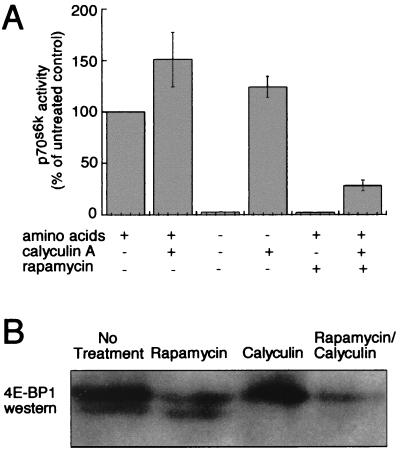
We used calyculin A, a molecule known to inhibit potently the serine/threonine protein phosphatases 1 and 2A (PP1 and PP2A), to test whether these phosphatase activities are required for rapamycin-induced, FRAP-mediated (3) dephosphorylation of 4E-BP1 and p70s6k. As shown in Fig. 1A, calyculin A treatment prevents the inactivation of p70s6k in Tag Jurkat cells after amino acid deprivation and reduces the extent of p70s6k inactivation after treatment with rapamycin. Rapamycin treatment also induces dephosphorylation of 4E-BP1 less effectively in the presence of calyculin A (Fig. 1B). Furthermore, like the p70s6k kinases (6), the 4E-BP1 kinase(s) appear to retain some activity in quiescent NIH 3T3 cells in the presence of rapamycin because calyculin A treatment causes phosphorylation of dephosphorylated 4E-BP1 in the presence of rapamycin (Fig. 1B, lane 4). These data suggest the involvement of a calyculin A-sensitive phosphatase in the rapamycin-induced dephosphorylation of p70s6k and 4E-BP1. The data do not exclude the possibility that a calyculin A-insensitive phosphatase or a rapamycin-sensitive p70s6k kinase may also be involved in p70s6k regulation. Indeed, although calyculin A prevents complete rapamycin-induced deactivation of p70s6k, some p70s6k dephosphorylation occurs in the presence of calyculin A as is consistent with results from different cell lines published previously (12, 13).
Figure 1.
Phosphatase inhibition interferes with FRAP-mediated dephosphorylation of 4E-BP1 and p70s6k. (A) The activity of endogenous p70s6k immunoprecipitated from TAg Jurkat cells after treatment for 45 min with or without amino acids, calyculin A, and rapamycin. Kinase activity is shown as a percentage of activity from cells grown in the presence of amino acids without calyculin A or rapamycin. Three experiments were performed for each condition; means and SD are shown. (B) α-4E-BP1 Western blot showing the relative quantities of hyper- (Upper) and hypophosphorylated (Lower) 4E-BP1 after various rapamycin and calyculin A treatments. Quiescent NIH 3T3 cells were either untreated, treated with 30 nM rapamycin or calyculin A for 60 min, or treated with 30 nM rapamycin for 30 min followed by addition of 30 nM calyculin A for an additional 30 min. Cell lysates were boiled and centrifuged to remove heat-unstable proteins before separation of heat-stable proteins by SDS/PAGE.
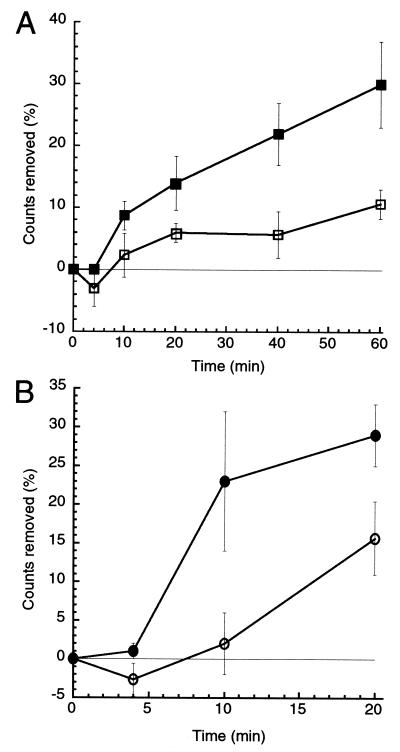
To determine whether FRAP regulates serine/threonine phosphatase activity, we tested the ability of cell lysates from rapamycin-treated and untreated cells to dephosphorylate 4E-BP1. Bacterially expressed GST–4E-BP1 fusion protein was purified and phosphorylated in vitro by incubation with [γ-32P]ATP and the mitogen-activated protein kinase Erk2, which phosphorylates 4E-BP1 at sites known to be phosphorylated in vivo (14). After labeling, excess Erk2 and free ATP were removed by thorough washing. TAg Jurkat cells were then treated with rapamycin or a control before detergent lysis. Clarified cell extracts were incubated with the labeled 4E-BP1 substrate, and 4E-BP1 dephosphorylation was measured over time. As shown in Fig. 2A, rapamycin-treated cells produce lysates with an increased level of serine/threonine phosphatase activity. The majority of this phosphatase activity was inhibited by addition of calyculin A or okadaic acid (data not shown). Although prevention of insulin-induced PP2A inactivation by rapamycin has been reported (15), a biochemical demonstration that phosphatase activity is induced on rapamycin treatment has not been shown previously. Serine/threonine protein phosphatases perform numerous functions in vivo, and their specificity is determined by such factors as subcellular localization and association with other proteins (16). It seems plausible that these factors contribute to localized phosphatase activation in vivo that is more robust than the modest effect observed in vitro with whole-cell lysates.
Figure 2.
Rapamycin increases serine-threonine phosphatase activity. Removal of 32P from exogenously phosphorylated and purified GST–4E-BP1 was measured. Cell lysates (A) or PP2A immune complexes (B) from untreated (open symbols) and rapamycin-treated (filled symbols) TAg Jurkat cells were incubated with 32P-labeled GST–4E-BP1. Aliquots of the reaction mix were removed at the times indicated, and the amount of 32P label remaining on the GST–4E-BP1 was determined by SDS/PAGE followed by phosphoimager quantification. Each point represents the percent decrease in GST–4E-BP1-bound 32P relative to 32P present at t = 0. Three experiments were performed; error bars reflect the SEM for each point.
Genetic and biochemical evidence suggest that serine/threonine phosphatases are affected by rapamycin in yeast. Rapamycin treatment or nutrient deprivation of Saccharomyces cerevisiae causes the PP2A homologs Pph21p and Pph22p, as well as a third phosphatase, Sit4p to dissociate from an essential protein Tap42p (17). Rapamycin-induced dissociation of mammalian PP2A from α4, the mammalian Tap42p homolog, has been reported (18, 19), although this effect was not observed in another study (20). To test whether PP2A is the calyculin A- and okadaic acid-sensitive phosphatase that is activated by rapamycin treatment, we measured the phosphatase activity of the PP2A catalytic subunit (PP2Ac) immunoprecipitated from rapamycin-treated and untreated TAg Jurkat cells. PP2Ac immunoprecipitated from rapamycin-treated cells exhibited increased phosphatase activity toward 32P-labeled 4E-BP1 (Fig. 2B), suggesting that PP2A is indeed regulated by FRAP. PP2Ac is associated with several distinct regulatory complexes in vivo (16). The identity of the specific PP2Ac-containing complex regulated by FRAP is unknown. Here, we have measured the activity of total cellular PP2A by immunoprecipitating the catalytic subunit. The modest effect of rapamycin treatment on total PP2A activity may reflect much stronger activation of a subpopulation of PP2A.
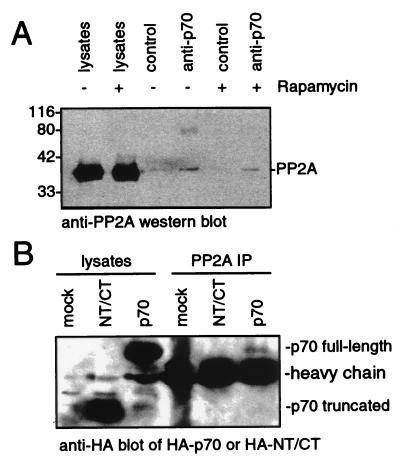
The involvement of a rapamycin-activated phosphatase in the actions of FRAP may explain the behavior of a rapamycin-resistant p70s6k mutant. Removal of the N and C termini of p70s6k (but not removal of the C terminus alone) yields a kinase that is readily phosphorylated and activated on serum stimulation but cannot be dephosphorylated and inactivated by rapamycin treatment in vivo (7, 8). This result suggests that the kinases that activate p70s6k remain active in the presence of rapamycin, or, less likely, that truncation permits phosphorylation by a promiscuous, mitogen-activated kinase. Furthermore, it suggests that the factor responsible for rapamycin-induced, FRAP-mediated (3) dephosphorylation of p70s6k interacts with the N terminus of p70s6k. This model is supported by the recent finding that PP2A associates with full-length p70s6k (ref. 21; Fig. 3A). As shown in Fig. 3B, PP2A exhibits a reduced ability to interact with the rapamycin-resistant, truncated p70s6k mutant. The reduced ability of the mutant p70s6k to interact with PP2A may be responsible for its resistance to rapamycin-induced, FRAP-mediated dephosphorylation.
Figure 3.
PP2A interacts with full-length p70s6k but not with a rapamycin-resistant p70s6k mutant. (A) Western blot of PP2A in cell lysates and p70s6k immune complexes. TAg Jurkat cells were treated with or without 25 nM rapamycin before lysis in buffer containing phosphatase inhibitors. They were then incubated with an α-p70s6k polyclonal antibody or a control polyclonal antibody. The immunoprecipitated proteins were separated by using SDS/PAGE, and PP2A was visualized by Western blotting. (B) Western blot of HA-tagged p70s6k in PP2A immune complexes. TAg Jurkat cells were mock-transfected or transfected with plasmids that express HA-tagged p70s6k or a rapamycin-resistant, HA-tagged p70s6k mutant that has been truncated at the N and C termini (NT/CT). PP2A was immunoprecipitated from the lysed cells, and proteins were separated by using SDS/PAGE. HA-tagged proteins were visualized by Western blotting.
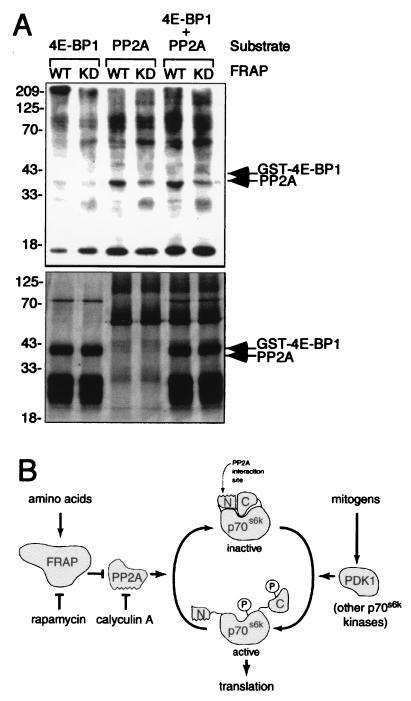
The interaction between p70s6k and PP2A may be analogous to the recently reported interaction of PP2A with Ca2+-calmodulin-dependent protein kinase IV (22). In both cases, only a small fraction of the cellular PP2A is complexed to the kinase, and the activity of neither the kinase nor the phosphatase is required for the interaction (Fig. 3A). Questions remain about the mechanism by which the association of FKBP12–rapamycin with FRAP leads to phosphatase activation. Because association of FRAP with the FKBP12–rapamycin complex causes inhibition of FRAP’s kinase activity (3, 23, 24), PP2A regulation may involve direct phosphorylation by FRAP. We tested the ability of FRAP to directly phosphorylate PP2A in vitro. Wild-type FRAP immunoprecipitated from baculovirus-infected insect cells phosphorylates purified PP2A more readily than kinase inactive FRAP (Fig. 4A). Parallel and competitive kinase reactions suggest that in vitro phosphorylation of PP2A is more efficient than in vitro phosphorylation of GST–4E-BP1, a purported direct FRAP substrate (10). At least 10 times more phosphate is incorporated into PP2A than into GST–4E-BP1, despite the greater molar abundance of GST–4E-BP1 in the reaction mixtures (Fig. 4A Lower). Whereas phosphorylation of PP2A by FRAP is more efficient in vitro than phosphorylation of 4E-BP1, total FRAP kinase activity remains low, as is characteristic for phosphatidylinositol kinase (PIK)-related kinases. Little is known about the effect of phosphorylation on PP2A activity, and further study is required to determine whether FRAP-mediated PP2A phosphorylation occurs in vivo and affects PP2A activity or localization.
Figure 4.
Possible mechanisms of PP2A regulation by FRAP. (A) FRAP phosphorylates PP2A in vitro. Kinase-active (WT) and kinase-dead (DA) FRAP expressed in insect cells by baculovirus infection were immunoprecipitated and incubated with GST–4E-BP1 and/or PP2A as substrates in the presence of [γ-32P]ATP. The products of the kinase reactions were separated by using SDS/PAGE before autoradiography (Upper). Relative quantities of kinase substrates were determined by silver staining (Lower). (B) A model of FRAP intervention in the p70s6k signaling pathway. Mitogenic stimuli promote phosphorylation of p70s6k which in turn promotes translation of a subset of cellular mRNAs. Inhibition of FRAP by amino acid deprivation or by rapamycin treatment interferes with p70s6k function by activating PP2A. PP2A dephosphorylates and inactivates p70s6k through its interaction with the N terminus of p70s6k.
Recent reports have led to the view that FRAP is a mitogen-activated p70s6k and 4E-BP1 kinase. Our data support a view of FRAP as a sensor of amino acid levels that is able to unleash a p70s6k and 4E-BP1 phosphatase after amino acid deprivation or rapamycin treatment (Fig. 4B). Phosphatase regulation offers distinct advantages over kinase regulation when there is a need for rapid dephosphorylation at multiple, disparate sites that are under the control of multiple kinases. In the case of translation initiation, several distinct kinases appear to be responsible for the phosphorylation of more than a dozen sites on 4E-BP1 and p70s6k (4). Activation of a phosphatase may allow rapid dephosphorylation of these disparate sites without necessitating inhibition of all of the kinases responsible for their phosphorylations.
The view of FRAP as an intracellular sensor that can intervene in critical regulatory pathways is consistent with the roles of other members of the PIK-related kinase family, including ATM, ATR, and DNA-PK. These proteins sense intracellular abnormalities and transmit signals that halt the cell cycle machinery. In this light, we note that ionizing radiation results in the dephosphorylation of p53 in an ATM-dependent manner (25). It will be interesting to see whether other PIK-related kinases use phosphatases while carrying out their roles as checkpoint proteins.
Acknowledgments
We thank N. Sonenberg for the anti-4E-BP1 antibody and the 4E-BP1 expression plasmid, J. Blenis for providing p70s6k mammalian expression plasmids, and R. Erikson and members of the Schreiber group for helpful discussions. J.S.H. and R.T.P. are recipients of a Leukemia Society of America postdoctoral fellowship and a Howard Hughes Medical Institute predoctoral fellowship, respectively. B.N.D. is supported by a National Institutes of Health predoctoral training grant (5T32CA09141-23). The research was funded by the National Institute of General Medical Sciences (GM-38627). S.L.S. is an investigator at the Howard Hughes Medical Institute.
ABBREVIATIONS
- FRAP
FKBP12-rapamycin associated protein
- 4E-BP1
eukaryotic initiation factor 4E-binding protein 1
- p70s6k
70–kDa S6 kinase
- PP2A
protein phosphatase 2A
- TAg
simian virus 40 large tumor antigen
- HA
hemagglutinin
- GST
glutathione S-transferase
References
- 1.Brown E J, Schreiber S L. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 2.Hara K, Yonezawa K, Weng Q-P, Kozlowski M T, Belham C, Avruch J. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 3.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Nature (London) 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 4.Thomas G, Hall M N. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Farmer J D, Jr, Lane W S, Friedman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 6.Dennis P B, Pullen N, Kozma S C, Thomas G. Mol Cell Biol. 1996;16:6242–6251. doi: 10.1128/mcb.16.11.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng Q P, Andrabi K, Kozlowski M T, Grove J R, Avruch J. Mol Cell Biol. 1995;15:2333–2340. doi: 10.1128/mcb.15.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheatham L, Monfar M, Chou M M, Blenis J. Proc Natl Acad Sci USA. 1995;92:11696–11700. doi: 10.1073/pnas.92.25.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 10.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C, Jr, Abraham R T. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 11.Burnett P E, Barrow R K, Cohen N A, Snyder S H, Sabatini D M. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albers M W, Brown E J, Tanaka A, Williams R T, Hall F L, Schreiber S L. Ann NY Acad Sci. 1993;696:54–62. doi: 10.1111/j.1749-6632.1993.tb17142.x. [DOI] [PubMed] [Google Scholar]
- 13.Chung J, Kuo C J, Crabtree G R, Blenis J. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 14.Haystead T A, Haystead C M, Hu C, Lin T A, Lawrence J C., Jr J Biol Chem. 1994;269:23185–23191. [PubMed] [Google Scholar]
- 15.Begum N, Ragolia L. J Biol Chem. 1996;271:31166–31171. doi: 10.1074/jbc.271.49.31166. [DOI] [PubMed] [Google Scholar]
- 16.Cohen P T. Trends Biochem Sci. 1997;22:245–251. doi: 10.1016/s0968-0004(97)01060-8. [DOI] [PubMed] [Google Scholar]
- 17.DiComo C J, Arndt K T. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 18.Murata K, Wu J, Brautigan D L. Proc Natl Acad Sci USA. 1997;94:10624–10629. doi: 10.1073/pnas.94.20.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inui S, Sanjo H, Maeda K, Yamamoto H, Miyamoto E, Sakaguchi N. Blood. 1998;92:539–546. [PubMed] [Google Scholar]
- 20.Chen J, Peterson R T, Schreiber S L. Biochem Biophys Res Commun. 1998;247:827–832. doi: 10.1006/bbrc.1998.8792. [DOI] [PubMed] [Google Scholar]
- 21.Westphal R S, Coffee R L, Jr, Marotta A, Pelech S L, Wadzinski B E. J Biol Chem. 1999;274:687–692. doi: 10.1074/jbc.274.2.687. [DOI] [PubMed] [Google Scholar]
- 22.Westphal R S, Anderson K A, Means A R, Wadzinski B E. Science. 1998;280:1258–1261. doi: 10.1126/science.280.5367.1258. [DOI] [PubMed] [Google Scholar]
- 23.Brunn G J, Fadden P, Haystead T A J, Lawrence J C., Jr J Biol Chem. 1997;272:32547–32550. doi: 10.1074/jbc.272.51.32547. [DOI] [PubMed] [Google Scholar]
- 24.Brunn G J, Williams J, Sabers C, Wiederrecht G, Lawrence J C, Jr, Abraham R T. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 25.Waterman M J, Stavridi E S, Waterman J L, Halazonetis T D. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]