Abstract
We have constructed a linear array of coupled, microscale patches of habitat. When bacteria are inoculated into this habitat landscape, a metapopulation emerges. Local bacterial populations in each patch coexist and weakly couple with neighbor populations in nearby patches. These spatially distributed bacterial populations interact through local extinction and colonization processes. We have further built heterogeneous habitat landscapes to study the adaptive dynamics of the bacterial metapopulations. By patterning habitat differences across the landscape, our device physically implements an adaptive landscape. In landscapes with higher niche diversity, we observe rapid adaptation to large-scale, low-quality (high-stress) areas. Our results illustrate the potential lying at the interface between nanoscale biophysics and landscape evolutionary ecology.
Keywords: biophysics, microbiology, landscape ecology, metapopulation biology
In nature, habitats are patchy, aggregating at several scales generating a discrete habitat landscape (1). The landscape ecology of such environments provides communities with a distribution of characteristic scales (temporal and spatial) that can be used to partition such habitat landscapes (2, 3) allowing for species coexistence. In general, a metapopulation (4) or “population of populations” develops over such habitat landscape and is characterized by local population extinctions and colonizations. It is known that the topological properties of the ensemble and the quality of the individual patches have deep implications for fitness (4, 5).
The environments in which single-celled organisms exist is no different. However, natural habitat landscapes populated with “big” organisms are difficult to approach experimentally. On the contrary, bacteria are great experimental systems, and the environments they populate are amenable to experimental manipulation. Bacteria, like other life forms, self-organize into sophisticated dynamic assemblages. Escherichia coli individuals are known to exhibit complex patterns of motility (6). Individual bacteria are known to associate even further into very complex communities (biofilms) that resemble a human metropolis (7) in which microbes communicate with each other (8) and work together toward common goals (9), exploiting what is called niche complementarity (10).
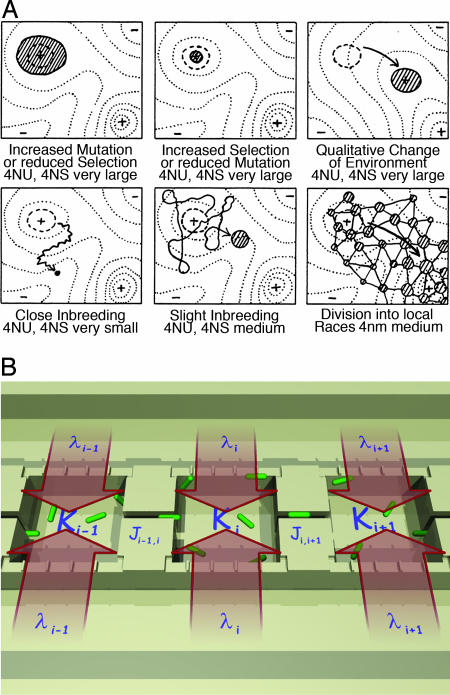
Sewall Wright (11) realized that a collection of interacting and interbreeding populations of organisms moving across patchy landscapes implemented a spatially distributed network, facilitating the flow of alleles across a fitness landscape. In Wright's view, spatially distributed populations adapted to different local environments but weakly coupled through population dispersal is the key to the dynamics of the evolution of coadapted complexes of genes. Wright's adaptive landscape (Fig. 1A) is a heuristic one. It changes with space and time, and is a function of many degrees of freedom, including the collective response of a population of interacting organisms as well as its intricate relationships with its habitat. The phenotype of an organism's genome consists of both individual aspects, such as cell growth and reproduction rates, and collective ones, such as the way the cells interact in mutually beneficial (or destructive) ways.
Fig. 1.
Sewall Wright's heuristic concept of an adaptive landscape (A) (adapted from ref. 11), and a 1D array of MHPs with landscape parameters [Ki, Ji,i+1, λi] (B).
Because the landscape ecology of the habitat distribution provides a proxy for fitness, micro- and nanofabrication techniques open up the possibilities of making spatially complex habitat landscapes that probe how microorganisms can adapt to both temporally and spatially varying challenges to fitness (adaptive landscape). Fig. 1B shows our solution to this problem: microhabitat patches (MHPs) that allow distinct local populations to fill a given habitat patch of quality parameters determined by nanoslits linking each MHP to external feeding channels. Individual MHPs have coupling corridors. Thus, a species can move from one patch to another, allowing the bacterial metapopulation to adapt to the different regions of our designed landscapes.
The chemostat was conceived by Novick and Szilárd (12) to provide us with an homogeneous ecology. Because chemostats lack spatial structure, they do not allow organisms to search out different niches in a spatially heterogeneous habitat. But, natural habitats are indeed heterogeneous. The nanofabricated habitat landscapes we can construct afford a variability in habitat structure, allowing us to experimentally explore Wright's adaptive landscape. There have been microchemostat systems created recently (13, 14), but the technology discussed here differs in a fundamental way. Microfabricated chemostats described so far (just as the macroscopic ones) do not allow for the emergence of a metapopulation [a spatially distributed network, of parallel populations adapted to different local conditions but weakly coupled with one another by dispersal (4)]. Moreover, in a “strict” chemostat, biomass is constantly removed and resources are added to reach a steady-state, yielding cells in exponential growth, rather than in other phases of growth (15). By adding spatial structure to the system, we can create heterogeneity in habitat structure and study how cells adapt to different regions of the landscape (ecotopes).
We constructed a one-dimensional (1D) array of coupled MHPs; the running index i here is used to denote the ith MHP. The corridors “coupling” MHPs are designed to be narrow enough so that each MHP can be viewed as a local niche in a much larger adaptive landscape generated by the heterogeneous array of habitat patches. There are three fundamental parameters that characterize the habitat in this array of coupled MHPs: (i) the local carrying capacity, Ki (patch size), of bacteria in the ith MHP; (ii) the coupling strength, Ji,i+1 (corridor structure), between adjacent MHPs; and (iii) the coupling strength, λi (number of nanoslits), between the MHP and feeding channels that allow food to diffuse into, and waste out of, a given MHP.
In general, vectors K, J, and λ (landscape parameters) can be made a strong function of the index i, so that nanoscale patchy environments can be designed to test the fitness of organisms to different ecotopes of the landscape. We address the question of how bacterial metapopulations behave when allowed to populate such landscapes.
Results
Fig. 3A shows the dynamics of an E. coli population in a single MHP [zero-dimensional (0D) device described in Fig. 2D] with all nanoslits open (λmax). Although initially the density of bacteria follows a pattern of exponential growth, the weak coupling to an external resource (via λ) results in some unusual behavior: oscillations occur at least at two distinctive frequencies (high and low).
Fig. 3.
Resource supply, space limitation, and parameter λ. (A) E. coli population dynamics in a single MHP. The black solid line corresponds to bacterial density in one of our 0D devices with a λmax niche. The blue dotted line represents the modeled resource (Eq. 2), and the red dash-dotted line is the modeled density (Eq. 1). A strategy's life-history [ε, τr, τm] and environmental (λ) parameters are here fitted to the bacterial data (see Supporting Appendix). (B) Theoretical dynamics of the scaled system (see Supporting Appendix): The solid red and dashed blue curves correspond to bacteria φ(τ) and food ω(τ), respectively.
Fig. 2.
Micro/nanofabricated devices. (A) One-dimensional patchy landscape; full device consisting of the array of 85 MHPs, each coupled (by 10 nanoslits) to two parallel feeder channels seen at the top and bottom. (B) Local neighborhood (scanning electron microscopy of adjacent MHPs). MHP connectivity and array topology is implemented through inter-patch corridors. (C) MHPs with three different supply niches. (D) Zero-dimensional (scanning electron microscopy of two uncoupled MHPs). (Scale bars: 100 μm.)
A simple analysis of diffusion through the nanoslit to the MHP gives us an expression for the contribution of a single nanoslit to the exchange rate between the MHP and the feeding channels:
where A is the nanoslit's total cross-sectional area, V is the MHP's volume, l is the length of the nanoslit, and Dw is the average diffusion coefficient of resources and waste. From the volume of a MHP (V = 3 × 105 μm3), the approximate diffusion constant of small molecules such as amino acids (Dw ∼ 10−5 cm2·s−1), the area of a 200-nm-deep and 20-μm-long nanoslit (4 μm2), and the width of the nanoslit (l = 15 μm), we find that λ* ∼ 10−3·s−1.
The population density ρ(t) of an MHP can be modeled by the logistic equation (16),
Here, the per capita growth rate is determined by two factors: space and resources. Space limitation is represented as the logistic (1 − ρ/K) environmental resistance (17), where the parameter K represents the carrying capacity of the MHP. Resource-based growth rate r(w) is a function of habitat quality 0 ≤ w ≤1 inside the MHP but relative to the concentration of resources in the feeding channels. Without these resources, cells cannot grow. Thus, following resource competition theory (18), we use
as our resource utilization function. Here, 1/τm represents the rate of cell death and 1/τr represents the birth rate achieved when the medium inside the MHP is fresh LB (w = 1). After the biomass of the cells starts growing and transforming the medium, w decreases. Waste-saturated medium means w = 0.
The feeder channels supply the MHP with fresh LB by λ-limited diffusion into (and waste out of) it through its nanoslits. The rate dw/dt at which resource quality changes inside the MHP is then the difference between inward diffusion and consumption by E. coli, normalized by the efficiency ε by which resources are converted into bacteria. Thus,
From the known volume of a MHP (V) and the approximate volume of a single E. coli (0.5 μm3) we can estimate (an upper limit) that a close-packed MHP can hold about K̄ ≡ 106 E. coli cells. In practice, however, a MHP typically saturates at about K* ∼ 104 E. coli cells.
The chief concern of our 0D theory is to understand the effects of space and resource limitation upon bacterial growth (Fig. 3A). There are three (see Supporting Appendix) scenarios: (i) an extinction solution (Fig. 3B Left), with the equilibrium values φ̂ = 0 and ŵ = w(0); (ii) a resource-limited solution (Fig. 3B Center), where φ̂ = λ × ([1/w*] − 1)/ε and ŵ = w*; and (iii) a space-limited solution (Fig. 3B Right), where φ̂ = 1 and ŵ = 1/(1 + [ε/λ]). Here, life-history parameter w* = τr/τm (dimensionless) corresponds to the total number of cell divisions in a cell's lifespan. Thus, a given strategy [ε, w*] can have a totally different fate, depending on the value of habitat parameter λ nanostructured onto a given MHP. On the other hand, for a fixed environment λ, organisms can adapt by changing their strategy [ε, w*].
When our microecosystem is in a regime of resource limitation, the picture goes like this; as food resources are depleted, Eqs. 1 and 2 predict that the growth rate w/τr will become less than the death rate 1/τm, and the population in the MHP will start going extinct. However, resources can diffuse in from the feeder channels and growth can reinitiate; this can give rise to oscillations in the population density due to the diffusional lag between consumption and supply. Although high-frequency spikes and lower-frequency bumps exist, our model cannot accommodate both at the same time. Only a single frequency basic oscillation for population density vs. time is expected from our model. For a fixed environment λ, the frequency of the oscillation is determined by an organisms life-history strategy [ε, τr, τm]. A consortium of phenotypes would be expected to exhibit more frequencies.
Metapopulation Dynamics in a Flat Landscape.
The next step up in complexity is a 1D array of equivalent MHPs, which we call a flat landscape. Fig. 4 shows the spatial dynamics of E. coli growing on a flat landscape where all MHPs have all their nanoslits open (λi = λmax ∼ 10λ*, ∀i). Fig. 4A Right consists of a time-ordered stack of epifluorescence images of all 85 MHPs. Here, we scanned the array every 10 min (δt) 300 times, sampling a total of 3,000 min (Δt = 2.1 days). Each row of images ψt(x, y) of the 85 MHP array represents the configuration of the array at time t (Fig. 4A and Movie 1, which is published as supporting information on the PNAS web site). Local (MHP) population density average ρi at any given time t can then be calculated by integrating epifluorescence intensity ψt for all pixels (x, y) within the ith MHP (see Fig. 7A in Supporting Appendix, which is published as supporting information on the PNAS web site).
Fig. 4.
E. coli in a flat landscape of habitat patches. (A) Photograph montage (see text) capturing the spatial dynamics of the metapopulation during the phase of spatial expansion of its range of occupancy (spatial-log-phase) in a flat, fully open (λi = λmax) landscape. White pixels (x, y) correspond to GFP fluorescence. δt = 10 min. (B) Global (dashed blue) vs. local (solid black) dynamics. Logistic growth (Eq. 3) fitted to the global average is also shown (dotted red; τT ∼ 250 min). Vertical (dashed gray) lines in B mark the spatial-log-phase shown in A.
The dynamics of the landscape average ρ(t) = Σiρi/85 (Figs. 4B and 7B), resemble what is seen in batch cultures; after a lag period of ≈400 min during which little growth occurs in the array, a period of growth (exponential phase) followed by landscape saturation (stationary phase) at 104 cells per MHP (K* ∼ 3 × 1010 per ml) is observed. Because the landscape is flat, we would expect that over time the bacteria would inhabit all of the MHPs. However, because coupling is weak (small Ji,i+1), a metapopulation emerges. Thus, whereas the population density of an individual MHP (local scale) shows sharp rises and falls in density, occupancy of the entire array (landscape scale) shows a much slower growth rate and smoother dynamics than the single MHP (Fig. 4B). The fit (dotted red) of the logistic map (Eq. 1) to the globally averaged occupancy/MHP (solid blue) shown in Fig. 4B yields a τT ∼ 250 min (≈4 h). The reason for this slow growth is clear from Fig. 4: there are localized E. coli populations distributed over the landscape, interacting through local extinction and colonization processes operating at multiple spatial and temporal scales (Figs. 4 and 7). Notice that although the density seems constant (stationary phase) at the global scale, at the local MHP scale there are clear dynamics (Fig. 4B). On the other hand, although the global averages are at exponential phase because of continuous range expansion, individual populations can be in stationary as well as in death phases.
In Fig. 4A Right, we show a (mesoscale) 5-MHP-wide, “parent” population giving “birth” to a new population spreading to the right and settling six MHPs away. The message: in a flat habitat landscape, E. coli aggregates its biomass at multiple scales satisfying a careful balance between vacancy and occupancy. These multiscale aggregates correspond to spatial versions of the classical phases of growth: lag, log, stationary, and death. Zooming into a particular MHP (Fig. 4B, black solid line), we notice pulses of exponential growth with a 10- to 20-min time constant (local colonization events), stationary-phase, and death-phase oscillations (local extinction events) occurring at multiple spatial and temporal scales. Unlike the zero dimensions case, in one dimension, the bacteria can migrate into near by MHPs, so growth can continue in a delayed fashion throughout the MHP array.
Adaptation in a Black and White Landscape.
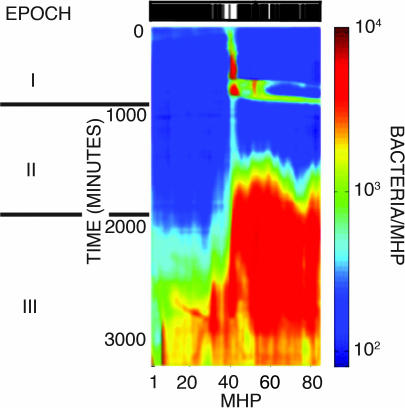
To induce a fitness pressure ∇λ on the E. coli metapopulation, we built a “black and white” (B&W) adaptive landscape by patterning two different ecotopes on each side of the landscape. Fig. 5 presents the basic idea of such adaptive landscape; the bar code (at the top) indicates the number of nanoslits that are opened in each MHP (indexed by i); in this case the nanoslits are fully closed on the left side (a stress-based black ecotope) and fully open (white ecotope) on the right side. The response of the bacteria to this landscape can be broken down into three basic epochs based on episodic expansions of its range of landscape occupancy (see Fig. 5 and Fig. 8A in Supporting Appendix).
Fig. 5.
Adaptation to a B&W landscape. Spatial dynamics ρi(t) of an E. coli metapopulation adapting to a simple, two-ecotope landscape. The black and white bar at the top shows the relative values of λi for the MHPs. White corresponds to λmax ∼ 10λ*, and black corresponds to λmin = 0 (see text). The log number of cells per MHP is color-coded as shown by the scale bar on the right. δt = 10 min. Epochs I–III defined by territorial expansions are indicated in gray. Solid gray bars mark the transition from one epoch to another.
Epoch I.
At first, there is a high increase in landscape average density (from 101 to 102 cells per MHP) due to invasion and growth of bacteria in the white ecotope. Initially, this growth is confined only to this ecotope, and it saturates at ≈102 cells per MHP. However, at around t ≈ 700 min (Fig. 5), the bacteria do probe into the black ecotope, thus expanding their range to the whole landscape. After this range expansion, the global average density achieved is larger but still ≈102 cells per MHP. Unfortunately for the bacteria, they quickly die out (death phase) in most of their range, going down to a very low spatially homogeneous density of ≈0.5 × 101 cells per MHP at the end of epoch 1, at about t ≈ 1,000 min after inoculation. Notice the large-scale correlation of the extinction event (Fig. 5); almost the whole metapopulation disappears from the landscape leaving very few colonizing cells.
Epoch II.
Average density is down to ≈0.5 × 101 cells × MHP. However, it is localized into a discrete set of few surviving populations (≈3 or 4). These populations compete for the landscape, increasing the average density to 0.4 × 102 cells per MHP. This growth is distributed across the whole landscape, unlike the localized growth observed in the first third of epoch I. Bacterial history repeats itself: another large-scale extinction event (Fig. 5; t ≈ 1,800 min) wipes out most of the population. This time, however, a larger number of local populations survive (≈8 or 10). Some of these populations are indeed inhabiting the low-nutrient, “stressful” region. Indeed, at least two of these stress-tolerant populations persist and ultimately merge with a new landscape-wide one (at ≈t ≈ 3,000 min). This time, the landscape-wide expansion in range is sustained for a longer period (1,000 min) than the one observed during epoch I (100 min); yet, it is also followed by massive extinction. However, this time more populations (≈10–15) survive. At the end of the epoch, a massive regrowth into the stress region is triggered from the right part of the landscape. Notice, however, that the speed of the dispersal front into the left of the array is much slower than the incursion of the second third of epoch I. As these populations expand to the left, they merge with other local ones. So far, densities nowhere in the landscape have >102 cells per MHP.
Epoch III.
At the beginning of epoch III (t ≈ 1,800 min), populations are expanding consistently to the left, into the stress domain of the landscape (densities are still low, at ≈2 × 101 cells per MHP). During the first third of the epoch (t < 2,500 min), most growth is widespread in the full supply region (λmax) while highly localized in the stress region (λmin) of the landscape. At around t = 2,500 min, populations from the λmax ecotope (right side) start expanding into stress territory (left side); until at t = 3,000 min they absorb the stress-tolerant population located around the 19th MHP of the landscape. After this event, two things seem to happen: (i) the territorial expansion into the stress region accelerates (Fig. 5), and (ii) the landscape average growth also accelerates (Figs. 5 and 8A). The following period (3,000 min < t ≤ 4,000 min) consists of vigorous growth in the λmax region and sustained growth in the λmin region. However, the bacteria in the stress and full-supply ecotopes have growth rates that differ by a factor of 2 (Fig. 8A Inset). At a late period of this epoch (t ≈ 4,000 min), a sharp density boundary has emerged between the two ecotopes, although the range of the metapopulation includes the whole landscape. This boundary divides the population in two different classes of abundance. On the left, populations reach densities of 102 cells per MHP, whereas on the right part of the landscape, populations reach densities of 103 to 104 cells per MHP. This boundary is similar to the boundary that separated populations at the beginning of epoch I. Then, the average densities were 101 cells per MHP on the left and 102 cells per MHP on the right. Notice the extinction events (around t ≈ 4,500 min), eliminating two (competing) nearby high-density (104 cells per MHP) populations located around the 50th and 60th MHPs. Landscape saturation is hard to reach with this (B&W) adaptive topology. Even after 5,000 min, the metapopulation is not fully adapted to life in the stress region.
Adaptation to a Rugged Landscape.
Evolutionary dynamics on adaptive landscapes are a function not only of the local fitness value but also a function of higher spatial derivatives of the landscape (19). Thus, the sharp fitness barrier (like the one in the B&W case) separating stress and full-supply ecotopes can be mixed with more “rugged” parts of the landscape where only fractions of nanoslits are closed or open, thus introducing more “intermediate” niches. To investigate the role of higher correlations in landscape parameters on adaptation, we developed a “rugged” landscape composed of three basic regions described by the black and white bar at the top of Fig. 6.
Fig. 6.
Adaptation to a rugged-landscape spatial dynamics ρi(t) of an E. coli metapopulation adapting to a complex, multiecotope landscape. The black and white bar at the top shows the relative values of λi for the ith MHP. The average number of cells in the local population ρi × K* of each MHP is color-coded as shown by the scale bar on the right. As before, δt = 10 min. Epochs I–III defined by territorial expansions are indicated in gray. Solid gray bars mark the transition from one epoch to another.
Notice that in this landscape only 10% of the MHPs are not stress MHPs; the MHP array as a whole then has less supply exchange-area than in the previous case (B&W landscape). However, the total amount of resources in storage in the feeder channels remains the same. The niche differences between this adaptive landscape and the previous (B&W) one are in the spatial localization and the rates of delivery, not in the total amount of resources. The adaptive topology of these two landscapes is therefore different: the number of local niches is higher in the rugged landscape because the B&W one only had two. Fig. 6 shows the response of bacteria to such a landscape. Here, as before, we once again break the bacterial response into epochs based on the range expansions of landscape occupancy.
Epoch I.
The first half (t < 500 min) of this epoch is characterized by the high localization of rapid growth around the central cluster (ecotope) of λmax niches. Bacteria grow to saturation at around K* ≈ 104 cells per MHP but remain localized to the center (around the 40th MHP), exploiting the better-supplied part of the landscape. Landscape average densities, however, are still below 3 × 102 cells per MHP because of the small scale of the range of occupancy. As these patches are exhausted at about t ≈ 500 min, the bacteria expand their range to the right side of the landscape where more clusters of opportunity (λi ≠ 0) are located (Fig. 6). When the bacteria start exploring these ecotopes, landscape average growth increases (Fig. 8B; t ≈ 600 min), a daughter population emerges around the 55th MHP, and global densities climb to 103 cells per MHP rapidly before a wide-range extinction takes place at about t ≈ 700 min (Fig. 6).
Epoch II.
After the population crash described above, we enter Epoch II. Landscape-wide average densities now are ≈0.5 × 102 cells per MHP. The few survivors are once again fully localized to the MHP cluster at the center of the landscape. Slow growth of bacteria is observed, with little further range expansion, increasing local density from post-crush levels back to 0.5 × 103 cells per MHP. At around t ≈ 1,500 min, this central population starts expanding its range once again to the rugged (right) side of the landscape. By t ≈ 1,800 min, the landscape begins to saturate (Fig. 8B) with most of the bacteria inhabiting the rugged (right) side of the landscape and reaching very high cell densities (Fig. 6). A huge-density barrier separates the left (102 cells per MHP) from the right (104 cells per MHP) demographic states of the metapopulation. As with the B&W landscape, bacteria in the different (stress, partial, and full supply) ecotopes, have growth rates that now differ by a factor of 3 (see Fig. 8B Inset). Even the supply to the MHPs is apparently less efficient. It indeed allows the metapopulation to adapt faster to the habitat landscape. After 1,800 min, populations saturate at 104 cells per MHP. Interestingly, even though the landscape is fully saturated on the right side, it holds plenty of capacity to the left. After a spatial lag, a large range expansion takes place at t ≈ 2,000 min.
Epoch III.
This new range expansion is slow. Vigorous growth of bacteria in the right region of the array during epoch II (while experiencing different, nearby variable λ ecotopes for at least five generations; Fig. 6) sets the conditions for the dynamics to come. Now, these populations start rapidly expanding into the left side (Fig. 8B) where the stress ecotope is located. Notice that the landscape-wide density remains constant at its saturation level (Fig. 6) The colonization of the stress (λmin) region of the landscape is completed by t = 2,500 min. Interestingly, at the same time the adaptive radiation is completed, there is a widely correlated decrease in density from 104 to 0.5 × 104 cells per MHP. This decrease in density coincides with the appearing of a less dense pattern of occupancy, propagating from the central region into both sides of the landscape. By t ≈ 3,000 min, a new pattern of occupancy spans the whole landscape. Metapopulation dynamics as in the case of the flat landscape are observed. In this state, MHPs fluctuate locally between having 103 cells per MHP or 0.5 × 104 cells per MHP. The landscape average density is stationary as expected. Now the bacteria are fully adapted to live on the entire landscape.
Discussion
The ecological corridors coupling MHPs are critical to our work: by weakly linking the habitat patches we are able to build heterogeneous landscapes as designed combinations [Ki, Ji,i+1, λi] of locally interacting MHPs while preserving the parallel and distributed nature of the (local) demographic process in each MHP, a spatially structured, adapting metapopulation (4). Under strong coupling our device behave like a giant 0D one, by loosing its patchy structure. In this case the population does not structure itself into localized demographic units.
Cell–cell communication through a signaling field Ct(x, y) determines chemotaxis-based bacterial movement (20) within each MHP. Keller–Segel flux (21) accounting for diffusion J0 (with strength D) and chemotaxis 𝒥1 (with strength X),
is usually used to describe the spatial dynamics ∂tψt = G + ∇𝒥 of bacterial densities ψt as a function of local growth G and chemotactic spatial coupling ∇𝒥. At the local scale, the balance between dispersive forces (𝒥0) and chemotaxis-based aggregation (𝒥1) is critically dependent on density (22). The oscillatory behavior of local density, which we learned from the 0D analysis (Fig. 3), leads us to conjecture that MHP demographics is to be shifting between two different regimes: (i) a 𝒥0-dominated regime (dispersive) when the average MHP density is below the critical value ρc, and (ii) a 𝒥1-dominated regime (attractive) when it is above ρc. These regimes are expected to be important in generating stochastic propagule dispersal events, implementing migration between nearby MHPs (see Movie 2, which is published as supporting information on the PNAS web site). As we have shown here within our patchy landscapes, density aggregates at more than one scale. So we would expect patterns of molecular response in the cellular assemblage to also match characteristic scales embedded within the interaction between the topology of the habitat and a strain's life-history strategy. The question is, what are the scales (1) at which spatial coupling ∇𝒥 and “local” growth G operate for a given strategy [ε, τr, τm] in a given landscape [Ki, Ji,i+1, λi]?
The “shifting balance” (11) between evolutionary forces (phenotypic plasticity, mutation, genetic drift, and selection) embedded in the adaptive gradients ∇λ across the landscape in conjunction with demographic process determine the evolution of competitive advantage and the struggle for existence (17) in our devices. Notice that the local adaptation observed could be the result of physiology or mutation; however, we do not yet know to which degree each of these processes are involved. Although conservative estimates of mutation rates are typically in the order of one mutation per 109 bp per generation, it is clear now that this number is highly variable depending on the various stresses imposed to the organism by our devices (23–25).
Growth advantage in stationary phase mutants are expected to take over (25) ecotopes with high density of (stress) λmin MHPs. Notice that stationary phase here can be induced not only by resource starvation (bottom-up) but also by space limitation (top-down). In this manner, the discrete nature of the environment allows for parallel stationary-phase states to be developed in a metapopulation that is otherwise expanding across the landscape. This slows down total growth but it increases the complexity of the assemblage by favoring stationary phase advantage phenotypes in different, but connected, local populations. Notice that space-limited vs. resource-limited patches can be used in a (niche) complementary fashion by two cooperating strategies exploiting different ecotopes across the landscape. Thus, in our devices the spatial implication of growth advantage in stationary phase phenomena in spatially explicit and heterogeneous settings as well as its responses to different adaptative topologies should be studied further. From the natural history comparison between rugged and B&W landscapes, preliminary results suggest that the number and complexity of ecotopes enhances the adaptive capacities of the metapopulation to high stress (λmin) territories.
Our arrays have an intrinsic dynamic variability due to the nonlinear nature of the demographic coupling. Weakly coupled logistic oscillators, are prone to spatiotemporal chaos (26) depending on the balance between local growth (G) and dispersal coupling Ji,i+1. A clear limitation of our study is its temporal extent Δt. Longer periods of culturing (as well as larger arrays) are needed to understand the evolution of ecological advantages under controlled adaptive topologies.
Conclusions
It is clear that the physics of evolutionary dynamics, although related to the general problem of multidimensional energy landscapes with dynamic local minima, is more complex because the peaks are not necessarily stable to perturbation in density. By designing more complex (temporal and spatial) gradients, the directed evolution of microbial populations should be possible if collaborations between landscape ecologists and experimental (nanoscale) physicists increases further.
Methods
The basic idea in our device is the creation (by etching in a Si wafer) of a row of microfabricated 100 μm × 100 μm × 30 μm MHPs that are weakly linked to each other and to a source of food (Fig. 2A). The MHPs are weakly linked together by 50-μm-long, 5-μm-wide, and 30-μm-deep corridors connecting adjacent MHPs (Fig. 2B). The final (1D) device consists of a chain of 85 MHPs (Fig. 2A). It is seeded from one end by bacteria from a larger “interface chamber” (see Supporting Appendix).
To make an ecosystem with a rate-limited supply of resources, we weakly link patches to two feeder channels for the supply of food (Fig. 2B). In this rate-limited scenario, organisms must adapt their demands on their environment. Each of the two feeder channels are connected to the MHPs via five nanoslits that are only 200 nm deep but 15 μm wide and 20 μm long. Thus, they act as weak links between the MHP and the feeder channels. These nanoslits allow nutrients (and waste) to diffuse into and out of the MHPs but are too thin for E. coli to pass through. They provide a critical role beyond the supply of food and removal of waste. By building a different number (m) of nanoslits feeding different MHPs, we introduce relative niche differences among collections of MHPs (Fig. 2C). We can build MHPs with no exchange, λmin = 0; intermediate exchanges, m × λ* for m ∈ {1, …, 9}; and maximum exchange, λmax = 10 × λ*. The value λ* here represents the contribution to the exchange rate by a single nanoslit. In this way, adaptive (fitness) landscapes (∇λ) can be created by patterning ecotopes (spatially connected collections of MHPs sharing the same λi) onto the habitat spatial distribution. The 1D experiments we describe in this article were conducted in three types of adaptive landscapes: (i) a flat one, consisting of a single ecotope of MHPs where all 10 nanoslits are open; (ii) a B&W landscape, consisting of two ecotopes (at the right of the array we place MHPs with all 10 nanoslits open, and on the left we put MHPs with all nanoslits closed); and (iii) a more complex “rugged” landscape, consisting of three zones [to the left a nutrient-limited “stress” domain made of MHPs with all nanoslits closed (no supply), at the center a high nutrient-supply zone, separating the stress zone from a rugged zone (to the right), made of clusters of all-open and partially open MHPs embedded on a desert of stressed MHPs].
Because metapopulations are expected to have a complex behavior, a simple 0D MHP was constructed (Fig. 2D) to allow simple studies of a single MHP.
Supplementary Material
Acknowledgments
We thank Ted Cox, Miguel Gaspar, Juliana Malinverni, Ned Wingreen, Pascal Silberzan, and Tom Silhavy for comments, discussions, and support, and Peter Wolanin for supplying the E. coli GFP construct. This work was supported by Air Force Office of Scientific Research Grant FA9550-05-01-0365, National Institutes of Health Grant HG01506, National Science Foundation Nanobiology Technology Center Grant BSCECS9876771, and the Cornell Nano-Scale Science and Technology Facility (National Science Foundation Grant ECS-9731293).
Abbreviations
- 0D
zero-dimensional
- 1D
one-dimensional
- B&W
“black and white”
- MHP
microhabitat patch.
Footnotes
The authors declare no conflict of interest.
References
- 1.Levin SA. Ecology. 1992;73:1943. [Google Scholar]
- 2.MacArthur R, Pianca ER. Am Nat. 1966;100:603–609. [Google Scholar]
- 3.Tilman D, Kareiva P. Spatial Ecology: The Role of Space in Population Dynamics and Interspecific Interactions. Princeton: Princeton Univ Press; 1997. [Google Scholar]
- 4.Hanski IA, Gilpin ME, editors. Metapopulation Biology: Ecology, Genetics, and Evolution. Burlington, MA: Elsevier; 1997. [Google Scholar]
- 5.Rousset F. Genetic Structure and Selection in Subdivided Populations. Princeton: Princeton Univ Press; 2004. [Google Scholar]
- 6.Budrene EO, Berg HC. Nature. 1991;349:630–633. doi: 10.1038/349630a0. [DOI] [PubMed] [Google Scholar]
- 7.Watnick P, Kolter R. J Bacteriol. 2000;182:2675–2679. doi: 10.1128/jb.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassler B. Cell. 2002;17:421–424. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg P. Nature. 2003;424:134. doi: 10.1038/424134a. [DOI] [PubMed] [Google Scholar]
- 10.Kinzig A, Pacala S, Tilman D. The Functional Consequences of Biodiversity. Princeton: Princeton Univ Press; 2001. [Google Scholar]
- 11.Wright S. Evolution: Selected Papers. Chicago: Univ of Chicago Press; 1986. [Google Scholar]
- 12.Novick A, Szilard L. Science. 1950;112:715–716. doi: 10.1126/science.112.2920.715. [DOI] [PubMed] [Google Scholar]
- 13.Balagadde FK, You L, Hansen CL, Arnold FH, Quake SR. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 14.Groisman A, Lobo C, Cho H, Campbell JK, Dufour YS, Stevens AM, Levchenko A. Nat Methods. 2005;2:685–689. doi: 10.1038/nmeth784. [DOI] [PubMed] [Google Scholar]
- 15.Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the Bacterial Cell: A Molecular Approach. Sunderland, MA: Sinauer; 1990. [Google Scholar]
- 16.Verhulst PF. Corresp Math Phys. 1839;10:113–121. [Google Scholar]
- 17.Gause GF. The Struggle for Existence. Baltimore: Williams & Wilkins; 1934. [Google Scholar]
- 18.Tilman D. Resource Competition and Community Structure. Princeton: Princeton Univ Press; 1982. [PubMed] [Google Scholar]
- 19.Kessler D, Levine H, Ridgway D, Tsimring L. J Stat Phys. 1996;87:519–544. [Google Scholar]
- 20.Park S, Wolanin PM, Yuzbashyan EA, Silberzan P, Stock JB, Austin RH. Science. 2003;301:188. doi: 10.1126/science.1079805. [DOI] [PubMed] [Google Scholar]
- 21.Keller E, Segel L. J Theor Biol. 1970;26:399–415. doi: 10.1016/0022-5193(70)90092-5. [DOI] [PubMed] [Google Scholar]
- 22.Park S, Wolanin PM, Yuzbashyan EA, Lin H, Darnton NC, Stock JB, Silberzan P, Austin RH. Proc Natl Acad Sci USA. 2003;100:13910–13915. doi: 10.1073/pnas.1935975100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taddei F, Halliday J, Matic I, Radman M. Mol Gen Genet. 1997;256:277–281. doi: 10.1007/s004380050570. [DOI] [PubMed] [Google Scholar]
- 24.Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 25.Zambrano M, Siegele D, Almiron M, Tormo A, Kolter R. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko K. Physica. 1989;37:60–82. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.