Abstract
At the center of the debate on the emergence of modern humans and their spread throughout the globe is the question of whether archaic Homo lineages contributed to the modern human gene pool, and more importantly, whether such contributions impacted the evolutionary adaptation of our species. A major obstacle to answering this question is that low levels of admixture with archaic lineages are not expected to leave extensive traces in the modern human gene pool because of genetic drift. Loci that have undergone strong positive selection, however, offer a unique opportunity to identify low-level admixture with archaic lineages, provided that the introgressed archaic allele has risen to high frequency under positive selection. The gene microcephalin (MCPH1) regulates brain size during development and has experienced positive selection in the lineage leading to Homo sapiens. Within modern humans, a group of closely related haplotypes at this locus, known as haplogroup D, rose from a single copy ≈37,000 years ago and swept to exceptionally high frequency (≈70% worldwide today) because of positive selection. Here, we examine the origin of haplogroup D. By using the interhaplogroup divergence test, we show that haplogroup D likely originated from a lineage separated from modern humans ≈1.1 million years ago and introgressed into humans by ≈37,000 years ago. This finding supports the possibility of admixture between modern humans and archaic Homo populations (Neanderthals being one possibility). Furthermore, it buttresses the important notion that, through such adminture, our species has benefited evolutionarily by gaining new advantageous alleles. The interhaplogroup divergence test developed here may be broadly applicable to the detection of introgression at other loci in the human genome or in genomes of other species.
Keywords: human evolution, introgression, admixture
Fossil records indicate that anatomically modern humans first emerged ≈200,000 years ago in Africa and since then spread throughout the world (1). For most of the period since their emergence, anatomically modern humans are known to have coexisted with several now-extinct Homo lineages, such as Neanderthals (Homo neanderthalis). This long period of coexistence, including cohabitation in the Middle East and Europe, raises the intriguing possibility of genetic admixture between anatomically modern humans and archaic Homo populations, which could have resulted in contributions by these extinct lineages to the modern human gene pool.
The extent to which anatomically modern humans admixed with archaic Homo has been the subject of repeated speculation, particularly in regards to Neanderthals (2–22). Thus far, the mainstream view from fossil and genetic studies leans toward a model where anatomically modern humans fully replaced archaic Homo lineages rather than admixed with them (2–8). However, a number of investigators have voiced opposition to this total replacement model on a number of grounds, and the debate has yet to be resolved (9–22). Particularly needed to settle this debate is the identification of genetic loci that show telltale signs of admixture. There have been several reports of loci in the human genome that display unusually deep genealogy (15, 16, 23, 24), and in some cases, admixture between humans and archaic Homo lineages has been invoked as a possible explanation. However, these studies cannot differentiate the admixture model from other possibilities, such as long-standing balancing selection, that also could contribute to deep genealogies (see Discussion). As such, proponents of the admixture scenario have yet to identify a concrete example of a genetic locus for which there is compelling evidence of admixture. Furthermore, most discussions of admixture tend to treat it as a selectively neutral event, one that happened simply as a byproduct of the geographical overlap between modern humans and archaic populations. Such discussions often overlook the possibility that admixture with archaic lineages, if it indeed occurred, might have brought adaptive alleles (along with the traits they determine) into the modern human gene pool, thus profoundly impacting the biological evolution of our species.
A major difficulty in looking for traces of ancient admixture in the modern human gene pool is that, under neutrality, very low levels of admixture are not expected to be readily detectable because of the effects of genetic drift. Indeed, simulations of ancient admixture showed that an archaic genetic contribution of <0.1% is unlikely to be detectable even in a very large set of polymorphism data (25). Thus, the absence at present of conclusive genetic data in support of the introgression scenario should not be taken as evidence against the possibility of any introgression.
If introgression of archaic lineages into the modern human gene pool indeed occurred, then genes that have been subject to recent positive selection in humans may be enriched for introgressed alleles. Although selectively neutral alleles introgressed from archaic lineages at low levels are likely lost by drift or swamped by the large influx of modern human DNA, an introgressed allele that is selectively advantageous could escape the effect of genetic drift and rise to high frequency. As such, these alleles might become detectable in the modern human gene pool.
The gene microcephalin is a critical regulator of brain size. In humans, loss-of-function mutations in this gene cause a condition known as primary microcephaly, which is characterized by a severe reduction in brain volume (by 3- to 4-fold) but, remarkably, a retention of overall neuroarchitecture and a lack a overt defects outside of the brain (26). The exact biochemical function of microcephalin has yet to be elucidated, but this gene likely plays an essential role in promoting the proliferation of neural progenitor cells during neurogenesis (26). microcephalin has been shown to be the target of strong positive selection in the evolutionary lineage leading from ancestral primates to humans (27, 28). This observation, coupled with the fact that this gene is a critical regulator of brain size, suggests the possibility that the molecular evolution of microcephalin may have contributed to the phenotypic evolution of the human brain (27, 28).
In a recent study, we found that the haplotype structure at the human microcephalin locus is consistent with the action of recent positive selection (29). Specifically, we found that a class of haplotypes at the locus, dubbed haplogroup D, has a remarkably young coalescence age (≈37,000 years) despite an exceptionally high worldwide frequency (≈70%). This observation implies a rapid rise in the frequency of haplogroup D in humans, which is incompatible with genetic drift and instead supports the notion that positive selection has operated on haplogroup D to drive up its frequency. In the present work, we examine the origin of haplogroup D. We provide evidence that haplogroup D may have originated from a lineage separated from modern humans for ≈1.1 million years and introgressed into the human gene pool by ≈37,000 years ago. We discuss the implications of our findings for the understanding of modern human origins and the biological adaptation of our species as it spreads around the globe.
Results
Highly Unusual Genealogy of the microcephalin Locus.
We have examined (29) microcephalin in a panel of 89 individuals assembled to approximate the worldwide diversity of major human populations. We resequenced the panel for a 29-kb region that spans exons 4–9 of the 14-exon microcephalin gene. This process led to the identification of 220 segregating sites delineating 86 distinct haplotypes (Table 1, which is published as supporting information on the PNAS web site). Of the 178 chromosomes we sampled, 124 (or 70%) belonged to haplogroup D, defined by the derived C residue at the G37995C diagnostic nonsynonymous polymorphic site. (For simplicity, we will refer to haplogroup D as the D allele and the non-D haplotypes as the non-D allele.) Despite the high worldwide frequency of the D allele, its coalescence age is merely ≈37,000 years, far younger than the non-D allele (29).
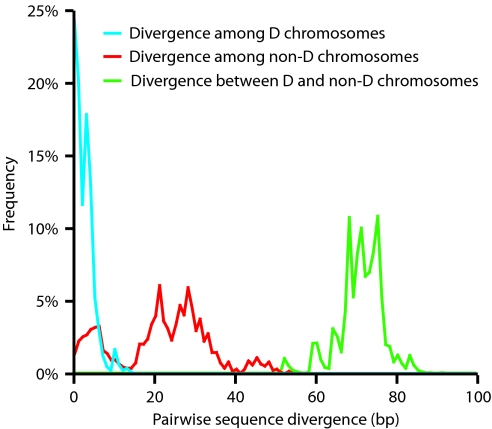
Closer scrutiny of the haplotype data in Table 1 revealed a rather peculiar pattern: the average pairwise divergence between D and non-D chromosomes across the 29-kb region is 3.3 times the divergence seen within non-D chromosomes and a striking 30 times the divergence seen within D chromosomes (Fig. 1). Further examination showed that this is because microcephalin has a highly unusual, lopsided, and deeply divided genealogy, which is schematized in Fig. 2A.
Fig. 1.
Distribution of pairwise sequence divergence between and within D and non-D chromosomes at the microcephalin locus.
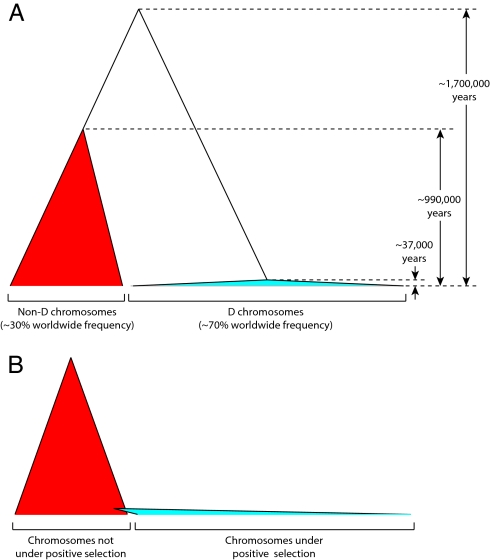
Fig. 2.
Comparison of the microcephalin genealogy with an idealized genealogy. Each filled triangle represents a genealogical clade, with the width of the triangle representing frequency in the population. (A) The genealogy consistent with the haplotype data at the microcephalin locus. The coalescence age of D chromosomes (≈37,000 years), non-D chromosomes (≈990,000 years), and between D and non-D chromosomes (≈1,700,000 years) are indicated. (B) The idealized genealogy of a partial positive selective sweep, wherein the adaptive allele first emerged by a mutational event on a random chromosome in the population.
Three features are prominent in this unusual genealogy. First, the D chromosomes coalesce to its most recent common ancestor (MRCA) at ≈37,000 years before present, whereas the non-D chromosomes coalesce at a far older ≈990,000 years before present. The much younger coalescence age of the D chromosomes, despite their much higher frequency, is consistent with the action of positive selection on the D allele as reported previously (29). Second, and more surprisingly, however, we found that the D and non-D chromosomes belong to two distinct, deeply divided clades connected by a single branch around the root of the tree (except for a few rare recombinants between the two clades, as discussed later). In other words, the D clade is a distant outgroup of the non-D clade, and vice versa. As shown in Fig. 3 and Table 1, the deep division between the D and non-D clades is due to a large number of segregating sites scattered throughout the 29-kb resequenced region that consistently differentiate the two clades, i.e., segregating sites for which the D chromosomes are characterized by one allele, whereas the non-D chromosomes are characterized by the other allele. For most of these segregating sites, the D chromosomes bear the derived allele, whereas the non-D chromosomes bear the ancestral allele (Fig. 3), an observation consistent with the fact that the internal branch from the MRCA of the D clade to the root of the tree is much longer than the internal branch from the MRCA of the non-D clade to the root (Fig. 2A). Such a tree topology does not resemble the expected genealogy of a recent selective sweep, as schematized in Fig. 2B, wherein the adaptive allele is introduced by a mutational event in a panmictic population. Fourth, whereas the coalescence age of ≈990,000 years for the non-D clade is similar to the human genome average, the D and non-D clades coalesce with each other at a much older ≈1,700,000 years before present, with virtually no genetic exchange between the two clades except for a few rare recombinants (discussed below). These unusual features of the microcephalin genealogy suggest the possibility that the MRCA of the D clade introgressed into humans from a divergent Homo lineage at or some time before ≈37,000 years ago. In the ensuing sections, we describe stringent statistical tests that support this introgression model.
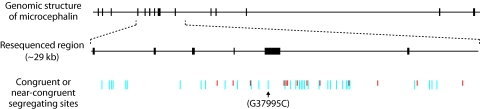
Fig. 3.
Distribution of congruent or near-congruent segregating sites in the 29-kb resequenced region of microcephalin. Congruent sites are defined as showing consistently different alleles between D and non-D haplotypes; near-congruent sites are defined as having no more than four differences from congruent sites. Sites for which the D chromosomes are characterized by the derived allele are indicated by long blue lines, whereas sites for which the D chromosomes are characterized by the ancestral allele are indicated by short red lines (for exact positions of these sites, see Table 1). Also indicated is the G37995C nonsynonymous site used to define the D chromosomes (bearing the derived C allele) and the non-D chromosomes (bearing the ancestral G allele).
Statistical Analysis of the Introgression Model.
When two populations are isolated from each other for a prolonged period and are then followed by admixture, a key signature at the affected genetic locus is the presence of two haplotype clades bearing an excessive level of sequence divergence from each other (25, 30). In particular, the two haplotype clades should differ consistently from each other at a large number of sites, where the two historically separated populations have each fixed (or nearly fixed) a set of population-specific alleles during the time of separation before admixture. One way to test for admixture is to examine whether there is an excess of congruent segregating sites (i.e., sites having the same segregation pattern in the set of chromosomes sampled, as would be expected of sites that consistently differentiate two deeply divided clades), as compared with the null expectation of a panmictic model (25). A limitation of this approach is that it does not consider near-congruent sites whose perfect congruency has been slightly eroded when occasional recombination and gene conversion events occurred between the two haplotype clades since their admixture. We therefore used a modified approach called the interhaplogroup divergence test, which examines whether the divergence between two haplotype clades relative to the divergence within a clade exceeds the null expectation (see Materials and Methods). Our specific goal was to use the test to examine whether, at the time of coalescence of the D allele, the level of divergence separating the MRCA of D chromosomes from the non-D chromosomes, when scaled to the divergence within the non-D chromosomes, exceeds the null expectation.
To perform the test, we used the coalescent process as described (31, 32) to generate a large set of random genealogies containing the observed number of segregating sites over a 29-kb region under the observed recombination rate of the locus (see Materials and Methods). For each genealogy, we calculated the mean divergence between a given chromosome and the rest of the chromosomes (π0), as well as the mean divergence among the rest of the chromosomes (π1). A wide range of demographic scenarios was used in the coalescent simulations, including constant population size, exponential growth, and severe population bottlenecks (see Materials and Methods), all showing that the π0/π1 ratio of 3.3 observed in the real data is highly unlikely (P = 0 with 10,000 replicas). We also performed a much more conservative test by assuming that the mutational event that created the MRCA of the D allele landed on a chromosome, which, by chance, happened to be the most divergent chromosome in the genealogy (i.e., the chromosome with the highest π0/π1 ratio in the genealogy). This test still produced highly significant results (P < 0.0006). Finally, we performed the test under the extreme condition of zero recombination, which makes the test highly conservative, because in the absence of recombination, the chance occurrence of an extreme genealogy will be shared by all nucleotide positions across the region. This also yielded significant results (P < 0.003).
These simulation results are incompatible with the null model of genetic drift under panmixia and suggest instead that the observed genealogy of microcephalin is likely the result of population subdivision. As depicted in Fig. 4, two scenarios of population subdivision may be compatible with the data. The first scenario is prolonged subdivision of two populations followed by complete admixture of the two populations recently (Fig. 4A). Under this scenario, the two populations are reproductively isolated for a prolonged period such that one population was fixed for the D allele, whereas the other population was fixed for the non-D allele. The two populations were then thoroughly admixed ≈37,000 years ago, and the high frequency of the D allele in the final admixed population is not the result of selection but rather because the D-bearing population contributed a significant faction to the admixture. However, to explain the young coalescence age of the D allele, one has to argue that the D-bearing population is genetically extremely homogeneous, presumably because of a severe and prolonged bottleneck in the recent history of this population that dramatically reduced diversity. If this is true, then other loci in the genome should show similar genealogies as observed for microcephalin. However, this is clearly not the case, as shown below and by other studies (33, 34). We therefore argue that this scenario is unlikely.
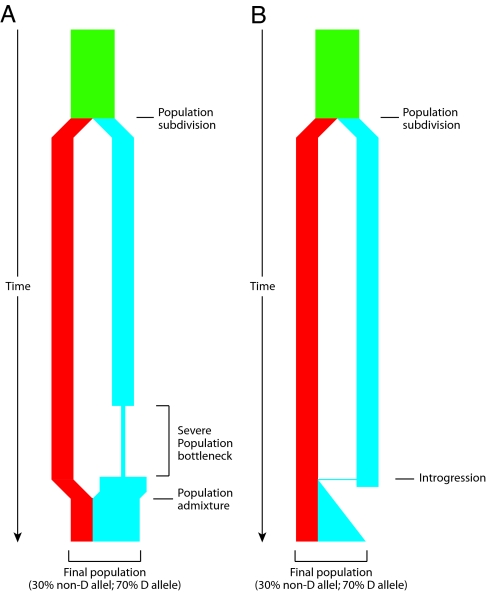
Fig. 4.
Schematic depiction of two demographic scenarios compatible with the observed genealogy of the microcephalin locus. In both scenarios, an ancestral population, depicted in green, was subdivided into two reproductively isolated populations. One population, depicted in red, fixes the non-D allele, whereas the other population, depicted in blue, fixes the D allele. (A) In the first scenario, the blue population went through a severe bottleneck that dramatically reduced genetic diversity. It then expanded and merged with the other population. (B) In the second scenario, a rare interbreeding event occurred between the two populations, bringing a copy of the D allele from the blue into the red population. This copy subsequently amplified to high frequency under positive selective pressure. The first scenario depends on demography only and does not require selection. This scenario should therefore affect all sites in the genome. The second scenario requires the action of positive selection on the introgressed allele and is therefore not expected to have a genome-wide effect. The observation that the genealogy of microcephalin is not representative of the genome is consistent with the second scenario.
The second scenario is introgression of an adaptive allele from an isolated population (Fig. 4B). Under this scenario, just as in the first scenario, two subdivided populations were reproductively isolated from each other for a prolonged period, such that one population was fixed for the D allele, whereas the other population was fixed for the non-D allele. Unlike the first scenario, however, the two populations did not admix completely. Rather, a rare interbreeding event occurred between the two populations ≈37,000 years ago, which resulted in the introgression of a copy of the D allele from the D-bearing into the non-D population. The D-bearing population subsequently went extinct, but the introgressed D allele spread to exceptionally high frequency in the remaining population because of positive selection. Because this scenario invokes positive selection specifically at the microcephalin locus, it is not expected to have a genome-wide effect. Other regions of the genome brought over by the interbreeding event are expected to be lost by genetic drift unless they also confer a selective advantage. As discussed below, the lopsided and deeply divided genealogy observed at the microcephalin locus is highly atypical of the genome, which is consistent with this introgression scenario.
To compare microcephalin with other loci of the genome, we applied the interhaplogroup divergence test to the polymorphism data in the Seattle SNP data set (35). We chose this data set because, like the microcephalin data, it is based on resequencing rather than genotyping, which allows for a more fair comparison. For each locus in the data set, we obtained its π0/π1 ratio, which showed microcephalin to be an outlier (see Materials and Methods). This result thus further argues that the haplotype pattern of microcephalin is atypical of the genome.
Balancing Selection Cannot Explain the Unusual Genealogy.
An alternative to population subdivision that could explain the unusual genealogy of microcephalin is long-standing balancing selection within a panmictic population. Balancing selection has the effect of creating deep genealogies, a well known example being the MHC genes, whose high levels of diversity are believed to be the result of balancing selection (36). When coupled with suppressed recombination by inversion, balancing selection can create two deeply divided haplotype clades across a large genomic region (36, 37), which could potentially explain the deep genealogies previously observed at a number of loci (refs. 15–17 and 23; see Discussion). For microcephalin, balancing selection could give rise to the lopsided and deeply divided genealogy, as schematized in Fig. 2A, if the following conditions are met. First, recombination between the D and non-D haplotypes is suppressed. Second, the D and non-D alleles persisted under balancing selection alongside each other for a prolonged period, creating a deep divergence between them. Third, the D allele experienced an extended period of very low frequency in the population, leading to loss of diversity within the D chromosomes. Finally, a recent change in the selective regime caused the D allele to rise to high frequency worldwide. Such a change could involve the introduction of positive selection on the D chromosomes or a shift in balancing selection toward favoring higher frequencies of the D allele.
We note, however, that one of the above requisite conditions, suppressed recombination, appears rather inconsistent with empirical observation. The main mechanism for recombination suppression is inversion. Yet, there is no evidence of inversion at the microcephalin locus. The 29-kb resequenced region of microcephalin is well within the 237-kb gene and encompasses 6 of the 14 exons in the gene. An inversion within the gene that encompasses the 29-kb region is not feasible, because it would completely disrupt the function of the gene. A larger inversion that encompasses the entire gene is also not a possibility, because, although the D chromosomes are in nearly perfect linkage disequilibrium across the 29-kb region relative to the non-D chromosomes (Table 1), the 5′ and 3′ ends of the gene are no longer in strong linkage disequilibrium with the 29-kb region (29). This indicates the occurrence of recombinations between D and non-D alleles within the 237-kb gene. Furthermore, within the 29-kb resequenced region, there are four recombinants between D and non-D alleles of the 178 chromosomes surveyed. Although this is a small number of recombinants, it is consistent with a genome-average rate of recombination operating at this locus, because the D allele first emerged in humans only by ≈37,000 years ago. Thus, the D and the non-D chromosomes at the microcephalin locus are evidently colinear with each other and engage in a normal rate of recombination.
Given that the above arguments are only qualitative, we performed quantitative tests to assess the extent to which a given rate of recombination between D and non-D alleles might be compatible with balancing selection. Assuming that balancing selection is indeed responsible for maintaining the coexistence of D and non-D alleles in a panmictic population since their coalescence ≈1,700,000 years ago, we calculated the probability of observing four or fewer recombinants between the two alleles within the 29-kb resequenced region among the 178 chromosomes sampled. To be conservative, we considered a range of recombination rates in our calculations. This probability is 10−184 if we assume the locus-specific recombination rate as previously estimated for microcephalin (29, 38) and 10−148 if we assume the genome-average recombination rate. The probability is still highly significant at 10−6, even if we assume an unrealistic recombination rate that is only 1% of the genome average. Thus, it appears highly unlikely that the D and non-D alleles, each defined by an extended block of linkage disequilibrium, could have been maintained by balancing selection alongside each other in a panmictic population for the last 1,700,000 years since their coalescence while engaging in so few recombination events. Instead, our data support the alternative model, i.e., the presence of population subdivision that kept the D and the non-D alleles in reproductive isolation from each other for a prolonged period such that the two alleles accumulated a large number of sequence differences from each other and were unable to recombine because of the reproductive isolation (the duration of the reproductive isolation is discussed below).
Estimating the Duration of Reproduction Isolation Between D and Non-D Alleles.
Next, we sought to estimate the duration of separation between the two populations before introgression. Even though the coalescence time between D and non-D alleles is ≈1,700,000 years, the time of reproductive isolation between them should be less because of the preexistence of polymorphisms within the population before its split into separate populations. Assuming negligible gene flow during separation, we simulated a range of separation times and calculated the π0/π1 ratio for each time point (note that a separation time of zero results in a demographic model that is the same as the null model of panmixia described above; see Materials and Methods). We found that a separation time of ≈1,100,000 years best recapitulated the observed π0/π1 ratio; that is, simulations conditioned on this separation time produced a mean π0/π1 ratio that closely resembles the observed value (Fig. 5; see Materials and Methods). We also calculated the 5% confidence lower-bound of the separation time, which is defined as the separation time for which 5% of the simulations produced π0/π1 ratios at or greater than the observed value. We placed this lower bound at ≈530,000 years. We note, however, that because these calculations are based on a number of assumptions, such as the size of historical populations, the results should be taken as rough estimates only that are broadly consistent with the data. Furthermore, the calculations assume complete reproductive isolation of the two populations. As an alternative, there could be a small amount of gene flow between the two populations, which would likely require an even longer separation time of the two populations to achieve the level of sequence divergence observed between D and non-D alleles. In this case, the amount of separation time estimated based on zero gene flow can be considered as conservative.
Fig. 5.
Relationship between the time separating two populations and the π0/π1 ratio. Each circle represents the average π0/π1 ratio of 1,000 simulations at a given separation time (see Materials and Methods). Dashed lines show that a separation time of ≈1,100,000 years produces the observed π0/π1 ratio. The average generation time is assumed to be 25 years.
Discussion
In this study, we investigate the origin of the microcephalin D allele in modern humans. We show that the D allele is unlikely to have arisen within a panmictic population. Instead, our data are consistent with a model of population subdivision followed by introgression to account for the origin of the D allele. By this model, schematized in Fig. 4B, the lineage leading to modern humans was split from another Homo lineage, and the two lineages remained in reproductive isolation for ≈1,100,000 years. During this period of reproductive isolation, the modern human lineage was fixed for the non-D allele at the microcephalin locus, whereas the other Homo lineage was fixed for the D allele. These two alleles are differentiated by a large number of sequence differences accumulated during the prolonged isolation of the two populations. At or sometime before ≈37,000 years ago, a (possibly rare) interbreeding event occurred between the two lineages, bringing a copy of the D allele into anatomically modern humans. Whereas the original D-bearing Homo population had since gone extinct, this introgressed copy of the D allele in humans had subsequently spread to exceptionally high frequency throughout much of world because of positive selection.
Several studies have reported loci in the human genome that are associated with unusually deep genealogies containing highly divergent clades (15, 16, 23, 24). Although this type of observation can result from the admixture of reproductively isolated populations, the observation is itself insufficient evidence for admixture. In particular, deep divergence can occur if two or more allele classes at a locus are maintained by balancing selection for a prolonged period. So, even for a locus whose genealogy is too deep to be statistically compatible with neutral evolution, it is essential to rule out balancing selection before the admixture model can be adopted. One notable example is the MAPT locus, which has two distinct haplogroups, H1 and H2, that diverged from each other ≈3 million years ago, whereas the coalescence age of H2 is much younger (24). One explanation for this unusual observation is that H2 introgressed into modern humans from an archaic Homo lineage (17). However, because H1 and H2 are inverted relative to each other and thus cannot recombine, deep divergence between them across an extend genomic region and the young age of H2 are also compatible with balancing selection that maintained H2 at low frequencies for extended periods (along with H1) followed by a recent rise in the frequency of H2 (24). For the microcephalin locus, in contrast, we were able to rule out the possibility of balancing selection with a high degree of confidence. As such, microcephalin shows by far the most compelling evidence of admixture among the human loci examined thus far.
Speculation about the identity of the archaic Homo population from which the microcephalin D allele introgressed into the modern human gene pool points to the Neanderthal lineage as a potential (although by no means only) candidate. Anatomically modern humans and Neanderthals shared a long period of coexistence, from as early as 130,000 years ago in the Middle East (39) to as late as 35,000 years ago in Europe (40), consistent with the estimated introgression time of the microcephalin D allele at or sometime before ≈37,000 years ago. Furthermore, the worldwide frequency distribution of the D allele, exceptionally high outside of Africa but low in sub-Saharan Africa (29), suggests, but does not necessitate, admixture with an archaic Eurasian population. Finally, our estimate of the separation time between D and non-D alleles (i.e., ≈1,100,000 years with a lower-bound confidence interval of ≈530,000 years) is largely consistent with the divergence time between modern humans and Neanderthals based on mitochondrial DNA (mtDNA) sequence difference (320,000–740,000 years; refs. 41 and 42) and with the earliest appearance of Neanderthals in the fossil record ≈500,000 years ago (43). It would be of great interest to sequence the microcephalin locus in Neanderthals or other archaic Homo lineages, should it become technically feasible to retrieve and analyze nuclear DNA from ancient hominid remains.
Our results not only provide genetic evidence in support of the possibility of admixture between modern humans and an archaic Homo lineage but also support the notion that the biological evolution of modern humans might have benefited from the contribution of adaptive alleles from our archaic relatives. In the case of microcephalin, it is all the more intriguing given the fact that the adaptive allele is associated with an important brain development gene. As anatomically modern humans emerged from Africa and spread across the globe, the “indigenous” Homo populations they encountered had already inhabited their respective regions for long periods of time and were, in all likelihood, better adapted to the local environments than the colonizing humans, at least in some biological domains. It is perhaps not surprising then that modern humans, although likely superior in their own way, could in theory benefit from adopting some adaptive alleles from the populations they replaced. That this might indeed be the case for the brain size-determining gene microcephalin should add an important new perspective to the discussion of human origins and the recent evolution of our species. Furthermore, any admixture between modern humans and archaic populations is likely to affect more than one locus in the genome. Our study thus provides a methodological template for identifying additional loci in the human genome that might harbor alleles from archaic populations through introgression and subsequent positive selection.
Materials and Methods
Resequencing of DNA Samples.
The resequencing data were obtained as described (29). Briefly, a panel of 89 DNA samples was obtained from the Coriell Institute that broadly represents the global diversity of major human populations. It included nine sub-Saharan Africans, seven North Africans, nine Iberians, seven Basques, nine Russians, nine Middle Easterners, nine South Asians, eight Chinese, one Japanese, eight Southeast Asians, six Pacific Islander, and seven Andeans. The region of microcephalin studied encompasses exons 4–9 of the 14-exon gene and was 29,027 bp in length. Of this, 23,416 base pairs were fully sequenced with double-stranded sequencing in all individuals of the Coriell panel and a common chimpanzee. Inference of haplotypes was performed with the PHASE 2.1 software as described (44).
Calculation of Coalescence Time.
The method for calculating coalescence time follows the described procedure (29), which is known to be unbiased by demographic history. First, the MRCA of all of the chromosomes in the sample was obtained by using chimpanzee sequence as the outgroup. The average sequence divergence separating the MRCA and each of the chromosomes was then calculated. Last, this average divergence was scaled to mutation rate as obtained from human–chimpanzee divergence in the region to produce coalescence time.
Coalescent Simulation.
All software programs developed for the study are available upon request. The coalescent process as implemented in the ms software (31, 32) was used to simulate genealogies under the following parameters. The number of chromosomes was 50 (corresponding to the number of chromosomes present at the coalescence of the D chromosomes); the number of segregating sites was 178 (corresponding to the number of segregating sites present at the coalescence of the D chromosomes); and recombination (38) and gene conversion (45) rates were as described for the locus (29). Several demographic models were considered, including a constant population with an effective size of 10,000 individuals; population growth models with 10,000 individuals growing to 100,000 over 1,000, 2,000, or 3,000 generations; 10,000 individuals growing to 1,000,000 over 3,000 or 5,000 generations; and 10,000 individuals growing to 10,000,000 over 5,000 generations; and bottleneck models with 10,000 individuals dropping to 1,000 at 5,000 generations ago, then staying constant until 2,000 generations ago, at which time it exponentially grows to 10,000 or 10,000,000 at present. For each genealogy, a chromosome was selected, and the ratio of π0 (average divergence between the chosen chromosome and the other chromosomes) to π1 (average divergence among the other chromosomes) was calculated. This was repeated for all of the chromosomes and the highest π0/π1 ratio was recorded. SeattleSNP data were downloaded from http://pga.gs.washington.edu/education.html. Loci with <10 kb or 100 segregating sites were not considered. The MAPT locus was excluded, because it is known to contain a large inversion with suppressed recombination (24).
Estimation of Separation Time.
A population with an effective size of 10,000 was split t generations ago into two subpopulations each with an effective size of 10,000. One subpopulation contributes one chromosome (i.e., the introgressing chromosome), and the other subpopulation contributes the remaining chromosomes. For any value of t, the average π0/π1 ratio for the introgressing chromosome was obtained from 1,000 simulations.
Supplementary Material
Abbreviation
- MRCA
most recent common ancestor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.McDougall I, Brown FH, Fleagle JG. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 2.Stringer CB, Andrews P. Science. 1988;239:1263–1268. doi: 10.1126/science.3125610. [DOI] [PubMed] [Google Scholar]
- 3.Lahr MM. J Hum Evol. 1994;26:23–56. [Google Scholar]
- 4.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 5.Armour JA, Anttinen T, May CA, Vega EE, Sajantila A, Kidd JR, Kidd KK, Bertranpetit J, Paabo S, Jeffreys AJ. Nat Genet. 1996;13:154–160. doi: 10.1038/ng0696-154. [DOI] [PubMed] [Google Scholar]
- 6.Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 7.Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL. Mol Biol Evol. 1998;15:427–441. doi: 10.1093/oxfordjournals.molbev.a025939. [DOI] [PubMed] [Google Scholar]
- 8.Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, et al. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 9.Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB. Am J Hum Genet. 1997;60:772–789. [PMC free article] [PubMed] [Google Scholar]
- 10.Wolpoff MH, Hawks J, Caspari R. Am J Phys Anthropol. 2000;112:129–136. doi: 10.1002/(SICI)1096-8644(200005)112:1<129::AID-AJPA11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Yu N, Zhao Z, Fu YX, Sambuughin N, Ramsay M, Jenkins T, Leskinen E, Patthy L, Jorde LB, Kuromori T, Li WH. Mol Biol Evol. 2001;18:214–222. doi: 10.1093/oxfordjournals.molbev.a003795. [DOI] [PubMed] [Google Scholar]
- 12.Templeton A. Nature. 2002;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
- 13.Harding RM, McVean G. Curr Opin Genet Dev. 2004;14:667–674. doi: 10.1016/j.gde.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Trinkaus E. Annu Rev Anthropol. 2005;34:207–230. [Google Scholar]
- 15.Garrigan D, Mobasher Z, Severson T, Wilder JA, Hammer MF. Mol Biol Evol. 2005;22:189–192. doi: 10.1093/molbev/msi013. [DOI] [PubMed] [Google Scholar]
- 16.Garrigan D, Mobasher Z, Kingan SB, Wilder JA, Hammer MF. Genetics. 2005;170:1849–1856. doi: 10.1534/genetics.105.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy J, Pittman A, Myers A, Gwinn-Hardy K, Fung HC, de Silva R, Hutton M, Duckworth J. Biochem Soc Trans. 2005;33:582–585. doi: 10.1042/BST0330582. [DOI] [PubMed] [Google Scholar]
- 18.Eswaran V, Harpending H, Rogers AR. J Hum Evol. 2005;49:1–18. doi: 10.1016/j.jhevol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Templeton AR. Am J Phys Anthropol. 2005;41(Suppl):33–59. doi: 10.1002/ajpa.20351. [DOI] [PubMed] [Google Scholar]
- 20.Smith FH, Jankovic I, Karavanic I. Q Int. 2005;137:7–19. [Google Scholar]
- 21.Plagnol V, Wall JD. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrigan D, Hammer MF. Nat Rev Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- 23.Harris EE, Hey J. Proc Natl Acad Sci USA. 1999;96:3320–3324. doi: 10.1073/pnas.96.6.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir VG, et al. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 25.Wall JD. Genetics. 2000;154:1271–1279. doi: 10.1093/genetics/154.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, et al. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YQ, Su B. Hum Mol Genet. 2004;13:1131–1137. doi: 10.1093/hmg/ddh127. [DOI] [PubMed] [Google Scholar]
- 28.Evans PD, Anderson JR, Vallender EJ, Choi SS, Lahn BT. Hum Mol Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- 29.Evans PD, Gilbert SL, Mekel-Bobrov N, Vallender EJ, Anderson JR, Vaez-Azizi LM, Tishkoff SA, Hudson RR, Lahn BT. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- 30.Nordborg M. Life Sci; Proceedings of the NATO ASI Workshop, “Genes, Fossils, and Behaviour: An Integrated Approach to Human Evolution,”; Amsterdam: IOS; 2001. [Google Scholar]
- 31.Hudson RR. Oxford Surv Evol Biol. 1990;7:1–44. [Google Scholar]
- 32.Hudson RR. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 33.Wang ET, Kodama G, Baldi P, Moyzis RK. Proc Natl Acad Sci USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voight BF, Kudaravalli S, Wen X, Pritchard JK. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, Smith JD, Eberle MA, Kruglyak L, Nickerson DA. Am J Hum Genet. 2004;74:610–622. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer D, Thomson G. Ann Hum Genet. 2001;65:1–26. doi: 10.1046/j.1469-1809.2001.6510001.x. [DOI] [PubMed] [Google Scholar]
- 37.Dobzhansky T. Genetics of the Evolutionary Process. New York: Columbia Univ Press; 1970. [Google Scholar]
- 38.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 39.Grun R, Stringer C, McDermott F, Nathan R, Porat N, Robertson S, Taylor L, Mortimer G, Eggins S, McCulloch M. J Hum Evol. 2005;49:316–334. doi: 10.1016/j.jhevol.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Gravina B, Mellars P, Ramsey CB. Nature. 2005;438:51–56. doi: 10.1038/nature04006. [DOI] [PubMed] [Google Scholar]
- 41.Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Paabo S. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 42.Krings M, Geisert H, Schmitz RW, Krainitzki H, Paabo S. Proc Natl Acad Sci USA. 1999;96:5581–5585. doi: 10.1073/pnas.96.10.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bermudez de Castro JM, Martinon-Torres M, Carbonell E, Sarmiento S, Rosas A, van der Made J, Lozano M. Evol Anthropol. 2004;13:25–41. [Google Scholar]
- 44.Stephens M, Donnelly P. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiuf C, Hein J. Genetics. 2000;155:451–462. doi: 10.1093/genetics/155.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.