Abstract
Prion propagation has been modeled in vitro; however, the low infectious titer of PrPSc thus generated has cast doubt on the “protein-only” hypothesis. Here we show that prion delivery on suitable nitrocellulose carrier particles abrogates the apparent dissociation of PrPSc and infectivity. Misfolded prion protein generated by protein misfolding cyclic amplification is as infectious as authentic brain-derived PrPSc provided that confounding effects related to differences in the size distribution of prion protein aggregates generated in vitro and consecutive differences in regard to biological clearance are abolished.
Keywords: clearance, nitrocellulose, protein misfolding cyclic amplication
Prion diseases such as Creutzfeldt–Jakob disease and bovine spongiform encephalopathy are caused by a unique infectious agent termed prion that seems to propagate without a nucleic acid genome (1). The infectious agent in prion diseases has been shown to consist mainly, if not exclusively, of an aggregated protease-resistant conformer of the cellular prion protein (PrPC) that is present in the brain of infected individuals and has been designated PrPSc (2). The structural conversion of PrPC into a misfolded, protease-resistant isoform of PrP (termed PrPres) has been modeled in vitro (3). However, because of the low yield, these experiments did not allow a direct proof of infectivity. Legname et al. (4) were recently able to induce prion disease in transgenic mice overexpressing PrP by inoculating amyloid-like aggregates (5) of recombinant PrP; however, the infectious titer obtained by this approach was very low.
The protein misfolding cyclic amplification (PMCA) reaction provides a unique opportunity to convert relatively large quantities of PrPC into PrPres (6). Exploiting a mechanism of seeded aggregation, minute amounts of PrPSc are incubated with an excess of PrPC in a cyclic process consisting of alternating incubation and sonication steps. By combining PMCA with serial passages of the reaction mixture, we have confirmed an autocatalytic self-propagation of PrPres as postulated by the “protein-only” hypothesis (7). Adopting this approach, Castilla et al. (8) could recently demonstrate the generation of new infectious units by serial PMCA (sPMCA). However, the phenomenon that PrPres generated in the test tube by sPMCA is associated with a considerably lower specific infectivity than authentic PrPSc generated in vivo has cast doubt on the protein-only hypothesis of prion propagation and has supported theories invoking additional species of infectious PrP or further agent-associated factors.
Our current experimental investigations have led us to understand this apparent discrepancy between the amount of abnormal PrPres and infectivity, which we could relate to the size distribution of newly generated prion aggregates. Based on this insight we were able to design an experimental PMCA setup yielding in vitro generated prions that are indistinguishable from prions isolated from scrapie hamster brain in terms of proteinase K (PK) resistance, autocatalytic conversion activity, and, most notably, specific biological infectivity.
Results and Discussion
In our previous sPMCA experiments (7) the apparent specific infectivity of sPMCA-derived PrPres was >10 times lower than for cerebral PrPSc. This finding is consistent with the data of Castilla et al. (8), who found a 10- to 100-fold-lower specific infectivity of PrPres obtained by sPMCA as compared with brain-derived PrPSc. Although the specific infectivity of sPMCA-derived PrPres is substantially higher than that of misfolded PrP obtained from recombinant PrP (4), it is still unclear whether the lower specific infectivity of sPMCA-generated PrPres in comparison to brain-derived PrPSc is due to fundamental differences in the structure and/or composition of this material. We reasoned that, apart from potential conformational differences and potential cofactors, the specific biological infectivity of PrPres preparations should, according to theoretical considerations (9) and experimental findings (10), also depend on the size of PrPres aggregates. In principle, breaking up infectious PrP aggregates into smaller units could either increase the infectious titer by increasing the number of infectious particles or decrease biological infectivity by reducing aggregate stability and facilitating clearance from the brain. To address this question, we analyzed the effect of sonication-induced changes in aggregate size distribution on the specific infectivity of brain-derived PrPSc in a murine bioassay using scrapie strain RML (11).
Effect of Aggregate Size on Specific Infectivity.
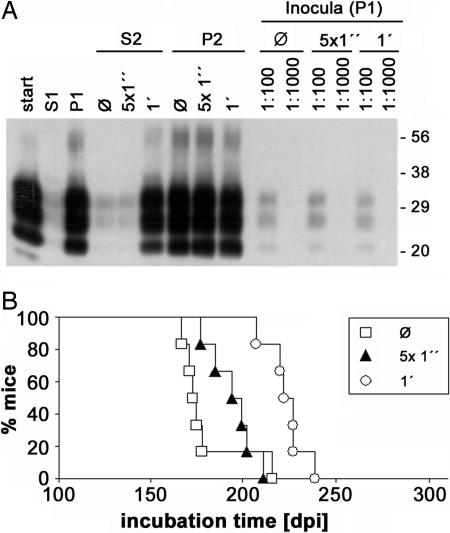
To obtain a robust measure of changes in aggregate size of PrPSc in brain homogenates, we quantified PrPSc in the pellet fraction and in the supernatant obtained by a centrifugation assay (Fig. 1A). This assay allowed direct analysis of crude brain homogenates and avoided a potential distortion of aggregate size distribution during sample processing. In conditions optimized for PrPSc analysis (see Fig. 6, which is published as supporting information on the PNAS web site), PrPSc was found almost exclusively in the pellet fraction. However, sonication induced a dose-dependent shift toward smaller aggregate size, as evidenced by the prominent presence of PrPSc in supernatant after 1 min of ultrasonic treatment (Fig. 1A, lane 6). When we compared the infectious titer of samples that contained the same amount of protease-resistant PrP, albeit with a different size distribution of PrPSc aggregates, we found that sonication-induced aggregate fragmentation was associated with a pronounced prolongation of incubation times (Fig. 1B). This finding indicates that, in the used murine bioassay, measured infectivity decreased with increasing fragmentation of PrPSc aggregates. This effect could be due to a lower stability and/or more efficient clearance of smaller aggregates. Similarly, the relatively low apparent specific infectivity of sPMCA-derived PrPres reported previously (7, 8) may have resulted from fragmentation of PrP aggregates by ultrasonication in the sPMCA reaction. To test this hypothesis, we used adsorption to suitable carrier particles to modify particle size distribution.
Fig. 1.
Correlation between aggregate size and biological infectivity of PrPSc in C57BL/6 mice. A 10% (wt/vol) brain homogenate prepared from RML-infected mice was digested with PK in the presence of 0.2% (wt/vol) SDS and then subjected to centrifugation. The supernatant (S1) was collected, and aliquots of the resuspended pellet (P1) were left unmodified or sonicated for 5 × 1 s or for 1 × 60 s, respectively, before further centrifugation. The supernatant (S2) was collected, and the pellet (P2) was resuspended. (A) Western blot analysis of PrPres in the raw homogenate (start), S1, P1, S2 of unmodified (Ø), 5 × 1 s (5 × 1”), and 1 × 60 s (1′) sonicated samples and in corresponding P2 samples using 6H4 antibody at a 1:2,000 dilution. In addition, 100-fold and 1,000-fold dilutions of untreated (Ø), 5 × 1 s (5 × 1”), and 1 × 60 s (1′) sonicated P1 used for bioassay inoculations are shown (Inocula). Positions of the molecular mass markers are given on the right (in kilodaltons). (B) Survival curves obtained for untreated (□), 5 × 1 s (▴), and 1 × 60 s (○) sonicated P1 samples corresponding to lanes 10, 12, and 14 in A.
Adsorption of Proteins to Nitrocellulose (NC) Particles Reduces Biological Clearance.
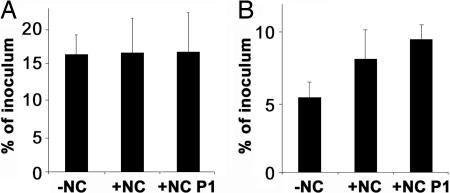
We reasoned that NC particles, which are known to firmly retain proteins, might be a suitable inert carrier for PrP aggregates and that adsorption to NC could reduce confounding effects on biological clearance that may result from differences in the size distribution of PrPSc. Using this approach, we could demonstrate that, in intracerebrally (i.c.) injected C57BL/6 mice in the presence of NC particles, clearance of proteins was efficiently reduced (Fig. 2). We compared 35S-labeled cell lysates extracted from RK13 cells expressing hamster PrP adsorbed to NC particles to control samples. Whereas exactly the same amount of radioactivity was retained in the brains of dead mice (Fig. 2A), in living animals clearance of radioactivity was strongly reduced in the presence of NC particles (Fig. 2B). This effect was even more pronounced in a pellet fraction enriched for NC-bound proteins.
Fig. 2.
Effect of NC particles on the biological clearance of proteins from mouse brains. A 35S-labeled cell lysate extracted from RK13 cells expressing hamster PrPC was centrifuged for 1 min at 1,000 × g. One aliquot of the 35S-labeled cell lysate was left untreated (−NC), and another aliquot was adsorbed to NC particles (+NC) Additionally, one NC-adsorbed aliquot was centrifuged for 15 min at 13,400 × g, and the resulting pellet was resuspended in PBS (+NC P1). Samples (−NC, +NC, and +NC1 P1) of 30 μl were inoculated i.c. into C57BL/6 mice. The radioactivity retained in the brain was measured in the brains of dead control mice killed 5 min before inoculation (A) and in the brains of mice killed 24 h after inoculation (B).
NC Adsorption and Size Distribution of PrPSc Aggregates in Hamster Brain Homogenates.
For our experiments regarding infectivity of PMCA-derived PrPres, we used the well established hamster system (6, 7), which in our experience provides a more efficient and robust amplification than the mouse system. To analyze NC adsorption and aggregate size distribution of PrPSc in hamster brain homogenates, we used the centrifugation assay described above. Similar to the results obtained for RML scrapie, hamster PrPSc was found almost exclusively in the pellet fraction (Fig. 3 A and B). This result was unchanged by incubation at 37°C. However, sonication at a dose similar to the conditions used during one cycle of PMCA caused a significant shift of PrPSc into the supernatant fraction, indicating that sonication breaks up aggregates into smaller units. This finding was even more pronounced in samples subjected to one cycle of PMCA, which clearly shows that PMCA causes a shift toward smaller aggregates recovered in the supernatant fraction. The observed effect may at least in part be due to a more efficient recruitment of newly formed PrPres by smaller aggregates during the PMCA reaction, in addition to the effect of sonication itself. Notably, in the presence of NC particles the sonication/PMCA-induced shift toward smaller aggregates was diminished (Fig. 3 A and C). Obviously, PrPSc adsorbed to NC particles, which thwarted the altered sedimentation behavior of small aggregates. Thus, we reasoned that adsorption to suitable carriers such as NC particles might abolish an increased biological clearance of sPMCA products caused by their relatively small aggregate size. To test this hypothesis, we used a bioassay of sPMCA-generated PrPres and appropriate control samples in the presence and absence of carrier particles.
Fig. 3.
Analysis of the size distribution of PrPSc aggregates in hamster brain homogenates before and after adsorption to NC particles. A 10% (wt/vol) brain homogenate from healthy hamsters was spiked 1:20 with a 10% (wt/vol) 263K brain homogenate and left untreated (start), incubated for 11 h at 37°C (incub.), subjected to PMCA, or sonicated for 50 × 1 s (sonic.). One aliquot of each sample was left on ice (unstabilized), and another aliquot was incubated with NC particles (NC-adsorbed). After centrifugation, the supernatant was collected, and the pellet was resuspended in PBS containing 0.05% (wt/vol) SDS and 0.5% (vol/vol) Triton X-100. Then, samples were treated with 100 μg/ml PK for 1 h at 37°C. (A) Western blot analysis of PrPres in the whole homogenate (Left) and in the supernatant and the pellet before and after adsorption to NC particles (Right). Positions of the molecular mass markers are given on the right (in kilodaltons). (B) Quantification of PrPres in the supernatant (open bars) and the pellet (filled bars) derived from unstabilized brain homogenates (means of two independent experiments + range). (C) Corresponding quantification in NC-adsorbed samples (means of two independent experiments + range).
Bioassay of PrPres Generated by sPMCA.
Using 15 cycles of sPMCA, we achieved a 200,000-fold total amplification of PrPres concurrently to a 2.514 (373,000-fold) dilution of the initial PrPSc used to seed the conversion reaction (ref. 12 and Fig. 4A). To investigate the infectious properties of the newly formed sPMCA-derived PrPres, we inoculated wild-type hamsters i.c. with 50-μl aliquots of (i) the initial reaction mixture before sPMCA (the “starting material” group); (ii) the reaction mixture after sPMCA (the sPMCA group); (iii) the starting material diluted 2.514-fold (the “dilution-only” group); and control samples resulting from sPMCA-like serial passages either (iv) without sonication (the “no sonication” group) or (v) without incubation steps (the “no incubation” group). Thus, groups ii–v all represent a 2.514-fold dilution of the starting material. However, because of autocatalytic amplification the amount of PrPres in the sPMCA sample was ≈43% of the amount of PrPSc in the starting material, whereas no PrPres was detectable in samples iii–v by Western blot analysis (Fig. 4A).
Fig. 4.
Western blot analysis of sPMCA-generated PrPres and incubation times in corresponding bioassay experiments. Aliquots (50 μl) of the starting material sample (start, □), the sPMCA sample (sPMCA, ●), the dilution-only sample (dilut., ▵), the no-sonication sample (no son., ▴), and the no-incubation sample (no inc., ○) were analyzed before (−) and after (+) digestion with PK. (A) Before inoculation, samples were left unmodified (Left) or adsorbed to NC particles (Right). All processed samples (indicated by black bars) represent a 2.514 (373,000-fold) dilution of PrPSc contained in the starting material. Positions of the molecular mass markers are given on the right (in kilodaltons). (B) Survival curves for unmodified samples. (C) Corresponding survival curves for NC-adsorbed samples.
The animals in the starting material group showed an attack rate of 100% (Fig. 4B) and a mean incubation time to terminal disease of 90 ± 3.7 days (mean ± standard deviation), which corresponds to a mean infectious titer in the inoculum of 1.3 × 104 50% i.c. lethal dose (LD50i.c.) calculated from a dose–incubation time calibration curve. Because these inocula each contained 2.5 × 10−5 g of homogenized scrapie-infected hamster brain, the measured titer is equivalent to 5 × 108 LD50i.c. per gram of scrapie-infected hamster brain. Only two of nine hamsters challenged with the dilution-only sample showed signs of scrapie 120 and 150 days after inoculation. One of the animals in the no-incubation group and none of the animals in the no-sonication group became ill. This finding is consistent with the titer measured for the starting material group and proves that diluted control samples, on average, contained well below one infectious unit per inoculum. The mean incubation time for the sPMCA sample was 104 ± 6.0 days, corresponding to 4.6 × 102 LD50i.c. per inoculum. Because this sample contained the same amount of scrapie-infected brain material as the diluted control samples, sPMCA resulted in a clear-cut increase in infectivity by more than three orders of magnitude. However, in line with previous data (7, 8), the apparent specific infectivity of sPMCA-derived PrPres was >10 times lower than for brain-derived PrPSc.
Bioassay of Stabilized PrPres Aggregates.
To analyze infectivity of newly formed sPMCA-derived PrPres with a modified size distribution and/or stability, we used chemical cross-linking on one hand and adsorption to suitable carriers on the other hand. Chemical cross-linking can be used to covalently stabilize aggregates without significantly affecting size distribution (13). Cross-linking of the samples did not abolish the difference in the infectious titers between the starting material and the sPMCA-derived samples. In contrast, after adsorption of the starting material and the post-PMCA fraction to metal beads, no significant difference in infectious titers was observed (see Table 1, which is published as supporting information on the PNAS web site). However, handling samples with metal beads is fraught with the problem of rapid sedimentation, which interferes with preparation of homogeneous samples for inoculation and resulted in a high standard deviation in these experimental groups.
In the hamster bioassay of inocula adsorbed to NC particles, the incubation times in the sPMCA group (101 ± 4.5 days, corresponding to 8.8 × 102 LD50i.c.) converged to the starting material group (96 ± 6.0 days, corresponding to 2.8 × 103 LD50i.c.) (Fig. 4C). Notably, the difference in incubation times was not statistically significant (P > 0.05, two-tailed t test), whereas for the corresponding unmodified samples incubation times differed significantly (P < 0.001). Thus, when using NC-adsorbed samples, sPMCA-derived PrPres exhibited virtually the same specific infectivity as brain-derived PrPSc. The incubation times for the starting material with and without NC were not significantly different. This important control shows that NC particles only improved the infectious titer of samples with smaller aggregate size, thus ruling out an unspecific general effect of NC particles on incubation times.
Characterization of Infected Hamsters.
Prion disease was confirmed in infected animals by Western blot analysis, and no differences in regard to biochemical strain characteristics such as electrophoretic mobility and glycoform pattern were found between the different experimental groups (Fig. 5A). Additionally, the clinical disease phenotype did not differ between the different experimental groups. These findings were confirmed by secondary passage of brain homogenates from affected animals, which resulted in incubation times and a disease phenotype indistinguishable from the 263K strain used to seed the sPMCA (Fig. 5B). Thus, there is no indication that modifications of prion strain properties were induced by sPMCA or by adsorption to NC particles. At 250 days after inoculation, none of the hamsters challenged with NC-adsorbed samples from the dilution-only and no-sonication groups had developed clinical disease, and only two animals in the no-incubation group became ill at 209 and 226 days after infection (Fig. 4C), demonstrating that adsorption to NC particles without PMCA is not sufficient to induce disease.
Fig. 5.
Western blot analysis of PK-treated brain homogenates from diseased wild-type hamsters. (A) Western blot analysis of brain homogenates from animals that developed clinical TSE symptoms after i.c. inoculation with the infectious starting material (start), PrPres generated by sPMCA, and the initial reaction mixture after 2.514-fold dilution without PMCA (dilut.), respectively, before (unstabilized) and after (NC-adsorbed) binding to NC particles. (B) Secondary passage from affected animals inoculated with the unstabilized starting material (start) and the sPMCA-derived PrPres (sPMCA) before (unstabilized) and after (NC-adsorbed) adsorption to NC particles, respectively. The brain samples of the examined animals contained large amounts of proteinase-resistant PrP identical to that of the 263K strain used to seed the PMCA reaction in regard to electrophoretic mobility and glycoform pattern. Positions of the molecular mass markers are given on the right (in kilodaltons).
Thus, we conclude that adsorption to NC particles reduces differences in bioassay-measured infectivity that are primarily due to variations in the size distribution of PrPres aggregates. When brain infectivity is assayed after i.c. inoculation, the major part of infectivity disappears rapidly from the brain (14). The rate of this clearance is highly variable and influenced by the particle size and the strain of the inoculum (9, 15). The finding that PMCA leads to a shift in the size distribution of PrPres aggregates (as compared with brain-derived PrPSc) toward smaller particles is in accord with the notion that these aggregates exhibit a higher rate of clearance. Therefore, our findings indicate that the decreased survival times in the NC-adsorbed sPMCA-derived samples are caused by reduced clearance from the brain because of an increase in the size and/or stability of PrPres aggregates rather than by intramolecular structural alterations of PrPres particles induced by adsorption to NC. This reasoning is supported further by the finding that neither sPMCA nor adsorption to NC altered the Western blot profile and the strain properties of PrPSc.
Conclusion
In summary, we have devised a cell-free technique to prepare and deliver misfolded PrP that exhibits properties indistinguishable from PrPSc derived from brains of diseased animals, including proteinase resistance, autocatalytic converting activity, and infectivity in vivo. Thus, in the setting of PrPSc amplified by PMCA, these findings resolve the apparent discrepancy observed in previous studies on the quantitative relation of infectivity and amount of PrPres without the need to invoke hypothetical molecular species such as PrP* (16), because all essential properties of the infectious agent can be related to molecular properties of PrPres, provided that confounding effects related to differences in the size distribution of PrPres aggregates and consecutive differences in regard to biological clearance are abrogated by prion delivery on suitable carrier particles. This observation in turn rebuts one of the main arguments against the protein-only hypothesis. Furthermore, our findings do not support the previously formulated hypothesis of Castilla et al. (8) regarding potential changes in strain properties during in vitro amplification of PrPres.
In addition, this technique provides a unique opportunity to further elucidate the structure and molecular mode of action of infectious prion particles and the exponential amplification of prions for diagnostic purposes. Furthermore, the use of NC carrier particles may provide a novel tool to study prion infectivity and potentially also the effect of other pathological protein deposits in a more efficient way in the bioassay.
Materials and Methods
Analysis of Size Distribution and in Vivo Infectivity of Mouse PrPSc After Sonication.
A 10% (wt/vol) brain homogenate in PBS with 0.2% (wt/vol) SDS from mice infected with mouse-adapted scrapie strain RML (11) was digested with PK (100 μg/ml) for 1 h at 37°C and subjected to centrifugation for 15 min at 13,400 × g. The supernatant (S1) was collected, and the pellet (P1) was resuspended in PBS with 0.2% (wt/vol) SDS. Aliquots of PK-digested P1 (300 μl each) were left untreated or were subjected to sonication for 5 × 1 s and 1 × 60 s, respectively, by using a microsonicator (Sonopuls; Bandelin Electronic, Berlin, Germany) with power setting at 40%.
For in vivo infectivity assays, samples were diluted 100-fold in PBS and aliquots of 30 μl were inoculated i.c. into C57BL/6 mice (Charles River, Sulzfeld, Germany). Control mice received uninfected C57BL/6 brain homogenate. Inoculated mice were monitored daily for clinical signs of disease according to the criteria of Dickinson et al. (17).
To analyze the size distribution of PrPSc aggregates in the untreated and sonicated P1 samples used for inoculation, aliquots of these samples were subjected to centrifugation for 15 min at 13,400 × g. The supernatant (S2) was collected and the pellet (P2) was resuspended in PBS containing 0.2% SDS. For the quantification of PrPres, 10-μl aliquots were subjected to Western blot analysis using the 4H11 (18) antibody at a dilution of 1:2,000. PrP was visualized by enhanced chemiluminescence reaction (Amersham Pharmacia, Freiburg, Germany).
Analysis of the Effect of NC Particles on the Biological Clearance of Proteins from the Brain.
RK13 cells (19) expressing hamster PrPC were metabolically labeled with [35S]methionine and [35S]cysteine (Amersham Bioscience, Freiburg, Germany). Subsequently, cells were scraped off the culture dish and lysed by passing through a syringe with a needle of 0.7-mm diameter. The lysate was centrifuged for 1 min at 1,000 × g, and the protein content of the supernatant was adjusted to that of a 1% mouse brain homogenate. One aliquot of the 35S-labeled cell lysate was left untreated. and another aliquot was adsorbed to NC particles (20) for 24 h under constant agitation at 4°C. Briefly, NC sheets were dissolved in DMSO (1.25 ml/cm2) at room temperature for 10 min. Subsequently, NC particles were precipitated by dropwise addition of 1 volume of double-distilled water. The particles were centrifuged at 1,620 × g for 8 min, washed three times with PBS, and finally resuspended in PBS (1.25 ml/cm2). NC-bound samples were diluted 10-fold in the NC suspension before inoculation.
Additionally, one NC-adsorbed aliquot was centrifuged for 15 min at 13,400 × g, and the resulting pellet was resuspended in PBS. Samples of 30 μl were inoculated i.c. into C57BL/6 mice. Brains were analyzed 24 h after inoculation by using a TRI-CARB 2900TR β-counter (PerkinElmer, Rodgau, Germany). As a control, dead mice were inoculated with the same aliquots.
Analysis of the Size Distribution of Hamster Scrapie Prion Aggregates in Brain Homogenates.
A 10% (wt/vol) brain homogenate from healthy Syrian golden hamsters was spiked 1:20 with 263K scrapie hamster brain homogenate. Two-hundred-microliter volumes of the reaction mixture were left untreated, incubated for 11 h at 37°C, sonicated for 50 × 1 s by using a microsonicator (Sonopuls), or subjected to one cycle of PMCA consisting of 10 rounds of 5 × 1-s sonication and 1-h incubation at 37°C. Subsequently, the reaction mixture was split, and one aliquot was left on ice whereas the other aliquot was incubated with NC particles (1 cm2/ml) for 2 h at room temperature under constant agitation. After centrifugation for 15 min at 13,400 × g the supernatant (S1) was collected and the pellet (P1) was resuspended in PBS containing 0.05% (wt/vol) SDS and 0.5% (vol/vol) Triton X-100. For quantification of PrPres, 20-μl aliquots were treated with 100 μg/ml PK for 1 h at 37°C and subjected to Western blot analysis as described above.
Serial Transmission PMCA Reactions.
Serial transmission PMCA experiments and control reactions were performed exactly as described in detail by Piening et al. (12). Briefly, brain homogenate (10% wt/vol) from 263K-infected Syrian hamsters in terminal stage of disease was prepared according to the protocol published by Saborio et al. (6) and diluted 1:20 with similarly prepared brain homogenate extracted from uninfected hamsters. Samples of 200 μl were subjected to 5 × 1 s of sonication by using an ultrasonic microtip probe at 40% power setting (Sonopuls 2070), followed by 1 h of incubation at 37°C. After 10 rounds of alternated sonication and incubation (completing one amplification cycle) the reaction mixture was passaged by diluting 2.5-fold into normal hamster brain homogenate. In total, 15 amplification cycles, including 10 PMCA rounds each, with 14 sequential 2.5-fold dilution steps were performed (150 rounds of PMCA). To avoid microbial contaminations, all experimental procedures were performed under sterile conditions. After the 15th passage, aliquots of 100 μl were collected and stored at −80°C until further use, avoiding any additional freeze–thaw cycles. For control reactions, either the sonication step (incubation only) or the incubation step (sonication only) was omitted. Additionally, 200-μl aliquots of the initial reaction mixture were subjected to 14 sequential 2.5-fold dilution steps (dilution only) and frozen immediately.
PK Digestion, Western Blotting, and Quantification of PrPres.
For the quantification of PrPres, samples were thawed, digested with PK (100 μg/ml; ratio of total protein:PK ≈ 1:60) for 1 h at 37°C, and subjected to Western blot analysis by using the 3F4 antibody at a dilution of 1:2,000 (21). PrP was visualized by enhanced chemiluminescence reaction (Amersham Pharmacia) and quantified by using a Diana II luminescence imaging system and the AIDA software package (Raytest, Straubenhardt, Germany). Amplification factors were determined as described (12).
In Vivo Infectivity Studies with Hamsters.
Infectivity of sPMCA-derived PrPres and the corresponding control samples was analyzed by hamster bioassay. Samples were either diluted 1:10 in PBS (unmodified samples) or adsorbed to NC carrier particles (20), which also resulted in a 1:10 dilution, before inoculation into the reporter animals. Six-week-old Syrian hamsters (nine animals per experimental group) were inoculated i.c. as described previously by Kimberlin and Walker (22) with 50-μl aliquots of the unmodified or NC-bound samples. Inoculation and clinical monitoring of hamsters were performed by using blinded sample aliquots. The amount of infectivity (LD50i.c.) in the inoculated samples was assayed as described (22) by the observed incubation times (t, in days) until terminal scrapie disease, using dose–incubation curves (23), when all of the inoculated animals succumbed to fatal disease. Alternatively, infectivity titers were calculated according to the method of limited dilution titration (24) if not all of the inoculated recipients developed lethal disease. For the calculation of infectivity from incubation times, the following empirical equation was used: log(LD50i.c.) = 0.0008 t2 − 0.2575 t + 20.7929 [mean error of assay: ±0.4 log(LD50i.c.)]. To confirm scrapie infection, brains from terminally diseased animals were collected, tested for PrPSc by Western blot analysis, and used for secondary passage inoculations.
Analysis of the Infectivity of Photochemically Cross-Linked and Metal-Bound Samples.
The in vivo infectivity of sPMCA-derived PrPres and the corresponding control samples was analyzed by hamster bioassay. Samples were diluted 1:10 in PBS and left unmodified (unstabilized samples), subjected to chemical crosslinking as described (13), or adsorbed to metal beads (316L; Hauner, Röttenbach, Germany). Briefly, 0.1-g metal beads and a 1-ml sample were incubated for 90 min at room temperature under constant shaking. Subsequently, 50-μl aliquots of unmodified and metal-adsorbed samples were inoculated i.c. into the reporter animals (nine animals per group). Inoculation and clinical monitoring of hamsters were performed by using blinded sample aliquots. The amount of infectivity (LD50i.c.) in the inoculated samples was assayed as described by Kimberlin and Walker by the observed incubation times (t, in days) until terminal scrapie disease, using dose–incubation curves (23). For the calculation of infectivity from incubation times, the following empirical equation was used: log(LD50i.c.) = 0.0008 t2 − 0.2575 t + 20.7929 [mean error of assay: ±0.4 log(LD50i.c.)].
Supplementary Material
Acknowledgments
We thank Salah Soliman and Marion Joncic for excellent technical assistance and Dominique Krüger for organizational help. This work was supported by State of Bavaria Grant Forprion LMU 8 and Deutsche Forschungsgemeinschaft Grant SFB 596-B13.
Abbreviations
- PK
proteinase K
- PrP
prion protein
- PrPC
cellular PrP
- PrPres
PK-resistant PrP
- PMCA
protein misfolding cyclic amplification
- sPMCA
serial PMCA
- i.c.
intracerebral(ly)
- LD50i.c.
50% i.c. lethal dose
- NC
nitrocellulose
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Prusiner SB. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Landsbury PT, Jr, Caughey B. Nature. 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 4.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 5.Baskakov IV. J Biol Chem. 2004;279:7671–7677. doi: 10.1074/jbc.M310594200. [DOI] [PubMed] [Google Scholar]
- 6.Saborio GP, Permanne B, Soto C. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 7.Bieschke J, Weber P, Sarafoff N, Beekes M, Giese A, Kretzschmar HA. Proc Natl Acad Sci USA. 2004;101:12207–12211. doi: 10.1073/pnas.0404650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castilla J, Saá P, Hetz C, Soto C. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Masel J, Jansen VAA. Biochim Biophys Acta. 2001;1535:164–173. doi: 10.1016/s0925-4439(00)00095-8. [DOI] [PubMed] [Google Scholar]
- 10.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler RL. Lancet. 1961;i:1378–1379. doi: 10.1016/s0140-6736(61)92008-6. [DOI] [PubMed] [Google Scholar]
- 12.Piening N, Weber P, Giese A, Kretzschmar H. Biochem Biophys Res Commun. 2005;326:339–343. doi: 10.1016/j.bbrc.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 13.Piening N, Weber P, Högen T, Beekes M, Kretzschmar H, Giese A. Amyloid. 2006;13:67–77. doi: 10.1080/13506120600722498. [DOI] [PubMed] [Google Scholar]
- 14.Safar JG, Kellings K, Serban A, Groth D, Cleaver JE, Prusiner SB, Riesner D. J Virol. 2005;79:10796–10806. doi: 10.1128/JVI.79.16.10796-10806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Safar JG, Cohen FE, Prusiner SB. Arch Virol Suppl. 2000;16:227–235. doi: 10.1007/978-3-7091-6308-5_22. [DOI] [PubMed] [Google Scholar]
- 16.Weissmann C. Cell. 2005;122:165–168. doi: 10.1016/j.cell.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson AG, Meikle VM, Fraser H. J Comp Pathol. 1968;78:293–299. doi: 10.1016/0021-9975(68)90005-4. [DOI] [PubMed] [Google Scholar]
- 18.Ertmer A, Gilch S, Yun SW, Flechsig E, Klebl B, Stein-Gerlach M, Klein MA, Schätzl HM. J Biol Chem. 2004;279:41918–41927. doi: 10.1074/jbc.M405652200. [DOI] [PubMed] [Google Scholar]
- 19.McCarthy K, Taylor-Robinson CH, Pillinger SE. Lancet. 1963;15:593–598. doi: 10.1016/s0140-6736(63)90393-3. [DOI] [PubMed] [Google Scholar]
- 20.Gulle H, Eibl MM, Wolf HM. J Immunol Methods. 1998;214:199–208. doi: 10.1016/s0022-1759(98)00058-1. [DOI] [PubMed] [Google Scholar]
- 21.Kascsak RJ, Rubenstein R, Merz PA, Tonna-DeMasi M, Fersko R, Carp RI, Wisniewski HM, Diringer H. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimberlin RH, Walker CA. J Gen Virol. 1986;67:255–263. doi: 10.1099/0022-1317-67-2-255. [DOI] [PubMed] [Google Scholar]
- 23.Kimberlin RH, Walker CA. J Gen Virol. 1977;34:295–304. doi: 10.1099/0022-1317-34-2-295. [DOI] [PubMed] [Google Scholar]
- 24.Gregori L, McCombie N, Palmer D, Birch P, Sowemimo-Coker SO, Giulivi A, Rohwer RG. Lancet. 2004;364:529–531. doi: 10.1016/S0140-6736(04)16812-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.