Abstract
The pressure-driven transport of individual DNA molecules in 175-nm to 3.8-μm high silica channels was studied by fluorescence microscopy. Two distinct transport regimes were observed. The pressure-driven mobility of DNA increased with molecular length in channels higher than a few times the molecular radius of gyration, whereas DNA mobility was practically independent of molecular length in thin channels. In addition, both the Taylor dispersion and the self-diffusion of DNA molecules decreased significantly in confined channels in accordance with scaling relationships. These transport properties, which reflect the statistical nature of DNA polymer coils, may be of interest in the development of “lab-on-a-chip” technologies.
Keywords: nanofluidics
Transport of DNA and proteins within microfluidic and nanofluidic channels is of central importance to “lab-on-a-chip” bioanalysis technology. As the size of fluidic devices shrinks, a new regime is encountered where critical device dimensions approach the molecular scale. The properties of polymers like DNA often depart significantly from bulk behavior in such systems because statistical properties or finite molecular size effects can dominate there. DNA confinement effects have been exploited in novel diagnostic applications such as artificial gels (1), entropic trap arrays (2), and solid-state nanopores (3, 4). These advances underline the importance of exploring the fundamental behavior of flexible polymers in fluid flows and channels (5–10) that underlie current and future fluidic technologies.
Most transport in microfluidic and nanofluidic separation applications is currently driven by electrokinetic mechanisms that result in a uniform velocity profile and low dispersion (11, 12). An applied pressure gradient, in contrast, generates a parabolic fluid velocity profile that is maximal in the channel center and zero at the walls. Many important aspects of pressure-driven flows as a transport mechanism remain unexplored despite their ease of implementation and their ubiquity in conventional chemical analysis techniques such as high-pressure liquid chromatography. Our understanding of an object's fundamental transport properties in parabolic flows, mobility and dispersion, is at present based mainly on models for rigid particles (13, 14) that explain several important effects such as the following: (i) hydrodynamic chromatography, the tendency of large particles to move faster than small particles because large particles are more strongly confined to the center of a channel, where the flow speeds are highest, and (ii) Taylor dispersion (15), the mechanism by which analyte molecules are hydrodynamically dispersed as they explore different velocity streamlines by diffusion, an effect that has discouraged the use of pressure-driven flows in microfluidic separation technology. The applicability of rigid-particle models as useful approximations to the transport of flexible polymers is dubious in the regime where the channel size is comparable with the characteristic molecular coil size, the radius of gyration (Rg), yet remains untested there.
In this work, we present an investigation of the pressure-driven mobility and dispersion of individual DNA molecules in microfluidic and nanofluidic channels that reveals how this behavior is rooted in the statistical properties of polymer coils. DNA mobility exhibits both length-dependent and -independent regimes, while both the Taylor dispersion and the self-diffusion of DNA are observed to be strongly reduced in confined channels, in accordance with scaling relationships.
Results and Discussion
Microfluidic and nanofluidic channels (illustrated in Fig. 1A and B) were filled with aqueous buffer containing fluorescently labeled DNA molecules that were imaged by epifluorescence video microscopy. The three types of linear DNA fragment studied had lengths, L, of 48.5 kbp (22 μm), 20.3 kbp (9.2 μm), and 8.8 kbp (4 μm). The corresponding equilibrium DNA coil sizes (16) (Rg = 0.73, 0.46, and 0.29 μm, respectively) lie within the 175 nm to 3.8 μm range of the channel height, h. DNA molecules were transported along the channel by means of an applied pressure gradient, p, that was controlled by adjusting the height difference between two fluidic reservoirs connected to either end of the channel. The condition p = 0 was established by eliminating the drift of a collection of molecules. The pressure-driven fluid flow profile in a slit-like channel has been treated in detail and is characterized by a parabolic, Poisseuille flow across the channel height and a plug-like flow in the wide, transverse direction, decreasing to zero within a distance h of the slit edges (17). Because h is much smaller than the 50-μm channel width in our experiments, the fluid velocity as a function of the height, z, from the channel midplane, U(z), is well approximated by the parabolic flow profile for a fluid between parallel plates,
where η is the fluid viscosity. The trajectories of a large number of identical molecules were recorded for a series of p in each channel.‡ The fluid temperature, T, was monitored to correct for viscosity variations.
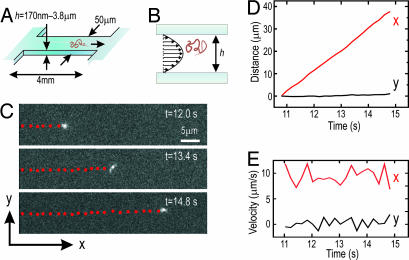
Fig. 1.
Experimental observation of pressure-driven DNA transport in microfluidic and nanofluidic channels. (A and B) Schematic illustrations of a rectangular, 50-μm-wide, 4-mm-long silica fluidic channel (A) and the channel cross-section over which an applied pressure gradient generates a parabolic fluid velocity profile (B). (C) Imaging a fluorescently labeled 48.5-kbp DNA molecule as it was transported through an h = 250 nm channel by an applied pressure gradient of 1.44 × 105 Pa/m. The red dots indicate the center-of-mass positions, recorded at a rate of 5 Hz. (D) The molecular trajectory along (x direction) and perpendicular to (y direction) the fluid flow, as a function of time. The linear increase in x position over time indicates a well defined average pressure-driven velocity. No net velocity is observed in the y direction. (E) The x and y components of the instantaneous molecular velocity as a function of time. The fluctuations along the flow are analyzed to study Taylor dispersion. The y direction fluctuations are independent of applied pressure and reflect thermal self-diffusion alone.
The trajectory of each DNA molecule's center of mass was tracked over a series of images by using custom-developed software, as shown in Fig. 1C and D. The average velocity of a molecular ensemble along the direction of flow, V̄, was calculated to be the mean of the instantaneous center-of-mass velocities for all molecules of a given length in each channel and at each p. The axial dispersion caused by velocity fluctuations within the ensemble (illustrated in Fig. 1E) was parameterized by the dispersion coefficient, D*, which is defined by 〈(Δx − V̄Δt)2〉 = 2D*Δt, where 〈(Δx − V̄Δt)2〉 is the mean square displacement of a molecule from its mean-velocity-shifted center of mass position in the time interval Δt. At p = 0, dispersion results only from the thermal self-diffusion of molecules, the diffusion coefficient for which we denote D0. Note that this situation represents DNA self-diffusion in a channel of height h and therefore includes hydrodynamic interactions with the walls and molecular confinement effects that are absent in bulk self-diffusion, the coefficient for which we denote Dbulk.
V̄ increased linearly with p for all DNA lengths in all channels. We found that V̄ was between the calculated maximum velocity of the fluid in the channel, Umax = h2p/8η, and the average fluid velocity, Ū = Umax (Fig. 2A Inset). To compare the transport of DNA in fluid of constant η, we first corrected for small temperature variations by rescaling velocities as§
We then defined the pressure-driven DNA mobility, ν, as the slope of V̄ vs. p, i.e., V̄ = νp. The standard deviation in the slope was taken to be the uncertainty in ν. The dependence of pressure-driven transport on DNA fragment length is best revealed by comparing the relative values of ν that were measured for each DNA length in the same channel and are therefore insensitive to microscopic channel irregularities. The ratios of ν for the 8.8- and 20.3-kbp DNA to νλ for 48.5-kbp DNA are plotted in Fig. 2A.
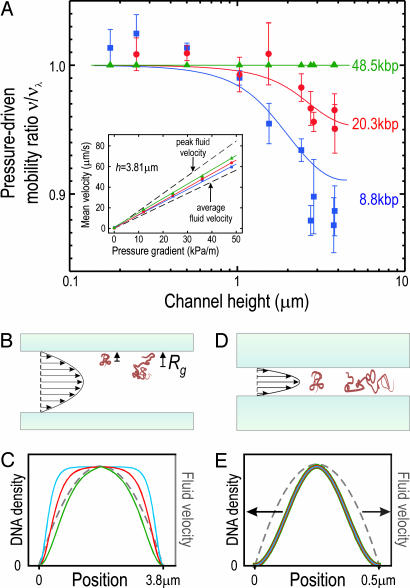
Fig. 2.
Dependence of pressure-driven DNA mobility on molecular length and channel height. (A) The average velocity of DNA molecules in an h = 2.73 μm channel increases linearly with applied pressure gradient (Inset). The slope of the curve defines the pressure-driven mobility, ν, which is observed to lie between the expected peak and average fluid mobility in the channel. The mobility ratio ν/νλ for 8.8- and 20.3-kbp-long DNA molecules, where νλ corresponds to 48.5-kbp-long λ-DNA molecules, is plotted as a function of the channel height. The solid lines indicate predictions of a transport model based on the equilibrium random-flight statistical behavior of DNA coils in a parabolic flow profile, as described in the text. (B) A schematic illustration of DNA configurations in a wide channel in which DNA mobility increases with molecular length. A molecule's center of mass is excluded from a region of length ≈Rg from the channel wall, inducing large molecules to spend a greater amount of time in the central, high-velocity region of the fluid flow. (C) The length-dependent DNA density profiles predicted in an h = 3.81 μm channel, with the parabolic fluid velocity profile indicated in gray. (D) A schematic illustration of DNA configurations in a narrow channel in which DNA mobility is independent of length. (E) The length-independent DNA density profile predicted for all three molecular lengths in an h = 500 nm channel with the parabolic fluid velocity profile indicated in gray.
Two distinct regimes of pressure-driven DNA transport can be clearly identified in Fig. 2A. In large channels (h > 2 μm), the mobility of DNA increased with molecular length. In the largest channels (h = 3.81 μm), ν for the 8.8-kbp (20.3 kbp) DNA fragments was reduced by 12% (5%) relative to that of the 48.5-kbp DNA. In small channels (h < 1 μm), ν was found to be independent of length within experimental error. The channel height corresponding to the cross-over between these two regimes increased with molecular length.
The observed pressure-driven mobility behavior can be explained by the statistical distribution of DNA molecules and modeled by using equilibrium random-flight statistics in the limit of low fluid shear rates. This approach therefore departs from naïve conventional models that approximate polymer coils as rigid objects (13, 14). We instead take the center-of-mass velocity of the DNA to be the average velocity of its segments, which travel at the local fluid velocity. V̄ can thus be expressed in terms of the average DNA segment concentration, ρ(z), and U(z), as
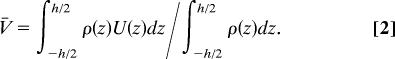 |
Consequently, the problem of determining the relative molecular speeds is reduced to determining ρ(z) as a function of h and DNA length.
We model a polymer coil as a random flight whose equilibrium conformation is described by the Edwards diffusion equation in a uniform potential field (19),
where P(z, s) is the probability that paths of contour length s end at z, and b is the mean independent step size, called the Kuhn length. The average concentration profile of DNA segments, ρ(z), for a molecule of length L is given by ρ(z) = (1/L)∫0LP(z, s)P(z, L − s)ds. The confinement of such a polymer to a narrow slit was first treated theoretically by Casassa (20) and Casassa and Tagami (19) by imposing noninteracting boundary conditions at the walls, setting P(±h/2) = 0. We have used Casassa's exact result,
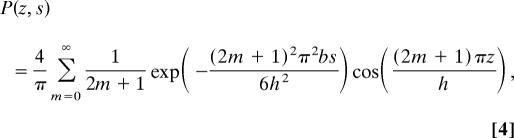 |
taking the equilibrium values of to numerically evaluate solutions to Eq. 1. The predicted ratios of ν for the three DNA lengths tested are plotted as solid lines in Fig. 2A.
Our polymer transport model predicts the length dependence of ν well over the full range of channel heights studied: A length-independent transport regime is predicted for thin channels, as well as length-dependent transport for sufficiently large channels. The predicted cross-over between these regimes agrees with our observations. In the length-dependent regime, the predicted reduction in ν/νλ corresponds perfectly to our data for the 20.3-kbp DNA and is only somewhat underestimated for the 8.8-kbp DNA in the highest channels. Note that our model contains no fitting parameters, relying instead on the well established bulk radii of gyration for DNA to parameterize their statistical behavior.
The physical origin of the two transport regimes is made clear by considering the channel size limits for which useful analytic approximations to ρ(z) exist (21): In large channels compared with the polymer coil size (h > 4Rg; illustrated in Fig. 2B), we find ρ(z) ∝ tanh2 . The DNA concentration profile is flat at the center of the channel where molecules can diffuse freely but is depleted in a region that extends ≈2Rg from the walls (Fig. 2C). Long molecules are therefore more strongly confined to the central, high-velocity region of the flow than short ones, explaining the observed length-dependent DNA mobility. This “hydrodynamic chromatography” transport regime for polymers has been proposed as a practical means to achieve size separation of long DNA molecules in microchannels because the mean separation between lengths increases linearly with Δt, whereas the width of a single-DNA-length distribution should only grow as because of dispersion (13, 22, 23). Our statistical polymer transport picture suggests that the observed velocity of a DNA molecule should approach the mean velocity on a measurement timescale, τ, that greatly exceeds both the diffusion time across the channel, i.e., τ ≫ 6η(h − 2Rg)2Rg/kBT, as well as the longest timescale for internal molecular reconfiguration, known as the Zimm time, i.e., τ ≫ 0.4ηRg3/kBT (24). Our model predicts an optimal channel height, h ≈ 10Rg, for separating DNA molecules where dv/dL is maximized. The resolving power of this technique would be limited in practice by constraints on a separation device's length, injection mechanism, separation time, the resolution, and the noise of DNA detection, among other considerations. A sensible comparison of DNA length separation by hydrodynamic chromatography to conventional technologies must therefore be made in the context of a complete device, which we do not attempt here.
In channels comparable with or smaller than the coil size (h < 2Rg; illustrated in Fig. 2D), we find ρ(z) ∝ cos2((πz)/h). In this situation the lateral distribution of a DNA molecule widens with length, but, importantly, its concentration profile across the channel height is length-independent (Fig. 2E), explaining why the ν are observed to be the same. This new “confined” transport regime is unique to flexible polymers, with no rigid-particle analog.
Our model successfully predicts pressure-driven DNA transport despite several assumptions that merit comment. We assume a constant η because the influence of polymer on the fluid viscosity is expected to be small for the low polymer concentrations and shear rates tested (25). For sufficiently high shear rates relative to the molecular relaxation times, i.e., for high Weissenberg numbers, Wi, fluidic shear forces could potentially distort DNA conformations from their assumed equilibria. In all our experiments, Wi was below 5, the onset of significant stretching in the flow direction (7). In addition, fluidic shear is concentrated near the channel walls, where the DNA concentration is depleted. The model also neglects forces normal to the flow direction that may arise from hydrodynamic coupling of the polymer segments to the channel walls (26, 27) or from the fluid inertia itself (28). Theory (29) and simulations (26, 27) predict that hydrodynamic interactions result in the migration of polymers toward the channel center to a degree that increases with Wi and h, an effect that has been experimentally confirmed for 48.5-kbp DNA in very large (h = 126 μm) channels (30). These effects are not expected to be significant for most of the range of small h tested in our experiments, but their onset at the highest Wi ∼ 5 and h ∼ 4 μm tested may explain the small discrepancy between our model predictions and the length-dependent ν/νλ observed there. Significant distortion of the equilibrium DNA concentration profile across the channel height by shear and hydrodynamic interactions, however, would result in a nonlinear V̄ vs. p curve, which was not observed.
DNA mobility in the low-pressure regime appears to be well explained by equilibrium polymer configurations; the dispersive behavior of DNA is of equal fundamental and practical significance and depends on molecular conformation fluctuations. Fig. 3A shows the dependence of D* for 48.5-kbp DNA in high (h = 2.73 μm) and low (h = 500 nm) channels on V̄. The Taylor dispersion of 48.5-kbp DNA was found to be greatly reduced in the thin, h = 500 nm channel relative to the h = 2.73 μm channel. Taylor dispersion theory (31) predicts that D* can be expressed as the sum of a component originating entirely from molecular self-diffusion (D0 in the present case) and a convective component that scales as V̄2. For all h, the dispersion of DNA can indeed be seen to obey D* = D0 + αTV̄2, where αT is a fit parameter that we call the Taylor time because it quantifies the hydrodynamic component of Taylor dispersion and is related to the time required for an object to explore all regions of the flow profile. The dependence of αT on h for all DNA lengths is plotted in Fig. 3B. A strong reduction in αT was observed with decreasing h for all but the smallest channels. For very small channels (h ≤ 250 nm), αT increased as h decreased, which we attribute to local fluid velocity variations caused by irregularities in the channel cross-section that gain increasing importance in the thinnest channels. αT generally decreased with increasing DNA length.
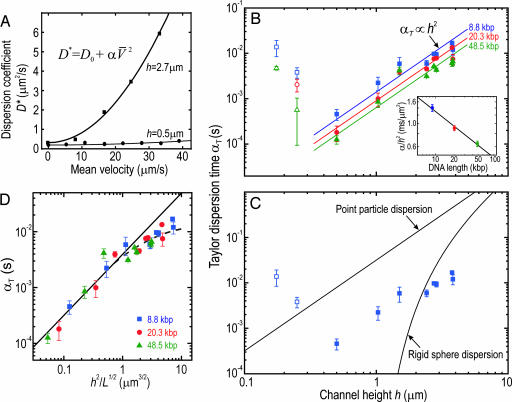
Fig. 3.
Taylor dispersion of DNA in microfluidic and nanofluidic channels. (A) The dispersion coefficient of λ-DNA molecules is plotted as a function of average molecular velocity for h = 2.73 μm and h = 500 nm. Solid lines indicate fits of D* = D0 + αTV̄2. The Taylor time, αT, quantifies the hydrodynamic component of dispersion. (B) The h dependence of αT is plotted for all DNA lengths. The αT values of the smallest two channels are likely dominated by irregularities in the channel cross-section and are consequently indicated with open symbols and excluded from further consideration. The data are well described by the power-law scaling relation, αT ∼ h2, which is fit for each DNA length and plotted with the solid lines. The fits provide a measure of the length dependence of Taylor dispersion through the average value of αT/h2, which is found to decrease as L−0.46±0.04 (Inset). (C) The observed values of αT for the 8.8-kbp DNA are compared with point-like and rigid-particle models of Taylor dispersion, highlighting the inadequacy of existing theories for describing the dispersion of polymers in confined channels. (D) Upon plotting αT vs. h2/Rg, we obtain a unique curve for all DNA lengths, suggesting a scaling relationship for DNA Taylor dispersion in small channels. The curve is linear in the thin channel limit (h2/Rg ≤ 1 μm) with a slope of 1.09 ± 0.11 and departs from this power-law scaling at higher h2/Rg.
The nearly constant slope of αT vs. h on the log–log scale of Fig. 3B for all DNA lengths suggests the power-law dependence αT ∝ h2.¶
The observed dispersion of DNA in small channels is striking when compared with the predictions of existing Taylor dispersion theory (Fig. 3C). Axial dispersion in a parabolic flow profile was first treated by Taylor (15) for a slow-moving point-like solute in a circular tube. On long timescales compared with the time for a particle to diffuse across the channel height, a condition that is satisfied in our experiments, the dispersion coefficient of a point-like solute in a thin, rectangular channel is given by D* = D0 + αT′V̄2, where αT′ = 0.038 × h2/D0 (32). The point-particle model therefore captures the observed αT ∝ h2 behavior. However, it fails to predict two important aspects of the observed DNA dispersion: First, the model overestimates αT by more than an order of magnitude for all h when D0 is taken to be the known bulk molecular diffusion constant. Second, the model predicts the opposite αT length dependence to what is observed: For longer DNA, D0 is smaller, and αT should increase as molecules spend more time on the same fluidic streamline. In fact, αT is observed to decrease with increasing DNA length.
Models approximating DNA behavior by rigid-particle dispersion also fail to describe the observed behavior of DNA in microfluidic and nanofluidic channels. In the simplest model, a polymer coil is treated as a free-draining, rigid sphere whose diameter is (13). Because the sphere cannot explore all streamlines equally, the h dependence of αT becomes modified relative to the point particle model as αT∝h2→αT∝h2 . The predictions of this rigid-particle model, which are plotted for 8.8-kbp DNA in Fig. 3C, overestimate αT in high channels and predict that αT should vanish as h approaches the sphere diameter, which is clearly not observed. More elaborate rigid-particle models that include hydrodynamic interactions (14) or particle size fluctuations (33) do not resolve these glaring inconsistencies. An analytical treatment of many interacting Brownian particles (34) is not tractable for large polymers, and computer simulations of polymer Taylor dispersion are lacking.
A heuristic description of DNA Taylor dispersion in microfluidic and nanofluidic channels is suggested by the observed L and h dependence of αT. In Fig. 3D, αT is plotted as a function of for all DNA lengths. Remarkably, we find that all points appear to lie on a common curve over two decades, with a slope of 1.09 ± 0.11 in the thin channel limit and with the data departing from the straight line at higher . Although DNA is an extended statistical object, the convective component of DNA dispersion in thin microfluidic and nanofluidic channels is consistent with a point-like solute description, i.e., αT = 0.038 × h2/Deff, where the effective DNA diffusion coefficient, Deff, scales as Deff ∝ . From the fit to our data, we find Deff ≈ 9.9 μm3/2·s−1 × . This result is in stark contrast to the bulk diffusion constant, Dbulk, which is known to obey Dbulk = 4.5 μm2+0.611·s−1 × 1/L(μm)0.611 (16). Deff therefore exceeds Dbulk for molecules larger than L ≃ 490 nm, a value corresponding to only a few DNA Kuhn segments. The surprising dispersion properties of DNA in small channels beg the development of a microscopic model. Flexible polymers differ crucially from point-like or rigid particles in that they possess many internal degrees of freedom. These may enable DNA to explore the parabolic flow more effectively, leading to an enhanced apparent diffusivity. These are also at the root of entropic elasticity (35), which would tend to confine the molecular center of mass to the center of the channel, thereby suppressing Taylor dispersion relative to that of point particles, while permitting fluctuations absent in rigid-particle models.
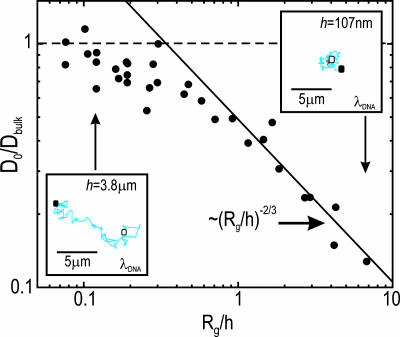
The high effective diffusion coefficient that can account for the reduced hydrodynamic dispersion of DNA in small channels is not caused by a high center of mass self-diffusion. Indeed, D0 is a measure of molecular self-diffusion and was found to decrease with decreasing h (Fig. 4). At low Rg/h, the ratio D0/Dbulk was nearly 1, and it decayed slowly with Rg/h for Rg/h ≥ 0.1. Above Rg/h ≈ 0.5, the self-diffusion of DNA decreased rapidly as D0/Dbulk ∝ (Rg/h)−2/3. This scaling relationship was predicted by Brochard and de Gennes (36) for sufficiently small channels in which highly confined molecules expand laterally, leading to a higher viscous drag and hence a reduced self-diffusion coefficient. First confirmed experimentally in a narrow tube geometry (37), this behavior has been modeled by computer simulations and experimentally verified for Rg/h as high as 1 in a slit geometry (26, 38). Here we see that D0/Dbulk ∝ (Rg/h)−2/3 to Rg/h values as high as 7.‖
Fig. 4.
Dependence of the normalized molecular self-diffusivity on the normalized channel height. The ratio D0/Dbulk is plotted as a function of Rg/h for all molecular lengths. (Insets) The self-diffusion trajectories of λ-DNA molecules in h = 3.8 μm and h = 107 nm channels, observed over a 20-s interval. The trajectories originate at the filled square and terminate at the open square.
In conclusion, we have shown how the pressure-driven transport behavior of DNA molecules in microfluidic and nanofluidic channels is dominated by the statistical properties of polymer coils. The distribution of a random-flight polymer across a channel leads to a pressure-driven mobility that increases with molecular length in large channels and remains independent of length in channels that are small compared with molecular coil size. The Taylor dispersion of DNA molecules is highly suppressed in confined channels and decays with channel height and molecular length according to a power-law scaling relationship. These polymer transport properties are of considerable significance to bioanalysis technology aimed at the separation of DNA by length or the uniform transport of DNA molecules through a fluidic system. An understanding of DNA transport characteristics can therefore guide the design of fluidic channels, the fundamental components of lab-on-a-chip technology.
Materials and Methods
Microfluidic and nanofluidic channels were prepared by using a sodium silicate bonding procedure (39). The 50-μm-wide and 4-mm-long channels were connected to large access holes at either end. The channel height, h, ranged from 175 nm to 3.8 μm. The channels were filled with buffer solution by capillarity and then electrophoretically cleaned of ionic impuries by applying 50 V across the channel for ≈10 min. A DNA solution was introduced into the channels via the access holes, which were then connected to open fluid reservoirs (10-ml glass syringe bodies) via Peek tubing, all filled with bubble-free buffer solution. The fluorescently labeled DNA molecules were imaged with an electron multiplication CCD camera (Andor, Belfast, Ireland) at a rate of 5 Hz by using an inverted oil-immersion fluorescence microscope (×100, 1.4 N.A.; Olympus, Tokyo, Japan) focused at the channel midplane.
The trajectories of DNA molecules were determined by using custom-developed molecular tracking software (Matlab; Mathworks, Natick, MA) that locates a molecule's center of mass as the first moment of the intensity distribution and follows it over a series of images. The integrated fluorescence intensity and the second moment of the intensity distribution (an estimate of Rg) were also calculated for each molecule and used as criteria to filter imaging noise, damaged DNA fragments, or overlapping molecules. Molecular trajectories were verified by eye to ensure faithful tracking. Ambiguous molecular trajectories that would intersect, divide (break), or irreversibly stick to the channel were manually excluded.
The three linear DNA fragments studied were as follows: 48,502-bp, unmethylated λ-phage DNA (λ-DNA; Promega, Leiden, The Netherlands); a 20,262-bp pBluescript 2× Topo plasmid construct (Stratagene, La Jolla, CA); and an 8,778-bp pBluescript+ 1,2,4 λ-DNA fragment plasmid construct (Stratagene). The DNA fragments were fluorescently labeled with YOYO-1 dye (Molecular Probes, Eugene, OR) using a base pair to dye ratio of 6:1 and suspended in an aqueous solution containing 50 mM NaCl, 10 mM Tris, 1 mM EDTA (pH 8.0), and 2% 2-mercaptoethanol by volume to minimize photobleaching. The concentration of DNA molecules was adjusted to introduce a convenient density (≈1–20 in an 80-μm-wide field of view) into each fluidic device tested.
Acknowledgments
We thank Peter Veenhuizen and Susanne Hage for preparing DNA fragments; Jerry Westerweel and Rene Delfos for assistance in calibrating fluid viscosities; Serge Lemay, Theo Odijk, and Cees Storm for useful discussions; and Andor Technology for the use of a camera. This work was supported by the Netherlands Organization for Scientific Research, Fundamenteel Onderzoek der Materie, and NanoNed.
Footnotes
The authors declare no conflict of interest.
The pressure gradients tested were limited by the ≈80 μm/s maximum molecular velocity to be reliably observable and by the maximum pressure gradient generated by an 80-cm-high fluid column.
The temperature-dependent value of η was parameterized by 1.05 × 10−3 Pa·s × 101.3272×(20−T)−0.001053×(20−T)2/(105+T), which was obtained by a fitting the temperature dependence of water's viscosity (18) and rescaling the absolute viscosity based on a measurement of the buffer solution viscosity by using a viscometer (Low Shear 40; Contraves, Zurich, Switzerland). We calculated p as gHφ/l, where g is the acceleration due to gravity (9.81 m/s2), φ is the buffer density (1 g/ml), H is the fluid column height, and l is the effective channel length (4.08 mm) that includes entrance effects.
The fit slopes were found to be 1.81 ± 0.26, 1.94 ± 0.20, and 1.60 ± 0.16 for the 48.5-, 20.3-, and 8.8-kb fragments, respectively.Linear fits of αT to h2, presented as solid lines in Fig. 3B, reveal that αT/h2 decreased with L as αT/h2 ∝ L−0.46±0.04 (Fig. 3B Inset).
An additional h = 107 nm channel was used to test the scaling of D0 to high degrees of confinement.
References
- 1.Volkmuth WD, Austin R H. Nature. 1992;358:600–602. doi: 10.1038/358600a0. [DOI] [PubMed] [Google Scholar]
- 2.Han J, Craighead H G. Science. 2000;288:1026–1029. doi: 10.1126/science.288.5468.1026. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 4.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Nat Mater. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 5.Perkins TT, Smith DE, Chu S. Science. 1997;276:2016–2021. doi: 10.1126/science.276.5321.2016. [DOI] [PubMed] [Google Scholar]
- 6.Smith DE, Chu S. Science. 1998;281:1335–1340. doi: 10.1126/science.281.5381.1335. [DOI] [PubMed] [Google Scholar]
- 7.Smith DE, Babcock HP, Chu S. Science. 1999;283:1724–1727. doi: 10.1126/science.283.5408.1724. [DOI] [PubMed] [Google Scholar]
- 8.Schroeder CM, Babcock HP, Shaqfeh ESG, Chu S. Science. 2003;301:1515–1519. doi: 10.1126/science.1086070. [DOI] [PubMed] [Google Scholar]
- 9.Quake SR, Babcock H, Chu S. Nature. 1997;388:151–154. doi: 10.1038/40588. [DOI] [PubMed] [Google Scholar]
- 10.Tegenfeldt JO, Prinz C, Cao H, Chou S, Reisner WW, Riehn R, Wang Y M, Cox EC, Sturm JC, Silberzan P, Austin RH. Proc Natl Acad Sci USA. 2004;101:10979–10983. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone HA, Stroock AD, Ajdari A. Annu Rev Fluid Mech. 2004;36:381–411. [Google Scholar]
- 12.Squires TM, Quake SR. Rev Mod Phys. 2005;77:977–1026. [Google Scholar]
- 13.Dimarzio EA, Guttman CM. Macromolecules. 1970;3:131–146. [Google Scholar]
- 14.Brenner H, Gaydos LJ. J Colloid Interface Sci. 1977;58:312–356. [Google Scholar]
- 15.Taylor G. Proc R Soc London Ser A; 1953. pp. 186–203. [Google Scholar]
- 16.Smith DE, Perkins TT, Chu S. Macromolecules. 1996;29:1372–1373. [Google Scholar]
- 17.Darnton N, Bakajin O, Huang R, North B, Tegenfeldt JO, Cox EC, Sturm J, Austin RH. J Phys Condens Matter. 2001;13:4891–4902. [Google Scholar]
- 18.Weast RC, editor. CRC Handbook of Chemistry and Physics. 63rd. Boca Raton, FL: CRC; 1982. [Google Scholar]
- 19.Casassa EF, Tagami Y. Macromolecules. 1969;2:14–26. [Google Scholar]
- 20.Casassa EF. J Polym Sci B Polym Lett. 1967;5:773–778. [Google Scholar]
- 21.Fleer GJ, Skvortsov AM, Tuinier R. Macromolecules. 2003;36:7857–7872. [Google Scholar]
- 22.Tijssen R, Bleumer JPA, van Kreveld ME. J Chromatogr. 1983;260:297–304. [Google Scholar]
- 23.Tijssen R, Bos J, van Kreveld ME. Anal Chem. 1986;58:3036–3044. [Google Scholar]
- 24.Doi M, Edwards SF. The Theory of Polymer Dynamics. Oxford: Oxford Univ Press; 1986. [Google Scholar]
- 25.Babcock HP, Smith DE, Hur JS, Shaqfeh ESG, Chu S. Phys Rev Lett. 2000;85:2018–2021. doi: 10.1103/PhysRevLett.85.2018. [DOI] [PubMed] [Google Scholar]
- 26.Jendrejack RM, Dimalanta ET, Schwartz DC, Graham MD, de Pablo JJ. Phys Rev Lett. 2003;91:038102. doi: 10.1103/PhysRevLett.91.038102. [DOI] [PubMed] [Google Scholar]
- 27.Jendrejack RM, Schwartz DC, de Pablo JJ, Graham MD. J Chem Phys. 2004;120:2513–2529. doi: 10.1063/1.1637331. and correction (2004) 120:6315. [DOI] [PubMed] [Google Scholar]
- 28.Segre G, Silberberg A. Nature. 1961;189:209–210. [Google Scholar]
- 29.Ma HB, Graham MD. Phys Fluids. 2005;17:083103. [Google Scholar]
- 30.Fang L, Hu H, Larson RG. J Rheol. 2005;49:127–138. [Google Scholar]
- 31.Brenner H. PhysicoChem Hydrodyn. 1980;1:91–123. [Google Scholar]
- 32.Dutta D, Leighton DT. Anal Chem. 2003;75:57–70. doi: 10.1021/ac020179r. [DOI] [PubMed] [Google Scholar]
- 33.Frankel I, Mancini F, Brenner H. J Chem Phys. 1991;95:8636–8646. [Google Scholar]
- 34.Brenner H, Nadim A, Haber S. J Fluid Mech. 1987;183:511–542. [Google Scholar]
- 35.Bustamante C, Marko JF, Siggia ED, Smith S. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 36.Brochard F, de Gennes PG. J Chem Phys. 1977;67:52–56. [Google Scholar]
- 37.Cannell DS, Rondelez F. Macromolecules. 1980;13:1599–1602. [Google Scholar]
- 38.Chen YL, Graham MD, de Pablo JJ, Randall GC, Gupta M, Doyle PS. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Top. 2004;70 doi: 10.1103/PhysRevE.70.060901. 060901. [DOI] [PubMed] [Google Scholar]
- 39.Stein D, Kruithof M, Dekker C. Phys Rev Lett. 2004;93 doi: 10.1103/PhysRevLett.93.035901. 035901. [DOI] [PubMed] [Google Scholar]
- 40.Blom MT, Chmela E, Oosterbroek RE, Tijssen R, van den Berg A. Anal Chem. 2003;75:6761–6768. doi: 10.1021/ac034663l. [DOI] [PubMed] [Google Scholar]