Abstract
Embryonic patterning and germ-cell specification in mice are regulative and depend on zygotic gene activities. However, there are mouse homologues of Drosophila maternal effect genes, including vasa and tudor, that function in posterior and germ-cell determination. We report here that a targeted mutation in Tudor domain containing 1/mouse tudor repeat 1 (Tdrd1/Mtr-1), a tudor-related gene in mice, leads to male sterility because of postnatal spermatogenic defects. TDRD1/MTR-1 predominantly localizes to nuage/germinal granules, an evolutionarily conserved structure in the germ line, and its intracellular localization is downstream of mouse vasa homologue/DEAD box polypeptide 4 (Mvh/Ddx4), similar to Drosophila vasa-tudor. Tdrd1/Mtr-1 mutants lack, and Mvh/Ddx4 mutants show, strong reduction of intermitochondrial cement, a form of nuage in both male and female germ cells, whereas chromatoid bodies, another specialized form of nuage in spermatogenic cells, are observed in Tdrd1/Mtr-1 mutants. Hence, intermitochondrial cement is not a direct prerequisite for oocyte development and fertility in mice, indicating differing requirements for nuage and/or its components between male and female germ cells. The result also proposes that chromatoid bodies likely have an origin independent of or additional to intermitochondrial cement. The analogy between Mvh-Tdrd1 in mouse spermatogenic cells and vasa-tudor in Drosophila oocytes suggests that this molecular pathway retains an essential role(s) that functions in divergent species and in different stages/sexes of the germ line.
Germ-line specification in many animals depends on the asymmetric partitioning of maternal determinants in oocytes. In Drosophila, females with homozygous loss-of-function mutations in posterior-group genes produce embryos that are defective in pole cells and abdominal formation, and several products of such genes, including oskar, vasa, and tudor, are localized to the pole plasm (1, 2). oskar triggers germ-line formation upstream of vasa and tudor (3, 4), and vasa regulates the intracellular distribution of Tudor protein (5). Interestingly, discrete granulofibrous structures, termed polar granules, are present in the pole plasm (6), and the products of oskar, vasa, and tudor accumulate in the polar granules (1, 2). Similar cytoplasmic structures are also observed in the germ line of diverse animals, such as P granules in Caenorhabditis elegans (7) and germinal granules in Xenopus (8). Based on morphological similarities including amorphous shape, the absence of surrounding membranes, and close association with mitochondria or nuclei, these ribonucleoprotein-rich cytoplasmic structures are collectively called nuage (9) or germinal granules. Nuage in oocytes and early embryos is thought to participate in the accumulation/partitioning of germ-line determinants, although the evidence is circumstantial, resting on the properties of mutants lacking nuage components and nuage (2).
In mice, germ-cell specification occurs among pluripotent epiblast cells depending on intercellular induction from somatic cells (10–13), and the maternal contribution to this specification has not been identified. Correspondingly, the presence of nuage during this determination process remains obscure. On the other hand, nuage in mice is discernible in differentiating germ cells at later stages, such as in spermatogonia and developing oocytes, and is most prominently observed in meiotic spermatocytes and haploid spermatids (14–17). In this study, we use the traditional term “intermitochodrial cement” for nuage observed among mitochondrial clusters in spermatogonia, spermatocytes, and in oocytes, and “chromatoid body” for larger solitary aggregates that are characteristic in spermatocytes and spermatids. Nuage in differentiating germ cells is not generally involved in asymmetric partitioning, and thus its developmental function may differ from that present during germ-line specification.
Interestingly, a homologue of Drosophila vasa, which encodes a DEAD box RNA helicase (18, 19), exists in mice (20). However, the mouse vasa homologue/DEAD box polypeptide 4 (Mvh/Ddx4) is expressed in differentiating germ cells rather than during germ-cell specification, and MVH protein has been shown to localize to chromatoid bodies in spermatids (21). The targeted disruption of Mvh leads to male-specific sterility because of postnatal defects in early spermatocytes (22), although Mvh is expressed in both male and female germ cells.
Tudor domain containing 1/mouse tudor repeat 1 (Tdrd1/Mtr-1; herein referred to as Tdrd1 following mouse genome informatics nomenclature) is a tudor-related gene in mice (23, 24). TDRD1 protein contains four Tudor domains and a zinc-finger MYND domain. The repeated copies of the Tudor domain were originally identified in Drosophila tudor protein, which is genetically downstream of vasa in regard to its intracellular localization (5, 25–28). We previously showed that Tdrd1 is expressed in differentiating germ cells, and that the protein predominantly localizes to intermitochondrial cement in spermatocytes and chromatoid bodies in spermatids (24). The germ-line expression, domain architecture, and intracellular localization to nuage are common properties shared by mouse TDRD1 and D. tudor.
In this study, we report that a targeted mutation of Tdrd1, a potential hypomorphic allele, leads to complete male sterility because of postnatal spermatogenic defects. Comparative analyses of the Tdrd1 and Mvh mutants revealed that the two genes have conserved properties and relationships analogous to those of tudor and vasa in Drosophila oocytes. Notably, Tdrd1 mutants lack, and Mvh mutants showed great reduction of, intermitochondrial cement in both spermatogenic cells and oocytes, although both mutants are male-specific sterile. This finding provides evidence that nuage assembly is not a critical prerequisite for oocyte development and female fertility in mice and suggests differing requirements for nuage and/or its components between male and female germ cells in mice.
Results
TDRD1 Is a Component of Mouse Nuage/Germinal Granules in both Male and Female Germ Cells.
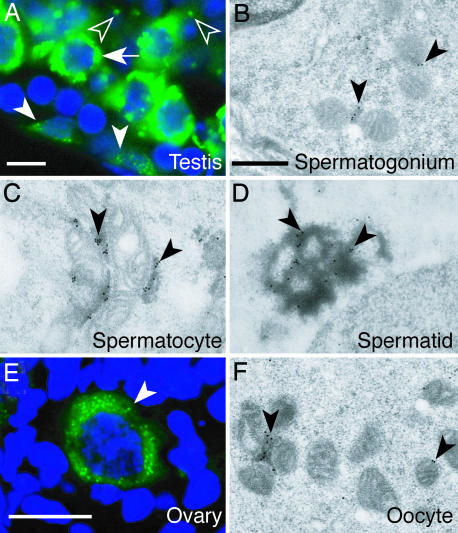
We previously reported that TDRD1 protein is present in fetal prospermatogonia, postnatal spermatocytes, and round spermatids, and that it localizes to intermitochondrial cement in spermatocytes and chromatoid bodies in round spermatids (24). In this study, we produced an anti-TDRD1 antibody, which was more sensitive than the previous one, and refined the immunostaining conditions by minimizing the tissue fixation strength to better retain the antigenicity. By these modifications, we found that TDRD1 was also detectable in postnatal spermatogonia (Fig. 1A, arrowheads) and in oocytes (Fig. 1E), with a fine granular appearance in the cytoplasm. By immunoelectron microscopy, TDRD1 in these cells were clearly localized to nuage, i.e., intermitochondrial cement (Fig. 1 B and F), like that in spermatocytes (Fig. 1C) and chromatoid bodies in spermatids (Fig. 1D). TDRD1 is the only molecule reported so far that localizes to nuage in spermatogonia and especially in oocytes in mice.
Fig. 1.
TDRD1 is a component of nuage in both male and female germ cells. (A) Immunostaining of a section of an adult testis with anti-TDRD1 antibody (green) counterstained with Hoechst dye (blue). Spermatogonia, pachytene-spermatocytes, and round spermatids are indicated by arrowheads, an arrow, and open arrowheads. (B–D) Immunoelectron microscopy of testis sections with anti-TDRD1 antibody. Intermitochondrial cement in a spermatogonium (B) and a spermatocyte (C) and a chromatoid body in a round spermatid (D) are indicated by arrowheads. (E) An adult ovary section immunostained for TDRD1 (green) and counterstained with Hoechst dye (blue). The arrowhead indicates a primary ooctye. (F) Immunoelectron microscopy of a primary oocyte for TDRD1. The arrowheads mark TDRD1 signals on intermitochondrial cement. (Scale bars: A and E, 10 μm; B–D and F, 1 μm.)
Targeted Mutagenesis of Tdrd1.
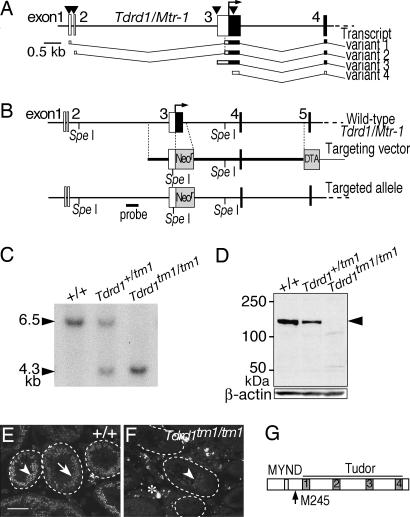
As a basis for designing a targeting vector of Tdrd1, we carried out 5′-RACE and obtained four transcript variants of Tdrd1 as summarized in Fig. 2A (for details, see Supporting Text, which is published as supporting information on the PNAS web site). The Tdrd1 gene consisted of 27 exons that spanned ≈50 kb on chromosome 19. Because the multiple domains of TDRD1 protein were encoded in separated exons, a Tdrd1 gene-targeting vector was designed to disrupt the first coding ATG of transcript variants 1–3. The drug selection cassette also replaced the promoter region of transcript variant 4 (Fig. 2B). Chimeric mice produced from homologous recombinant ES cells (29) were mated with wild-type females, and heterozygous mice were intercrossed to derive homozygous mice. Genotypes of the offspring were confirmed by Southern blotting (Fig. 2C) and by sequencing PCR products around the recombination junction (data not shown).
Fig. 2.
Targeted mutagenesis of Tdrd1. (A) The 5′ regions of four transcript variants of Tdrd1 are depicted under the exon structure. The arrowheads indicate transcription initiation sites, the open and filled boxes represent untranslated and translated regions, and the arrow marks the first coding ATG of transcript variants 1–3. (B) The targeted allele of Tdrd1. The neomycin-resistant cassette replaced the first coding ATG of transcript variants 1–3 and the promoter region of transcript variant 4. (C) Southern blot analysis of tail genomic DNAs digested with SpeI and hybridized with the probe shown in B. (D) Western blotting of testis lysates probed with anti-TDRD1 (Upper) and anti-β-actin (Lower) antibodies. The arrowhead indicates wild-type TDRD1. The two faint bands seen in the Tdrd1tm1/tm1 lane are nonspecific signals, because they are not detected in the Tdrd1tm1/+ lane. (E and F) Immunostaining of sections of wild-type (E) and Tdrd1tm1/tm1 (F) testes with anti-TDRD1 antibody. Spermatocytes and round spermatids are indicated by arrowheads and arrows. The dotted lines demarcate seminiferous tubules, and autofluorescence is labeled with an asterisk. (G) The domain composition of wild-type TDRD1. A potential first ATG in the truncated Tdrd1tm1 mRNAs (see text) corresponds to methionine 245 of the wild-type TDRD1 (arrow). (Scale bar: 100 μm.)
TDRD1 protein was undetectable in homozygous mutant testes by Western blotting probed with anti-TDRD1 C-terminal antibody (Fig. 2D). However, immunostaining of sections of homozygous testes with the antibody exhibited weak signals (Fig. 2 E and F). RT-PCR analyses of Tdrd1 transcripts revealed that homozygous testes produced truncated forms of transcript variants 1 and 2 in which exons 1 and 2 were directly joined to exon 4, skipping the 5′-UTR of exon 3. The truncated transcripts had a potential ORF of 928 aa (Fig. 2G) with the first ATG in exon 8, which was the same ORF in the wild-type transcript variant 4. The 5′-UTRs were >400 bp long, and two ORFs preceded the first ATG in exon 8. The translation efficiency from this potential ORF was quite low if present at all, because no truncated product was detected by Western blotting of heterozygous testes that contained the normal composition of spermatogenic cells (Fig. 2D). However, low levels of leaky scanning or reinitiation of translation (30) could account for the weak signals detected by immunostaining of homozygous testes (Fig. 2 E and F) and in ovaries (data not shown). This potential hypomorphic allele, targeted mutation 1 (Tdrd1tm1), caused clear phenotypes as follows.
Spermatogenic Defects in Tdrd1tm1/tm1 Mice.
Tdrd1tm1/tm1 mice were viable and showed no abnormalities in gross appearance. However, Tdrd1tm1/tm1 males were sterile, whereas females were fertile. Testes of Tdrd1tm1/tm1 adult mice were much smaller (≈30% by weight) than wild-type testes (Fig. 6A, which is published as supporting information on the PNAS web site), and spermatogenesis was severely disorganized (Fig. 6 B–D). In contrast, Tdrd1tm1/tm1 ovaries showed no histological abnormalities (Fig. 6 E and F).
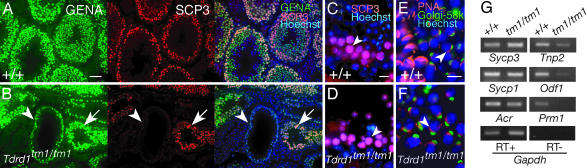
Immunostaining for germ-cell-specific nuclear antigen (31) and for meiotic synaptonemal complex protein 3 (SYCP3/SCP3; ref. 32) showed that spermatogonia and early spermatocytes at the leptotene-zygotene stages were normally present in Tdrd1tm1/tm1 testes (Fig. 3 A and B). However, the differentiation of spermatocytes later than the pachytene-diplotene stages was blocked in a subset of seminiferous tubules (Fig. 3B, arrowhead), whereas in others, meiosis progressed further (Fig. 3B, arrow, and C and D), and haploid round spermatids up to step 7 were differentiated (Fig. 3 E and F). At 8 weeks after birth, ≈20% of Tdrd1tm1/tm1 seminiferous tubules contained only spermatogonia and leptotene-zygotene spermatocytes, 65% contained up to pachytene-diplotene spermatocytes, and the remaining 15% harbored round spermatids. Cellular elongation or nuclear condensation of spermatids did not occur in Tdrd1tm1/tm1 testes, and mature spermatozoa were lacking, consistent with male sterility.
Fig. 3.
Differentiation defects among meiotic spermatocytes and in haploid round spermatids in Tdrd1tm1/tm1 mutants. (A and B) Sections of wild-type (A) and Tdrd1tm1/tm1 (B) testes stained for germ-cell-specific nuclear antigen (GENA; green), SYCP3/SCP3 (red) and with Hoechst 33258 (blue). In a Tdrd1tm1/tm1 seminiferous tubule (B, arrowhead), only GENA-positive but SCP3-negative spermatogonia are observed, whereas in another (B, arrow), pachytene-zygotene spermatocytes are differentiated. (C and D) Higher-magnification views of the sections of wild-type (C) and Tdrd1tm1/tm1 (D) testes stained for SYCP3 (red) and with Hoechst 33258 (blue). Meiotic synaptonemal complexes are normally seen in Tdrd1tm1/tm1 spermatocytes (arrowheads). (E and F) Sections of wild-type (E) and Tdrd1tm1/tm1 (F) spermatids stained with Rhodamine-PNA (red), anti-Golgi-58k antibody (green), and Hoechst 33258 (blue). Round spermatids with acrosomes are present in Tdrd1tm1/tm1 testis (F, and arrowhead), but further elongation of spermatids was not observed. (G) RT-PCR analyses of wild-type and Tdrd1tm1/tm1 testes for spermatocyte and spermatid-marker genes. RT, reverse transcriptase; +/+, wild-type; tm1/tm1, Tdrd1tm1/tm1. (Scale bars: A and B, 50 μm; C–F, 10 μm.)
RT-PCR analyses confirmed the above histological results (Fig. 3G). Sycp3, Sycp1, and Acr, which are expressed in meiotic spermatocytes (32–34), were detected in Tdrd1tm1/tm1 testes at approximately the same levels as in wild type. Odf1/RT7 and Tnp2, whose expression begins in round spermatids (35, 36), were also detected but at reduced levels, and Prm1, which is expressed at the latest stage of round spermatid differentiation (37), was undetectable in Tdrd1tm1/tm1 testes.
The defects among Tdrd1tm1/tm1 spermatocytes could arise through an intrinsic requirement of Tdrd1 function or through an indirect effect after the lack of elongated spermatids and spermatozoa, which could cause the detachment of spermatocytes from the seminiferous epithelium. To distinguish between these possibilities, we examined the first occurrence of spermatocyte defects in prepubertal testes (for details, see Supporting Text and Fig. 7, which is published as supporting information on the PNAS web site). At 14 days postpartum (dpp), when the first population of spermatocytes reaches the pachytene-diplotene stages but before the appearance of round spermatids, apoptosis was already increased among Tdrd1tm1/tm1 spermatocytes compared with wild type. Then, elevated apoptosis was seen among both spermatocytes and round spermatids at 20 dpp and continually in adult Tdrd1tm1/tm1 testes. Thus, spermatogenic defects in Tdrd1tm1/tm1 testes first occur in a subset of spermatocytes, and the remaining spermatocytes further develop up to round spermatids, whose differentiation is then completely blocked, followed by apoptotic cell death.
Mvh Regulates the Intracellular Localization of TDRD1.
MVH/DDX4 is another component of mouse nuage that functions in postnatal spermatogenesis (21, 22). To reveal a possible relationship between Tdrd1 and Mvh, we compared the localization pattern of these two proteins (data are presented in Fig. 8, which is published as supporting information on the PNAS web site, and Supporting Text). In brief, TDRD1 showed a fine granular appearance that corresponds to intermitochondrial cement in spermatogonia and developing oocytes, whereas MVH was diffusely observed in the cytoplasm in these cells. During spermatocyte differentiation, MVH showed granular distribution that merged with TDRD1, but such localization of MVH was gradual and occurred later than TDRD1. In round spermatids, the two proteins precisely colocalized to larger solitary aggregates that correspond to chromatoid bodies (21, 24). In all, TDRD1 more consistently localized to nuage than MVH during germ cell differentiation, whereas MVH translocated from the cytoplasm to nuage in spermatocytes.
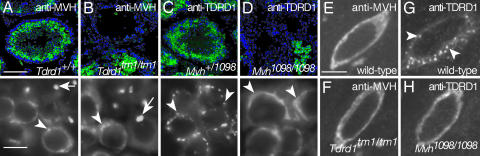
The expression and localization of TDRD1 and MVH were next examined in each reciprocal mutant. In Tdrd1tm1/tm1 testes, MVH protein was detected, and cytoplasmic granules in spermatocytes and spermatids were similarly seen as in wild type (Fig. 4 A and B). On the other hand, in Mvh1098/1098 testes whose spermatogenesis ceases during the meiotic prophase of spermatocytes (22), TDRD1 protein in spermatogonia and surviving spermatocytes did not exhibit characteristic granular localization. Instead, TDRD1 was diffusely distributed in the cytoplasm or more notably showed a crescent-like accumulation around the nucleus in spermatocytes (Fig. 4 C and D).
Fig. 4.
TDRD1 localization is regulated downstream of Mvh. (A–D) Sections of wild-type (A) and Tdrd1tm1/tm1 (B) testes immunostained for MVH, and of Mvh+/1098 (C) and Mvh1098/1098 (D) stained for TDRD1. The arrowheads and arrows indicate signals in spermatocytes and spermatids. Note that TDRD1 exhibits crescent-like accumulation in Mvh1098/1098 spermatocytes (D) instead of the granular distribution seen in the control (C). (E–H) Confocal images of sections of wild-type (E) and Tdrd1tm1/tm1 (F) primary oocytes immunostained for MVH, and of wild-type (G) and Mvh1098/1098 (H) stained for TDRD1. Granular localization of TDRD1, seen in wild type (G, arrowheads), is disrupted in Mvh1098/1098 oocytes. Nuclei were stained with Hoechst 33258 (blue). (Scale bars: A–D Upper, 100 μm; A–D Lower, 10 μm; E–H, 5 μm.)
In Tdrd1tm1/tm1 oocytes, MVH was diffusely present in the cytoplasm as in wild type (Fig. 4 E and F). In contrast, the localization of TDRD1 was altered in Mvh1098/1098 oocytes, and the characteristic granular distribution, seen in wild type (Fig. 4 G and H), was disrupted. Therefore, the intracellular localization of TDRD1 in both spermatogenic cells and oocytesis is downstream of Mvh, whereas Tdrd1tm1/tm1 mutation does not affect MVH distribution. Considering Mvh1098/1098 mutants are male-specific sterile, this result revealed that TDRD1 localization to nuage is not directly associated with female fertility.
Tdrd1 and Mvh Are Essential for the Assembly of Intermitochondrial Cement in both Male and Female Germ Cells.
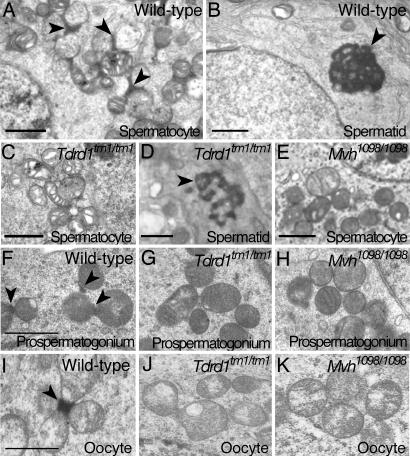
We then examined whether Tdrd1tm1/tm1 and Mvh1098/1098 mutants showed any specific changes at the ultrastructural level. In Tdrd1tm1/tm1 testes, spermatocytes and round spermatids frequently showed degeneration, such as intracellular disorganization and vacuolation. Meanwhile, in surviving spermatocytes and round spermatids, subcellular structures such as synaptonemal complexes, acrosomes, and flagella were seen as in wild type (data not shown). However, intermitochondrial cement in Tdrd1tm1/tm1 spermatocytes was lacking despite the clusters of mitochondria having formed (Fig. 5 A and C). Intermitochondrial cement is observed as minute amorphous material that stains densely among clustered mitochondria. Such staining is sometimes difficult to distinguish from artifacts, other cellular components like ruptured mitochondria, or chromatoid bodies adjoining mitochondria. Very occasionally, Tdrd1tm1/tm1 spermatocytes showed structures indistinguishable from intermitochondrial cement, but quantitative analysis established that the frequency of such stainings in Tdrd1tm1/tm1 spermatocytes was very low, if not completely absent, compared with wild type (Table 1, which is published as supporting information on the PNAS web site). In contrast, chromatoid bodies were clearly observed at similar frequencies in both wild type and Tdrd1tm1/tm1 round spermatids, although those in Tdrd1tm1/tm1 spermatids were smaller and less organized than in wild type (Fig. 5 B and D; see also Table 2, which is published as supporting information on the PNAS web site). This observation suggests that chromatoid bodies, whose origin has been controversial, likely have a precursor independent of or additional to intermitochondrial cement. In the wild-type testis, intermitochondrial cement is also seen in a subset of postnatal spermatogonia and most of fetal prospermatogonia. However, in Tdrd1tm1/tm1 mutants, intermitochndrial cement was again virtually absent in these spermatogonial cells (Fig. 5 F and G for prospermatogonia; see also Tables 3 and 4, which are published as supporting information on the PNAS web site).
Fig. 5.
Tdrd1 and Mvh are essential for the assembly of intermitochondrial cement in both male and female germ cells. Electron microscopy of adult testes (A–E), fetal testes (F–H), and adult ovaries (I–K) from mice with the indicated genotypes. (A and B) A wild-type spermatocyte (A) and round spermatid (B). The arrowheads mark intermitochondrial cement (A) and a chromatoid body (B). (C) A Tdrd1tm1/tm1 spermatocyte in which intermitochondrial cement is not discernible or quite diminutive, although a cluster of mitochondria is observable. (D) A Tdrd1tm1/tm1 round spermatid. A chromatoid body is clearly seen (arrowhead), whereas it appears sparse compared with wild type (B). (E) A morphologically unimpaired Mvh1098/1098 spermatocyte. Intermitochondrial cement is not detected. (F–H) Mitochondrial clusters in wild-type (F), Tdrd1tm1/tm1 (G), and Mvh1098/1098 (H) fetal prospermatogonia at 17.5 days postcoitum. Intermitochondrial cement, seen in wild type (F, arrowheads), is not discernible in the two mutant cells (G and H). (I–K) Primary oocytes of wild-type (I), Tdrd1tm1/tm1 (J), and Mvh1098/1098 (K) mutants. Intermitochondrial cement, present in wild type (I, arrowhead), is again not observed in the two mutant oocytes (J and K). Note that a small subset of Mvh1098/1098 prospermatogonium and oocyte sections show intermitochondrial cement as summarized in Tables 3 and 5. (Scale bars: 1 μm.)
In Mvh1098/1098 testes, spermatogenesis ceases at the prophase of meiosis, and most of the spermatocytes were undergoing degeneration. In surviving spermatocytes, intermitochondrial cement was not observed among clustered mitochondria (Fig. 5E and Table 1). We did not find round spermatids in electron microscopy sections examined in this study. On the other hand, intermitochondrial cement was seen in a small subset of Mvh1098/1098 fetal prospermatogonia, although the number of the structure was greatly reduced compared with wild type (Fig. 5 F and H and Table 3).
Both Tdrd1tm1/tm1 and Mvh1098/1098 mutants are male-specific sterile, and females are fertile. Unexpectedly, however, intermitochondrial cement was not observed in Tdrd1tm1/tm1 oocytes and was markedly reduced in Mvh1098/1098 oocytes (Fig. 5 I–K; see also Table 5, which is published as supporting information on the PNAS web site), whereas no other structural changes were identified in both mutants.
Taken together, Tdrd1 and also Mvh are essential for the assembly of intermitochondrial cement in male and female germ cells. This observation provides intriguing evidence that intermitochondrial cement is not a direct prerequisite for oogenesis and female fertility in mice, and it contrasts to polar granules/nuage, whose formation is closely correlated with proper oocyte development and patterning in Drosophila. Intermitochondrial cement is also likely dispensable for fetal and postnatal spermatogonia, contrastingly to spermatocytes, whose differentiation was severely impaired in Tdrd1tm1/tm1 and Mvh1098/1098 mutants.
Discussion
The properties and functions of mammalian nuage remain elusive. The chromatoid body in spermatids has been relatively well described because of its prominence by light and electron microscopy. It is implicated in RNA storage and regulation based on the abundance of RNA and RNA-binding proteins such as transition protein 2 mRNA (38) and p48/52 (39). Recently, Kotaja et al. (40) reported that the chromatoid body in spermatids contains Dicer and microRNAs, proposing that the structure is involved in a microRNA pathway. Intracellular and intercellular movement of chromatoid bodies also suggests that the structure functions in RNA trafficking and gene dosage compensation (17). Intermitochondrial cement, on the other hand, is much less studied, although it is more consistently present during germ-cell differentiation. We showed that TDRD1 specifically localizes to both intermitochondrial cement and chromatoid bodies. It is the only identified protein that constitutes nuage in both male and female germ cells and thus would provide a valuable clue to mark and study properties of mammalian nuage.
Tdrd1tm1/tm1 mutants showed male sterility with postnatal spermatogenic defects. The observed phenotypes may reflect a hypomorph or antimorph of this gene. However, heterogenous phenotypes among spermatocytes and spermatids, similar to those observed in Tdrd1tm1/tm1 mutants, were also reported for the null mutant of Grth/Ddx25, which encodes an RNA helicase that localizes to chromatoid bodies (41). We also found that Mvh1098/1098 testes harbor a small population of round spermatids, although most spermatocytes are degenerated during the meiotic prophase (data not shown). Thus, heterogenous defects during postnatal spermatogenesis may represent a common loss-of-function phenotype of a group of nuage components, including TDRD1. Although further entire deletion of the Tdrd1 gene would help examine whether additional phenotypes exist, the Tdrd1tm1 allele revealed that the tudor-related gene plays essential roles in postnatal spermatogenesis in mice. This Tdrd1tm1 allele also provides a precious mutant that lacks intermitochondrial cement in both male and female germ cells.
The molecular function of the repeated copies of the Tudor domain remains elusive. The single Tudor domain in the Survival Motor Neuron protein binds to Sm proteins of small nuclear ribonucleoproteins and functions in the assembly of the spliceosomal complex (42, 43). Consistently, TDRD1 and Sm proteins form complexes and colocalize in chromatoid bodies (24). Recently, 53BP1 containing two neighboring Tudor folds that comprise a single globular domain was shown to bind methylated histone H3 (44, 45). Multiple Tudor domains may function to assemble proteins that associate with each Tudor domain into macromolecular complexes. In addition, D. tudor is required for the localization of mitochondrial ribosomal RNAs to polar granules (46), implying that it also acts in RNA assembly. In agreement with the presumed scaffold function of multiple Tudor domains, hypomorphic and null mutants of D. tudor greatly reduce the number and size of polar granules in oocytes (25, 28), and Tdrd1tm1/tm1 mutants lacked intermitochondrial cement in both male and female germ cells. In contrast, chromatoid bodies in spermatids were clearly observed in Tdrd1tm1/tm1 mutants, although they were smaller and less organized compared with wild type. The origin of chromatoid bodies has been obscure and controversial. The structure has been suggested to arise from intermitochondrial cement or nuclei based on morphological similarities and topographical relations (14). We propose that the chromatoid body incorporates components from intermitochondrial cement, as exemplified by TDRD1 that localizes to both structures, but it should also have another source of precursors such as from the nucleus. In mice, there are other Tudor-related proteins, TDRD6 and TDRD7/TRAP, which localize to chromatoid bodies (M.H., unpublished work). It is conceivable that these proteins compensate TDRD1 for its spermatogenic function and the assembly of chromatoid bodies in Tdrd1tm1/tm1 mutants. Recently, RNF17 was also shown to contain multiple Tudor domains in addition to a RING finger domain. Interestingly, Rnf17 gene-targeted mice exhibit male sterility, but RNF17 localizes to cytoplasmic structures that are distinct from intermitochondrial cement and chromatoid bodies (47). The RNF17 granules may represent a novel type of nuage that has an essential function in spermatogenesis in mice.
We showed that Mvh is upstream of TDRD1 with respect to intracellular localization. The perinuclear accumulation of TDRD1 in Mvh1098/1098 spermatocytes suggests a possibility that TDRD1 takes part in trafficking molecules from the nucleus to the intermitochondrial regions, and that this TDRD1 activity depends on Mvh. The results for Tdrd1 and Mvh are comparable to those reported for D. tudor and vasa in several aspects: (i) both genes in Drosophila and mice are expressed and function in the germ line (25, 48, 49), (ii) the protein products of these genes localize to nuage in common (5, 50, 51), (iii) polar granules are not formed or greatly reduced in vasa and tudor mutants similarly to intermitochondrial cement in Mvh and Tdrd1 mutants (25, 28, 48, 49), and (iv) the intracellular distribution of Tudor/TDRD1 is regulated downstream of vasa/Mvh (5). One clear difference, on the other hand, is that Drosophila vasa-tudor maternally function in oocytes, whereas mouse Mvh-Tdrd1 zygotically play roles in spermatogenic cells. Thus, a group of genes and their molecular pathway that are essential for germ-cell specification and early embryonic patterning in Drosophila are likely conserved and analogously function in the germ line in mice but during the postnatal differentiation of male germ cells. Nuage is a site of assembly for these evolutionarily conserved molecules and probably retains a function(s) that is essential and independent of the differentiation stage, sex, and species of the germ line.
One particular and intriguing question raised in this study is whether mouse oocytes lacking intermitochondrial cement, as those in Tdrd1tm1/tm1 and Mvh1098/1098 mutants, are not substantially affected at the molecular level, or whether certain aberrations actually occur, but overall catastrophe does not follow. The latter possibility is supported by the notion that oocytes are less susceptible to meiotic checkpoint controls than spermatogenic cells in mice (52). This may also explain the sexual dimorphisms in Tdrd1tm1/tm1 and Mvh1098/1098 phenotypes. Similar questions would also apply to fetal and postnatal spermatogonia. The identification of molecular changes underlying these two mutant oocytes and spermatogonial cells would help to further reveal cellular and developmental features associated with nuage, an enigmatic ribonucleoprotein structure specifically conserved in the germ line.
Materials and Methods
Targeted Mutagenesis.
A 1.5-kb Tdrd1 genomic fragment upstream of the first coding ATG and a 6-kb fragment downstream of exon 3 were ligated to the neomycin resistance gene cassette of pKO SelectNeo (Lexicon Genetics, The Woodlands, TX) and to the diphtheria toxin A fragment gene cassette from pND3. The Tdrd1 targeting vector was used to derive homologous recombinant ES cells, and chimeric mice were produced from two recombinant ES clones. Male chimeras of one line were mated with wild-type females to obtain heterozygous mice.
For further details, see Supporting Text.
Supplementary Material
Acknowledgments
We thank A. Nagy (University of Toronto, Toronto, ON, Canada) for R1 ES cells; Y. Nishimune (Osaka University, Osaka, Japan) for antigerm-cell-specific nuclear antigen (GENA) antibody TRA98/104; T. Tada, H. Suemori, M. Shoji, T. Tanaka, E. Moribe, and M. Tanaka for experimental advice and assistance; and A. B. Cowan for comments on the manuscript. This study was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
References
- 1.Lehmann R, Ephrussi A. Ciba Found Symp. 1994;182:282–96. doi: 10.1002/9780470514573.ch16. discussion 296–300. [DOI] [PubMed]
- 2.Saffman EE, Lasko P. Cell Mol Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ephrussi A, Lehmann R. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 4.Smith JL, Wilson JE, Macdonald PM. Cell. 1992;70:849–859. doi: 10.1016/0092-8674(92)90318-7. [DOI] [PubMed] [Google Scholar]
- 5.Bardsley A, McDonald K, Boswell RE. Development (Cambridge, UK) 1993;119:207–219. doi: 10.1242/dev.119.1.207. [DOI] [PubMed] [Google Scholar]
- 6.Mahowald AP. J Exp Zool. 1962;151:201–216. [Google Scholar]
- 7.Strome S, Wood WB. Proc Natl Acad Sci USA. 1982;79:1558–1562. doi: 10.1073/pnas.79.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czolowska R. J Embryol Exp Morphol. 1969;22:229–251. [PubMed] [Google Scholar]
- 9.Eddy EM. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 10.Lawson KA, Hage WJ. Ciba Found Symp. 1994;182:68–84. doi: 10.1002/9780470514573.ch5. discussion 84–91. [DOI] [PubMed] [Google Scholar]
- 11.Tam PP, Zhou SX. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- 12.Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaren A. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 14.Fawcett DW, Eddy EM, Phillips DM. Biol Reprod. 1970;2:129–153. doi: 10.1095/biolreprod2.1.129. [DOI] [PubMed] [Google Scholar]
- 15.Eddy EM. Anat Rec. 1974;178:731–757. doi: 10.1002/ar.1091780406. [DOI] [PubMed] [Google Scholar]
- 16.Russell L, Frank B. Anat Rec. 1978;190:79–97. doi: 10.1002/ar.1091900108. [DOI] [PubMed] [Google Scholar]
- 17.Parvinen M. Int J Androl. 2005;28:189–201. doi: 10.1111/j.1365-2605.2005.00542.x. [DOI] [PubMed] [Google Scholar]
- 18.Hay B, Jan LY, Jan YN. Cell. 1988;55:577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- 19.Lasko PF, Ashburner M. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Proc Natl Acad Sci USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Mech Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang PJ, McCarrey JR, Yang F, Page DC. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 24.Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamune K, Nakatsuji N. Mech Dev. 2003;120:979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 25.Boswell RE, Mahowald AP. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 26.Golumbeski GS, Bardsley A, Tax F, Boswell RE. Genes Dev. 1991;5:2060–2070. doi: 10.1101/gad.5.11.2060. [DOI] [PubMed] [Google Scholar]
- 27.Ponting CP. Trends Biochem Sci. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- 28.Thomson T, Lasko P. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- 29.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka H, Pereira LA, Nozaki M, Tsuchida J, Sawada K, Mori H, Nishimune Y. Int J Androl. 1997;20:361–366. doi: 10.1046/j.1365-2605.1998.00080.x. [DOI] [PubMed] [Google Scholar]
- 32.Lammers JH, Offenberg HH, van Aalderen M, Vink AC, Dietrich AJ, Heyting C. Mol Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C. EMBO J. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kashiwabara S, Arai Y, Kodaira K, Baba T. Biochem Biophys Res Commun. 1990;173:240–245. doi: 10.1016/s0006-291x(05)81047-2. [DOI] [PubMed] [Google Scholar]
- 35.Shih DM, Kleene KC. Mol Reprod Dev. 1992;33:222–227. doi: 10.1002/mrd.1080330216. [DOI] [PubMed] [Google Scholar]
- 36.van der Hoorn FA, Tarnasky HA, Nordeen SK. Dev Biol. 1990;142:147–154. doi: 10.1016/0012-1606(90)90158-f. [DOI] [PubMed] [Google Scholar]
- 37.Mali P, Kaipia A, Kangasniemi M, Toppari J, Sandberg M, Hecht NB, Parvinen M. Reprod Fertil Dev. 1989;1:369–382. doi: 10.1071/rd9890369. [DOI] [PubMed] [Google Scholar]
- 38.Saunders PT, Millar MR, Maguire SM, Sharpe RM. Mol Reprod Dev. 1992;33:385–391. doi: 10.1002/mrd.1080330404. [DOI] [PubMed] [Google Scholar]
- 39.Oko R, Korley R, Murray MT, Hecht NB, Hermo L. Mol Reprod Dev. 1996;44:1–13. doi: 10.1002/(SICI)1098-2795(199605)44:1<1::AID-MRD1>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 40.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. Proc Natl Acad Sci USA. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai-Morris CH, Sheng Y, Lee E, Lei KJ, Dufau ML. Proc Natl Acad Sci USA. 2004;101:6373–6378. doi: 10.1073/pnas.0401855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buhler D, Raker V, Luhrmann R, Fischer U. Hum Mol Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 43.Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 44.Charier G, Couprie J, Alpha-Bazin B, Meyer V, Quemeneur E, Guerois R, Callebaut I, Gilquin B, Zinn-Justin S. Structure (London) 2004;12:1551–1562. doi: 10.1016/j.str.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 46.Amikura R, Hanyu K, Kashikawa M, Kobayashi S. Mech Dev. 2001;107:97–104. doi: 10.1016/s0925-4773(01)00455-5. [DOI] [PubMed] [Google Scholar]
- 47.Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ. Development (Cambridge, UK) 2005;132:4029–4039. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schupbach T, Wieschaus E. Roux's Arch Dev Biol. 1986;195:302–317. doi: 10.1007/BF00376063. [DOI] [PubMed] [Google Scholar]
- 49.Nusslein-Volhard C, Frohnhofer HG, Lehmann R. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- 50.Hay B, Ackerman L, Barbel S, Jan LY, Jan YN. Development (Cambridge, UK) 1988;103:625–640. doi: 10.1242/dev.103.4.625. [DOI] [PubMed] [Google Scholar]
- 51.Liang L, Diehl-Jones W, Lasko P. Development (Cambridge, UK) 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 52.Morelli MA, Cohen PE. Reproduction. 2005;130:761–781. doi: 10.1530/rep.1.00865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.