Abstract
Trip230 is a novel coactivator of the thyroid hormone receptor that is negatively regulated by the retinoblastoma tumor-suppressor protein. In an examination of its subcellular distribution, Trip230 localized predominantly to the vicinity of the Golgi instead of the nucleus, as other nuclear hormone receptor coactivators. Using a series of deletion mutants, a critical region identified for Golgi area targeting coincided with a previously defined thyroid hormone receptor-binding domain of Trip230. During cell cycle progression, the expression level of Trip230 is constant and a significant portion is imported into the nucleus at S phase. Within an hour of treating cells with T3, Trip230 immunofluorescence transiently colocalized with TR in prominent subnuclear structures. T3-dependent nuclear import of Trip230 does not require new protein synthesis. Coincident with T3 treatment and nuclear import, newly phosphorylated residue(s) appeared in Trip230, suggesting that phosphorylation may be involved in its nuclear import. These findings provided a novel mechanism for the regulation of nuclear hormone transcription factors by hormone-responsive phosphorylation and nuclear import of cytoplasmically located coactivators.
Steroid, retinoic acid, and thyroid hormone receptors function as ligand-activated transcription factors that control multiple biological functions, including cell proliferation, differentiation, morphogenesis, programmed cell death, and metabolic homeostasis. Understanding the molecular and cellular mechanisms underlying the ability of these receptors to regulate the transcription of specific target genes is a major challenge. In recent years, a number of interacting proteins have been identified that are critical for the transcriptional responses transduced by nuclear hormone receptors (reviewed in refs. 1 and 2). These coactivators or corepressors form multiprotein complexes to regulate the expression of hormone-responsive target genes. Different cell types may have distinct compositions of associated factors in the nuclear hormone receptor complexes, individual members of which may integrate distinct physiological signals (2).
How a small lipophilic molecule, such as 3,3′,5-triiodo-l-thyronine (T3), coordinates and activates transcription of target genes remains unclear. Thyroid hormone receptor (TR) bound to positive thyroid hormone-responsive elements in the absence of ligand decreases transcription. This silencing is due to the interactions of the nuclear corepressors NcoR and SMRT (3, 4). Ligand-dependent activation of the TR results in removal of the corepressors that are replaced with coactivator complex [pCIP/SRC1/NcoA-1/pCAF and CBP/p300 (reviewed in refs. 1 and 2)]. One such coactivator, Trip230, binds TR in a T3-dependent manner and potentiates transcription of T3-activated receptor (5). Trip230 originally was identified as a protein that binds the retinoblastoma protein, which, interestingly, can down-regulate Trip230/TR-mediated transcription (5). These results concerning modulation of hormonal signaling provided additional evidence of the pleiotrophic, but overlapping, functions of retinoblastoma protein (Rb) in cellular proliferation, differentiation, and development (6).
How coactivators are recruited coordinately to the activated TR is unclear. One possibility is that hormones also are involved in this process by modifying the receptor, their cofactors, or both. Nuclear localization is a prerequisite for nuclear hormone receptors and their cofactors to function as transcriptional regulators. Thus, translocation of these factors from cytoplasm to nucleus also may be regulated. Based on hormone-binding assays, it was proposed that TR is nuclear regardless of ligand status (7). Although its cellular localization is not entirely clear, cellular partitioning of TR may be, in part, dependent on ligand binding. How compartmentalization of nuclear receptors is regulated in conjunction with their coactivators is not known.
To study further the coactivator function of Trip230, we investigated its cellular partitioning in relation to TR in the presence or absence of thyroid hormone. Interestingly, we found that Trip230 is localized mainly at the Golgi complex region in the cells cultured in T3-free medium. Translocation into the nucleus and colocalization with TR are T3-dependent and correlate with phosphorylation of additional residues of Trip230.
MATERIALS AND METHODS
Cell Culture, Synchronization, and DNA Transfections.
CV1 monkey kidney cells were grown on plastic surfaces at 37°C in a humidified 10% CO2-containing atmosphere in DMEM (GIBCO) supplemented with 10% heat-inactivated FCS (Flow Laboratories). Each 10-cm dish of cells grown to 60% confluence then was cultured in medium containing charcoal-striped serum for 36 hr. In the case of hormone treatments, T3 was added to the medium to a final concentration of 10−6 M. Cyclohexamide at 10 μg/ml, in addition to T3, was included in the treatment of some cells shown in Fig. 4C. At the various times indicated, cells were fixed for immunostaining (see below) or lysed for Western blot analysis. Human bladder carcinoma cells, T24 (American Type Culture Collection, Manassas, VA), grown in DMEM/10% FCS, were synchronized at G1 by density arrest as described previously (8). The transfection experiments in Fig. 3 were done with 1 × 106 cells by using conventional calcium phosphate/DNA (10 μg) coprecipitation. Twelve hours posttransfection, the precipitates were removed, and the cells were refed fresh medium and observed under a fluorescence microscope (Nikon) 36 hr posttransfection.
Figure 4.
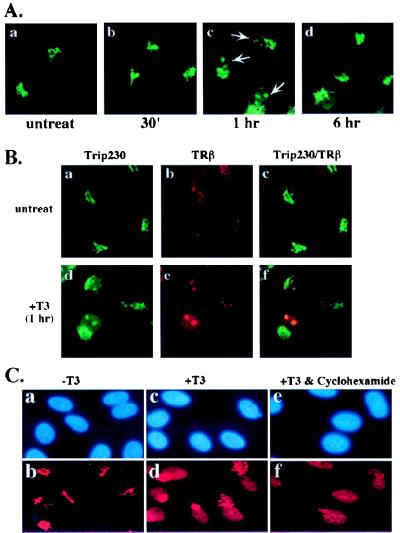
T3-responsive nuclear import of Trip230 and colocalization with TR. (A) CV1 cells treated with 10−6 M T3 for the indicated lengths of time were fixed and stained by indirect immunofluorescence by using antibodies specific for Trip230 and were analyzed by fluorescent microscopy. Treatment of cells with T3 for 1 hr demonstrates nuclear import of a portion of Trip230 immunoreactivity that appears as speckles or larger aggregates (arrows in c); compare a (untreated) and b (treated for 30 min) with c (treated for one hour). Anti-Trip230 nuclear immunofluorescence is not detectable 6 hr posttreatment (d). (B) Colocalization of Trip230 and TR in the nucleus after T3 treatment. One hour after CV1 cells were treated with T3, cells were fixed and analyzed for immunolocalization of Trip230 and TR by using antibodies specific for each protein in the absence (a–c) or presence (d–f) of T3. a and d show the pattern of immunofluorescence in the region of the Golgi (a and d) and nucleus (d) by using anti-Trip230 polyclonal serum and FITC-conjugated anti-mouse IgG secondary antibodies. b and e illustrate nuclear staining by using rabbit anti-TRβ antiserum and Texas red-tagged secondary antibodies. c and f are digital overlays of the images in a and b and d and e, respectively. The images were obtained by laser-scanning confocal microscopy. (C) Protein synthesis is not required for Trip230 nuclear import. CV1 cells (a, c, and e, 4′,6-diamidino-2-phenylindole-stained) either untreated (a and b), treated with T3 alone (c and d), or treated simultaneously with T3 and cyclohexamide (e and f) were immunostained with anti-Trip230 antibodies and imaged by fluorescence microscopy. Note the presence of anti-Trip230 nuclear speckling (f).
Figure 3.
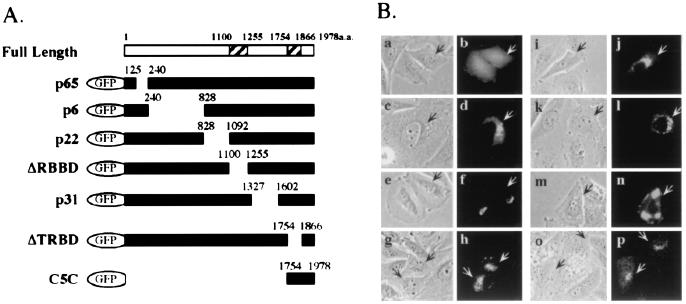
Mapping of Trip230 Golgi-localization domain. (A) Diagram of serial deletion constructs of GFP-Trip230 fusion proteins with the residues that were deleted indicated above the solid bars. ΔRBBD indicates deletion of the RB-binding domain (amino acids 1100–1255); ΔTRBD indicates deletion at TR-binding domain (amino acids 1754–1866). C5C shows the GFP fusion containing Trip230 residues 1754–1978 fused to GFP. (B) Localization of GFP and GFP-Trip230 fusion proteins in transiently transfected Saos-2 cells. Phase-contrast images (a, c, e, g, i, k, m, and o) show cell morphology for each transfection. GFP autofluorescence (green, b, d, f, h, j, l, n, and p) shows the subcellular location of the various GFP-Trip230 fusion proteins. a and b, fluorescence of GFP alone; c and d, p65; e and f; p6; g and h; p22; i and j; ΔRBBD; k and l; p31; m and n; ΔTRBD; and o and p, C5C. Arrows indicate transfected cells.
Immunoprecipitation and Western Blot Analysis.
Cells resuspended in Lysis 250 buffer were subjected to three freeze/thaw cycles (liquid nitrogen/37°C) and then centrifuged at 14,000 rpm for 2 min at room temperature. The supernatants were used for immunoprecipitation as described (9). Briefly, mouse polyclonal anti-C5 (Trip230) antisera (1 μl) were added to each supernatant. After a 1-hr incubation, protein-A Sepharose beads were added and incubation continued for another hour. Beads then were collected, washed five times with lysis buffer containing 250 mM NaCl, and then boiled in SDS-loading buffer for immunoblot analysis as described (8).
Immunostaining.
Cells grown on coverslips in tissue culture dishes were washed in PBS and fixed for 30 min in 4% formaldehyde in PBS with 0.5% Triton X-100. After treating with 0.05% Saponin in water for 30 min and washing extensively with PBS, the cells were blocked in PBS containing 10% normal goat serum. An overnight incubation with antibody diluted in 10% goat serum at 4°C was followed by three washes and then by another 1-hr incubation with fluorochrome-conjugated secondary antibody. Colocalization of Trip230 and mannosidase II or TR was performed by using a polyclonal (mouse) anti-C5 antibody mixed with polyclonal (rabbit) anti-ManII antisera (10) or polyclonal (rabbit) anti-TR antibody (TR-β117; Affinity BioReagents, Golden, CO). The respective antigens were visualized with goat anti-rabbit IgG conjugated to Texas red and goat anti-mouse IgG conjugated to FITC. After washing extensively in PBS with 0.5% Nonidet P-40, cells also were stained with the DNA-specific dye 4′,6-diamidino-2-phenylindole and mounted in Permafluor (Lipshaw-Immunonon, Pittsburgh). Images from cells obtained with a standard fluorescence microscope (Axiophot Photomicroscope, Zeiss) were recorded on Ektachrome P1600 film. Confocal microscopy was done by using a Zeiss LSM310 (Ar and NeNe lasers).
Two-Dimensional Gel Electrophoresis and Phosphopeptide Mapping.
CV1 cells were labeled with 32P in the presence or absence of T3 for 2 hr. After immunoprecipitation using anti-Trip230 antiserum, two-dimensional gel electrophoresis was performed. Tryptic phosphopeptide mapping (11) of the sites for Trip230 T3-responsive phosphorylation was performed on the anti-Trip230 immune complexes after their separation by SDS/PAGE and transfer to Immobilon-P (Millipore) membrane. After overnight exposure to x-ray film, the band corresponding to Trip230 was excised from the membrane. After rewetting with methanol for 1 min followed by soaking in H2O for 5 min, the excised membrane then was soaked with 0.5% polyvinylpyrrolidone-360 and 100 mM acetic acid at 37°C for 30 min. After five washes with H2O (1 ml each), the membrane then was washed twice with freshly prepared 0.05 M NH4HCO3. The treated membrane then was digested with 10 μg l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin in 150–200 μl freshly made 0.05 M NH4HCO3 for 2 hr at 37°C. An additional 10 μg of trypsin then was added to reaction followed by another 2-hr incubation at 37°C. At the end of the digestion, 300 μl of H2O was added to the sample and centrifuged at 14,000 rpm for 5 min. After drying in a Speed-Vac, the pellet was oxidized in 50 μl performic acid for 1 hr. After the addition of 400 μl H2O, the samples were vortexed, frozen on dry ice, and dried repeatedly in a Speed-Vac until a cotton ball-like dry residue was obtained. The dried samples then were dissolved in 10 μl electrophoresis buffer (pH 1.9) and spotted on a cellulose TLC plate (Sigma). After electrophoresis in the first dimension at 1.0 kV for 25 min, the TLC plates were dried by an electric fan and then chromatographed with pyridine/butanol/acetic acid/water (3:3:5:1:4) in the second dimension. The resulting TLC plates then were autoradiographed by exposure to x-ray film.
RESULTS
Expression of Trip230 During the Cell Cycle.
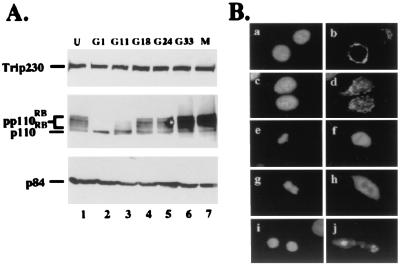
Trip230 interacts with TR and has been shown to be a coactivator for thyroid hormone (T3)-mediated transcription (5). Because T3 is important for growth regulation, especially through the insulin-like growth factor I pathway (12, 13), we were interested in determining whether Trip230 expression is cell cycle-regulated. Bladder carcinoma T24 cells were arrested at G1 by contact inhibition at high density and then released to enter the cell cycle by plating at low density (8, 14). M phase cells were enriched by an additional treatment with nocadozol. Cells were harvested at different stages of cell cycle, and protein expression was analyzed by Western blotting by using specific antibodies. As shown in Fig. 1A, the amount of Trip230 is not altered during the cell cycle. Rb, whose expression and phosphorylation is cell cycle-regulated, served as a marker for the cell cycle stages as described previously (14). p84, a nuclear matrix protein whose expression is not cell cycle-regulated, served as loading control. This result suggests that the regulation of Trip230 may be at the posttranslational level.
Figure 1.
Expression pattern and dynamics of subcellular localization of Trip230 in the cell cycle. (A) T24 cells were either unsynchronized (lane 1) or synchronized at G1 (lane 2) and released for various periods of time (G11 = 11 hr after release, etc.). Hypophosphorylated Rb protein (p110RB) and various phosphorylated forms (pp110RB) marked the stages of the cell cycle: G1 (lanes 2 and 3), G1/S boundary (lane 4), S (lane 5), G2/M (lane 6), and M (lane 7). Total cellular protein from different time points was separated by SDS/PAGE and immunoblotted with anti-Trip230 antiserum (Top), mAb 11D7, anti-Rb antibody (Middle), or mAb 5E10, anti-N5 antibody (Bottom). p84 served as an internal control for protein loading. (B) Immunocytochemical localization of Trip230 during different phases of the cell cycle (b, d, f, h, and j) in T24 cells (4′,6-diamidino-2-phenylindole-stained, a, c, e, g, and i). a and b are T24 cells fixed in early G1 phase (G11) showing perinuclear cytoplasmic staining (original magnification, ×1000); c and d illustrate representative cells in S phase (G24) showing Trip230 immunofluorescence in the perinuclear cytoplasm and nucleus; e and f show a representative cell in metaphase; g and h show a cell in telophase with more concentrated Trip230 staining surrounding the DNA; and i and j show a cell at cytokinesis in which the Trip230 immunofluorescence is concentrated to the region of the re-formed Golgi apparatus.
Cell Cycle Dynamics of Trip230 Cellular Partitioning.
Because Trip230 is proposed to be a coactivator for TR (5), a transcription factor that functions in the nucleus, we were interested in determining the cellular partitioning of Trip230 in relation to the cell cycle. Accordingly, we performed immunostaining with cells synchronized at different stages of the cell cycle by using specific anti-Trip230 antisera. Surprisingly, the majority of Trip230 immunofluorescence localized to the region of the Golgi apparatus during interphase (Fig. 1B, a–d). In S phase, there is also nuclear localization of speckled immunofluorescence in addition to Golgi staining (Fig. 1B, c and d), indicating partial transport of Trip230 into the nucleus during this stage. In metaphase, the diffuse immunostaining is localized to regions peripheral to the chromosomal DNA (Fig. 1B, e and f). During telophase, Trip230 immunofluorescence becomes more concentrated in the perichromosomal regions (Fig. 1B, g and h). After cytokinesis and reformation of the nuclear envelop, the partitioning of Trip230 is fully restricted to the region of the newly formed Golgi apparatuses (Fig. 1B, i and j). In Fig. 1B (i and j), although the immunofluorescence appears to be associated with the daughter nuclei, an examination of Z sections of these cells by confocal microscopy (data not shown) shows that the stained areas are adjacent to, but separate from, the daughter nuclei. This morphology is similar to that described previously for Golgi reformation after cell division (15).
Trip230 Localizes in the Region of the Golgi Complex.
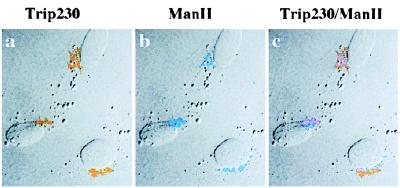
Based on its immunostaining pattern, Trip230 appears to colocalize with the Golgi complex. To address this possibility, we performed immunocytochemical colocalization by using antibodies specific for the Golgi stack membrane-associated protein, mannosidase II, as a marker for the Golgi complex (10), together with antibodies specific for Trip230. As shown in Fig. 2, Trip230 (a, orange) and mannosidase II (b, blue) colocalize to the Golgi apparatus. An overlay of two images of Trip230 and mannosidase II (Fig. 2c) shows substantial but incomplete overlap of the staining patterns. During mitosis, Trip230 does not appear as a dot pattern in regions peripheral to the chromosomes (Fig. 1B, e–h) as shown by the Golgi stack membrane-bound proteins (15). The localization data combined with the fact that Trip230 does not appear to be a secreted or plasma membrane-associated protein suggest that Trip230 is partitioned to the region of the Golgi but may not be cisternae-associated.
Figure 2.
Colocalization of Trip230 and mannosidase II to the region of the Golgi complex. Cells were fixed and double-immunostained with anti-Trip230 and antimannosidase II antisera. Shown in this representative field is the indirect immunofluorescence antibody staining imaged by laser-scanning confocal microscopy. a, pattern of immunofluorescence using anti-Trip230 polyclonal serum (1:1,000 dilution) and FITC-tagged anti-mouse IgG secondary antibodies that localize Trip230 to perinuclear cytoplasm (pseudocolored orange); b, rabbit antimannosidase II (a Golgi enzyme) antiserum and Texas red-tagged anti-rabbit secondary antibody labeling of the Golgi complex (pseudocolored blue); c, digital overlay of anti-Trip230 and mannosidase II antiserum images showing that while in the same vicinity of the Golgi, the overlap of Trip230 immunofluorescence is incomplete.
Domain Mapping for Trip 230 Localization to Golgi Regions.
To determine which region of the Trip230 protein is required for its localization to the region of the Golgi, several deletion constructs of Trip230 were generated as green fluorescent protein (GFP) fusion proteins (Fig. 3A). All the constructs were introduced into Saos2 cells by the calcium phosphate coprecipitation method, and live cells were observed with a fluorescence microscope. GFP alone does not colocalize with the Golgi area (Fig. 3B, a and b). As shown in Fig. 3B, the sequential deletion of regions located from amino acid 125 to 1602 in the GFP/Trip230 fusions (p65, p6, p22, ΔRBBD, and p31, Fig. 3A) had little, if any, effect on the ability of Trip230 to localize in the region of the Golgi (Fig. 3B, c–l). Interestingly, deletion of residues 1754–1866 (ΔTRBD, Fig. 3A), which contain a region that was demonstrated previously to be required for Trip230 binding to TR (5), resulted in abnormal aggregate partitioning of the GFP-ΔTRBD fusion to non-Golgi areas (Fig. 3B, m and n). To examine further the role of the TR-binding domain in targeting Trip230 to the region of the Golgi, residues 1754–1978 were translationally fused to GFP (C5C, Fig. 3A). When cultured in complete serum, cells transfected with C5C show immunofluorescence localized in regions of the Golgi and, interestingly, demonstrated limited nuclear staining (Fig. 3B, o and p). These data indicate a potentially interesting relationship between the TR-binding domain in Trip230, T3, the ligand for TR, and the subcellular partitioning of Trip230.
Nuclear Localization of Trip230 Induced by T3 Treatment.
To determine the effects of T3 on subcellular localization of Trip230, CV1 cells cultured on cover glasses were treated with hormone, fixed at different time points subsequent to treatment, and immunostained with anti-Trip230 antisera. As shown in Fig. 4A, 1 hr after T3 addition, Trip230 immunofluorescence moved from the area of the Golgi into the nucleus as speckles or prominent dots, as shown in Fig. 4A (c, arrows). However, this staining pattern was not detectable in cells treated with T3 for 6 hr (Fig. 4A, d), suggesting that either nuclear Trip230 is degraded, exported, or sufficiently modified to mask antibody epitopes.
TR Colocalizes with Nuclear Trip230 after T3 Treatment.
To examine the relationship between TR and Trip230 nuclear localization, we performed double immunostaining of cells treated with or without T3. In the absence of hormone, TR staining is located in the nucleus in a homogenous staining pattern (Fig. 4B, b), whereas that of Trip230 is in the vicinity of the Golgi complex (Fig. 4B, a). Thus, in the absence of T3 treatment, TR and Trip230 showed no overlap in their pattern of staining (Fig. 4B, c). One hour after treatment with T3, a portion of Trip230 immunofluorescence is evident in the nucleus in the familiar dot pattern observed previously (Fig. 4B, d). Interestingly, a significant portion of TR immunofluorescence also appears to be reorganized into the nuclear dot pattern (Fig. 4B, e) that overlaps with Trip230 (Fig. 4B, f). This result suggests that Trip230 and TR are repartitioned in response to T3 treatment of cells. To determine whether the observed changes in Trip230 and TR partitioning are dependent on protein synthesis stimulated by T3 induction, the cells were treated simultaneously with T3 and cyclohexamide. As shown in Fig. 4C, inhibition of new protein synthesis did not alter the nuclear import of Trip230 (c and f) and may have increased its nuclear retention (compare c and f with c and d).
Treatment with T3 Induces Further Phosphorylation of Trip230.
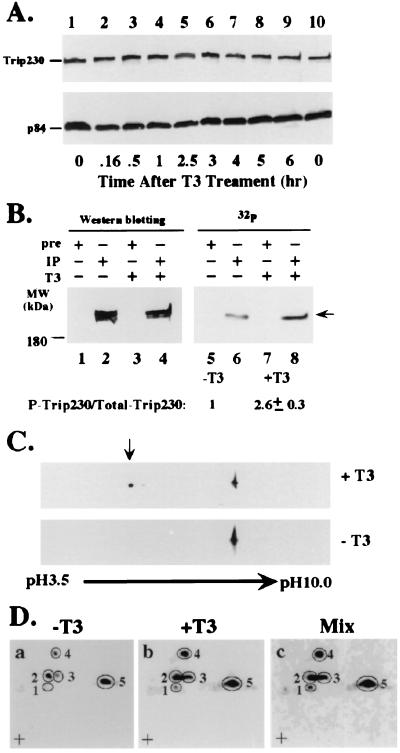
Because nuclear import often is regulated by protein phosphorylation (reviewed in rev. 16), we next examined the phosphorylation status of Trip230 in response to T3 treatment. To understand the regulation of Trip230 by T3 treatment, we first checked the overall levels of Trip230 protein before and after T3 treatment. As shown in Fig. 5A, the amount of Trip230 is unchanged after T3 treatment. p84, a nuclear matrix protein (17) that does not respond to T3 treatment, served as an internal control for protein loading. We next examined the phosphorylation of Trip230 in response to T3 treatment by metabolically labeling cells with 32P in the presence or absence of T3. The clarified labeled lysates were immunoprecipitated with anti-Trip230 antiserum and washed extensively with lysis buffer, and the resulting immunocomplexes were subjected to SDS/PAGE. As shown in Fig. 5B, the amount of the phosphorylated form of Trip230 increased in cells treated with T3 (compare lanes 6 and 8), whereas the total amount of Trip230 again remained unchanged (Fig. 5B, lanes 2 and 4). The calculated increase in the ratio of phosphorylated to total Trip230 in T3-treated cells is 2.6 ± 0.3 relative to that in untreated cells. This observation also is confirmed by two-dimensional gel electrophoresis that showed a change in the mobility toward acidic pH of a significant portion of Trip230 in response to T3 treatment (Fig. 5C Upper, arrow). Trip230 is constitutively phosphorylated (Fig. 5B, lane 6). To test whether T3 treatment recruits additional residues in Trip230 for phosphorylation, tryptic phosphopeptide mapping was performed by using Trip230 immunoprecipitated from 32P-metabolically labeled cells treated with or without T3. An analysis of the relative intensities of the spots in Fig. 5D indicates that spots 2, 3, 4, and 5 increased in response to T3 treatment relative to untreated cells. However, the intensity of spot 1 increased dramatically in lysates from cells treated with T3, suggesting a new site(s) of phosphorylation in Trip230 in response to hormone treatment. This is consistent with the two-dimensional gel analysis in Fig. 5C that showed the mobility of a part of Trip230 shifted toward acidic pH in response to T3 treatment.
Figure 5.
T3-responsive modification of Trip230 by nascent phosphorylation. (A) The pattern of Trip230 protein expression after T3 treatment. Cells (5 × 105 CV1) were treated with 10−6 M T3 for various lengths of time, indicated along the bottom of A. Proteins were extracted and immunoblotted with anti-Trip230 antiserum (Upper) or mAb 5E10 anti-N5 (p84) antibody (Lower). (B) Increase in the phosphorylated form of Trip230 after T3 treatment. CV1 cells, labeled with 32P-labeled phosphoric acid in the presence or absence of T3 for 2 hr, were lysed and the clarified lysate was immunoprecipitated with anti-Trip230 antiserum followed by immunoblot analysis with the same antiserum. The total amount of Trip230 was measured by densitometry, and labeled Trip230 was quantitated by PhosphorImager (Molecular Dynamics). A 2-fold increase in 32P-labeled Trip230 was obtained in each of three independent experiments (see text for further discussion). (C) Two-dimensional gel analysis of phosphorylated Trip230. Cells (5 × 105 CV1) were untreated (Lower) or treated with 10−6 M T3 for 1 hr (Upper). Proteins were separated by isoelectric focusing followed by SDS/PAGE. Trip230 was immunoblotted by using polyclonal mouse anti-Trip230 antibodies. Note the increase in Trip230 at acidic pH (arrow) in T3-treated cells. (D) Tryptic phosphopeptide mapping of Trip230. 32P-labeled anti-Trip230 immune complexes (prepared as in A) were fractionated by electrophoreses and blotted, and Trip230 was excised from the membrane and digested with l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin. The tryptic digests then were separated by electrophoresis (left to right) and chromatography (bottom to top, + is the origin). a, a digest from untreated cells; b, from cells treated with T3; and c, an equal mix of the radioactivity from treated and untreated cells. Note the increase in labeled phosphopeptides encircled in 3 and 4 and the new spot (1) appearing in b, indicating phosphorylation of residues separate from constitutive sites (represented by spots 2, 3, 4, and 5) in lysates from untreated cells in a.
DISCUSSION
Trip230 is a T3-dependent transcription coactivator that, unlike other nuclear receptors, appears to be specific for TR (5). It was surprising to discover that it is predominantly a cytoplasmic protein in cells not treated with T3. Cytoplasmic localization of transcription factors such as NF-κB and STATs and their nuclear transport in response to cell signaling have been well studied (reviewed in refs. 18 and 19). Cytoplasmic localization of a transcription coactivator has not, to our knowledge, been described and suggests the possibility that subcellular partitioning may play a role in T3 signaling.
The localization of Trip230 during the cell cycle revealed subtle, but reproducible changes in partitioning from the Golgi to the nuclear compartment during S phase, indicating that Trip230 could have coactivator functions related to this stage of the cell cycle. Trip230 localization to the Golgi is intriguing, although the biological significance of this compartmentalization is not understood. Based on two lines of evidence, Trip230 is not likely to be a Golgi cisternae-associated protein. The first is incomplete overlap in the staining pattern with mannosidase II, a membrane-associated protein. The second is the difference between the diffuse pattern of Trip230 staining compared with the speckled distribution of a Golgi enzyme such as GFP-NAGTI (15) during metaphase. The carboxyl-terminal, TR-binding domain of Trip230 is sufficient to target GFP to the region of the Golgi. It will be very interesting to elucidate the molecular basis of this partitioning mechanism.
The T3-dependent translocation of Trip230 to the nucleus is consistent with its proposed function as a coactivator of TR-mediated transcription (5). The subnuclear partitioning of Trip230 to variable-sized aggregate structures that are apparent 1 hr after treatment of T3 is intriguing. What is equally interesting is the apparent nuclear reorganization of the TR staining from a highly dispersed, speckled pattern in cells devoid of T3, to a pattern that includes aggregates that completely overlap the Trip230 dots. The possibility that the nuclear localization and subpartitioning of Trip230 is due to T3-mediated synthesis of additional proteins was ruled out by cyclohexamide treatment, which apparently increased the stability of the nuclear aggregates.
Recent studies using GFP translationally fused to TR showed about an equal distribution of the GFP-TRβ fusions between the nucleus and cytoplasm of living CV1 cells in the absence of T3 (20). Treatment with T3 shifted GFP-TRβ partitioning predominantly to the nucleus. The discrepancy between the localization of GFP-TRβ and nuclear immunostaining of endogenous TRβ in Fig. 4 could be due to differences in partitioning of the GFP fusions and/or overexpression overwhelming the nuclear capacity for either transport or retention.
What signals Trip230 to translocate from the area of the Golgi to the nucleus subsequent to treatment of cells with T3? Although the answer to this question is not known, it is intriguing that an increase in the phosphorylation of Trip230 is correlated with nuclear entry. Phosphorylation of transcription factors as a method to regulate nuclear import of proteins is well known (reviewed in ref. 16). For the NF-κB transcription factor, it is the phosphorylation of its cytoplasmically associated partner IκB by the IκB kinases (IKK-1 and IKK-2) and subsequent degradation of IκB that regulates NF-κB nuclear import. The IKKs respond to various inducers, including growth factors, lymphokines, cytokines, UV, and stress (reviewed in ref. 18). For the STAT family transcription factors, it is phosphorylation by the Jak kinases in response to various cytokine receptor signaling that prompts nuclear import, DNA-binding, and activation of target genes (reviewed in ref. 19). The histone acetyltransferase activity of the cAMP response element-binding protein, CBP, that coactivates transcription mediated by diverse transcription factors, including nuclear hormone receptors (reviewed in ref. 2), was reported to increase upon phosphorylation by a cell cycle-regulated kinase (21).
To our knowledge, phosphorylation of a transcription coactivator in response to T3 signaling is a unique observation. Tryptic peptide mapping indicates that additional new site(s) are phosphorylated in response to T3 treatment. The kinase responsible for T3-induced phosphorylation is unknown at present. Kinases that are potential candidates include IKK-1/2, discussed earlier, human cyclin B2 that localizes to the Golgi (22), and cdc2 kinase that phosphorylates Golgi matrix proteins and is required for fragmentation during mitosis (23). The identification of the T3-responsive kinase will be essential to elucidate this interesting signal transduction pathway.
Acknowledgments
We thank Paula Garza for her excellent assistance in antibody production. This work was supported by National Institutes of Health Grant EY05758 to W.H.L.
ABBREVIATIONS
- Rb
retinoblastoma protein
- TR and TRβ
thyroid hormone receptor
- T3
3,3′,5-triiodo-l-thyronine
- GFP
green fluorescent protein
References
- 1.Torchia J, Glass C, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 2.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O’Malley B W. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 3.Lee J W, Choi H S, Gyuris J, Brent R, Moore D D. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 4.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 5.Chang K H, Chen Y, Chen T T, Chou W H, Chen P L, Ma Y Y, Yang-Feng T L, Leng X, Tsai M J, O’Malley B W, Lee W H. Proc Natl Acad Sci USA. 1997;94:9040–9045. doi: 10.1073/pnas.94.17.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riley D J, Lee E Y, Lee W H. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 7.Spindler B J, MacLeod K M, Ring J, Baxter J D. J Biol Chem. 1975;250:4113–4119. [PubMed] [Google Scholar]
- 8.Chen Y, Farmer A A, Chen C-F, Jones D C, Chen P-L, Lee W-H. Cancer Res. 1996;56:3168–3172. [PubMed] [Google Scholar]
- 9.Chen P-L, Riley D J, Chen Y, Lee W-H. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 10.Novikoff P M, Tulsiani D R, Touster O, Yam A, Novikoff A B. Proc Natl Acad Sci USA. 1983;80:4364–4368. doi: 10.1073/pnas.80.14.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyle W J, van der Geer P, Hunter T. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 12.Mosier H D J. In: Endocrine Control of Growth. Daughaday W H, editor. New York: Elsevier; 1981. pp. 25–66. [Google Scholar]
- 13.Froesch E R, Zapf J, Audhya T K, Ben-Porath E, Segen B J, Gibson K D. Proc Natl Acad Sci USA. 1976;73:2904–2908. doi: 10.1073/pnas.73.8.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P L, Scully P, Shew J Y, Wang J Y, Lee W H. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 15.Lowe M, Nakamura N, Warren G. Trends Cell Biol. 1998;8:40–44. doi: 10.1016/s0962-8924(97)01189-6. [DOI] [PubMed] [Google Scholar]
- 16.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 17.Durfee T, Mancini M A, Jones D, Elledge S J, Lee W-H. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma I M, Stevenson J. Proc Natl Acad Sci USA. 1997;94:11758–11760. doi: 10.1073/pnas.94.22.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X G, Hanover J A, Hager G L, Cheng S Y. J Biol Chem. 1998;273:27058–27063. doi: 10.1074/jbc.273.42.27058. [DOI] [PubMed] [Google Scholar]
- 21.Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, et al. Nature (London) 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 22.Jackman M, Firth M, Pines J. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin D J, Warren G. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]