Abstract
Microsporidia are intracellular parasites that infect a variety of animals, including humans. As highly specialized parasites, they are characterized by a number of unusual adaptations, many of which are manifested as extreme reduction at the molecular, biochemical, and cellular levels. One interesting aspect of reduction is the mitochondrion. Microsporidia were long considered to be amitochondriate, but recently a tiny mitochondrion-derived organelle called the mitosome was detected. The molecular function of this organelle remains poorly understood. The mitosome has no genome, so it must import all its proteins from the cytosol. In other fungi, the mitochondrial protein import machinery consists of a network series of heterooligomeric translocases and peptidases, but in microsporidia, only a few subunits of some of these complexes have been identified to date. Here, we look at targeting sequences of the microsporidian mitosomal import system and show that mitosomes do in some cases still use N-terminal and internal targeting sequences that are recognizable by import systems of mitochondria in yeast. Furthermore, we have examined the function of the inner membrane peptidase processing enzyme and demonstrate that mitosomal substrates of this enzyme are processed to mature proteins in one species with a simplified processing complex, Antonospora locustae. However, in Encephalitozoon cuniculi, the processing complex is lost altogether, and the preprotein substrate functions with the targeting leader still attached. This report provides direct evidence for presequencing processing in mitosomes and also shows how a complex molecular system has continued to degenerate throughout the evolution of microsporidia.
Keywords: Antonospora, Encephalitozoon, inner membrane peptidase
Microsporidia are a large and diverse group of eukaryotic intracellular parasites that infect a wide variety of animal lineages, including humans (1, 2). They were long thought to be “primitive” eukaryotes but are now widely recognized as being fungi that have undergone reductive evolution as so to appear primitive (3–7). The apparent absence of mitochondria in microsporidia attracted much attention, and microsporidia and other “primitive eukaryotes” were accordingly proposed to have evolved before the endosymbiosis that gave rise to the organelle (8). However, molecular traces of mitochondria have since been found in microsporidia (9–13), and a highly reduced, cryptic organelle called the mitosome has now been identified (14), as in other investigated amitochondriates such as Giardia intestinalis (15), Trichomonas vaginalis (16), and Entamoeba histolytica (17, 18).
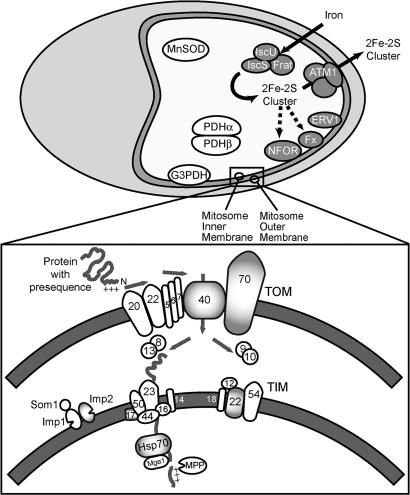
In microsporidia, this organelle is highly reduced from the perspective of both physical size and biochemical complexity. The number of proteins estimated to be acting within yeast mitochondria is 800-1,000 (19, 20). In contrast, the number of putative mitochondrial proteins identified within all microsporidian species amounts to 21 thus far (21, 22). Based on the whole genome sequence of Encephalitozoon cuniculi, the only clear function of the organelle from these proteins is iron-sulfur cluster (ISC) assembly, and it seems to have lost the capacity for ATP production through oxidative phosphorylation (21). The identified proteins can largely be categorized into those that act in protein and metabolite import (TOM70, TIM22, TOM40, Imp2, mitochondrial Hsp70, and ATM1-ABC transporter proteins) and those involved in ISC assembly and export (frataxin, ferredoxin, ISCU, ISCS, ERV1, and ferredoxin NADPH oxido-reductase [FNR]). The two subunits of pyruvate dehydrogenase, PDHα and -β, and mitochondrial glycerol-3-phosphate dehydrogenase (mtG3PDH) are involved in metabolic processes, and manganese-containing superoxide dismutase (MnSOD) is involved in protection against oxidative stress (Fig. 1).
Fig. 1.
Schematic of the microsporidian mitosome, a double-membrane-bounded reduced mitochondrion. Putative mitosomal matrix proteins associated with ISC assembly and export are shaded in gray. Generalized protein translocation is shown in the Inset, with proteins identified in microsporidia shaded in gray.
The microsporidian mitosome has no genome and is now completely reliant on import of nuclear encoded proteins for the functioning and maintenance of the organelle. In yeast, the proteins targeted to mitochondria possess targeting information defined by hydrophobicity and an amphiphilic α-helical conformation with a positively charged and an apolar side (23, 24). These features are usually present in N-terminal targeting presequences or associated with the transmembrane domains of membrane proteins (25–28). This targeting information allows recognition of the protein by the import machinery that consists of complexes in both the outer and inner mitochondrial membrane. An N-terminal targeting sequence enables import via the TOM complex (translocase of the outer membrane) before being recognized by the TIM23 complex (translocase of the inner membrane), which facilitates translocation through the inner membrane into the matrix. There, the presequence is removed by the mitochondrial matrix-located processing peptidase (MPP) (29, 30).
Of the identified proteins in microsporidia, few of the mitochondrial import and processing proteins are present (Fig. 1). In addition, very few of the microsporidian mitosomal proteins have N-terminal sequences with characteristics expected of a targeting sequence, so it is unclear how import is achieved. Here we have tested to what extent the common mitochondrial targeting system has been retained in microsporidia and how the complexity of the system is degenerating. We show that many mitosomal proteins are targeted appropriately in yeast, confirming that homologous elements of the ancestral system are still used and that the N terminus is at least partly responsible for encoding targeting information. Using the inner-membrane peptidase (IMP) as a model, we also show the complexity of the system has been progressively reduced during microsporidian evolution. IMP usually consists of two catalytic subunits (Imp1 and Imp2) and a noncatalytic regulator (Som1), but this complex is apparently reduced to a single functional protein in Antonospora locustae and lost altogether in E. cuniculi. The protein substrates of this complex are processed in A. locustae but not in E. cuniculi, where they function as unprocessed preproteins. The reduction of the IMP complex is typical of how complex molecular systems have degenerated in microsporidia.
Results and Discussion
Conserved and Diverged Presequence Features in Mitosomal Proteins.
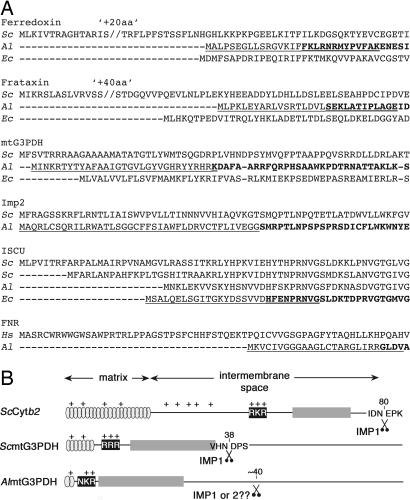
In yeast, N-terminal presequences typically consist of ∼20–60 residues with no consensus sequence (31). They are characterized by an overall positive charge, the propensity to form an amphiphilic α-helix (31–33), and the presence of an arginine residue at the −2 position from the MPP cleavage site (31, 34). This arginine residue is only found in ∼65% of investigated yeast presequences (35), so it seems likely that further structural elements are also important for MPP cleavage. In comparison, the N termini of microsporidian mitosomal proteins have no consistent characteristics (Fig. 2A). At the extreme, many proteins lack identifiable extensions beyond the expected start of the mature protein (e.g., Alferredoxin, AlHsp70, AlPDHα and -β; Al denotes a protein from A. locustae), whereas others have extended presequences (e.g., AlmtG3PDH and EcATM1; Ec denotes a protein from E. cuniculi), with few of these predicted to form basic, amphipathic helices (Table 1, which is published as supporting information on the PNAS web site). The absence of available data from other microsporidian parasites limits predictions to A. locustae and E. cuniculi.
Fig. 2.
N-terminal regions of microsporidian mitosomal proteins. (A) A. locustae and E. cuniculi mitosomal proteins in comparison with their yeast (S. cerevisiae) or Homo sapiens homologues. N-terminally truncated versions used in Fig. 3B are indicated by bold. N-terminal signals fused to GFP are underlined. (B) Schematic representation of the presequence of AlmtG3PDH in comparison to yeast ScCytb2 and ScmtG3PDH with regions corresponding to a basic amphipathic helix containing the matrix targeting information (gray) and the hydrophobic intermembrane sorting sequence (dark gray) preceded by a cluster of positively charged residues (black) important for recognition of the sorting sequence. For AlG3PDH a possible processing site is indicated around residue 40, an estimate from deletion experiments (see Fig. 4C).
Targeting Signals and Their Functional Conservation in Mitosomes and Mitochondria.
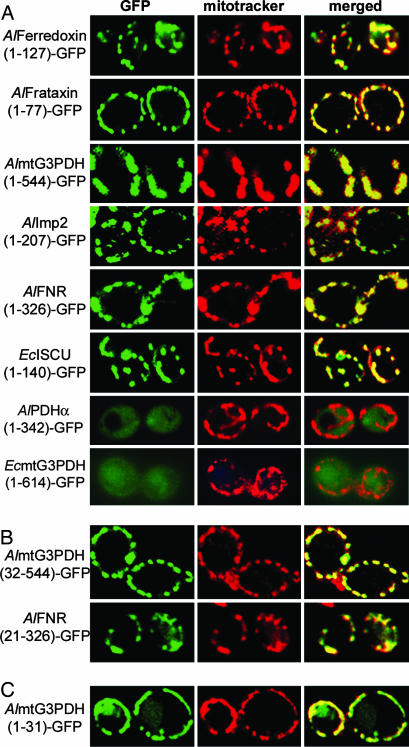
Because there is no genetic system for transforming and expressing proteins in microsporidia, we used another fungal system, the more tractable and well studied Saccharomyces cerevisiae. To determine whether the properties of mitosomal proteins from microsporidia can be recognized in yeast, GFP was C-terminally fused to full-length predicted mitosomal proteins and localized in yeast cells costained with MitoTracker Red. Many mitosomal proteins (AlFerredoxin, AlFrataxin, AlmtG3PDH, AlImp2, AlFNR, and EcISCU) are targeted to yeast mitochondria (Fig. 3A). The difference in shape and size of the mitochondria observed, ranging from tubular- to punctate-shaped structures, has been observed previously and is due to different morphologies of the organelle during fission and fusion events (36).
Fig. 3.
Analysis of microsporidian protein import into yeast mitochondria by confocal microscopy. (A) Yeast cells expressing GFP fusions to complete microsporidian preproteins were costained with the fluorescent dye MitoTracker Red and viewed by confocal microscopy. Filters selective for the green fluorescence of GFP (Left) or the red fluorescence of MitoTracker Red (Center) were used. Merged green and red fluorescence images are shown in Right. (B) AlmtG3PDH and AlFNR GFP-fusions lacking N-terminal amino acids are still targeted to mitochondria when expressed in yeast. (C) N-terminal extensions from AlmtG3PDH attached to GFP delivered a significant proportion of this fusion protein into mitochondria.
In contrast, several mitosomal proteins (AlATM1, AlHsp70, AlISCS, AlISCU, AlMnSOD, AlPDHα, AlPDHβ, EcATM1, EcmtG3PDH, and EcErv1) were not recognized by the yeast import machinery, with fusion proteins showing no mitochondrial localization. Two typical examples (AlPDHα and EcmtG3PDH) of cytosolic mislocalization are shown in Fig. 3A (bottom two rows). It cannot be excluded that this decreased mitochondrial specificity is due to the addition of a GFP molecule. Normal growth was observed in all cells, and toxic effects of the overexpressed GFP-fusion proteins seem unlikely.
In conclusion, there are no obvious correlations between proteins that are targeted to mitochondria and those predicted to have presequences based on Mitoprot or PsortII prediction protocols (Table 1).
The successful targeting of proteins with no distinguishable leader (e.g., EcISCU) raises questions about where targeting information is located in these proteins. To determine whether targeting is due to the presence of information at the N terminus, we constructed reciprocal deletion mutants and compared their targeting activity with that of the intact protein. No mitochondrial labeling was observed in cells expressing AlFerredoxin (17–127)-GFP, AlFrataxin (20–77)-GFP, AlImp2 (40–207)-GFP, and EcISCU (13–140)-GFP (data not shown), suggesting N-terminal targeting information is present in these cases. However, some other N-terminally truncated proteins were delivered to mitochondria [AlmtG3PDH (32–544)-GFP and AlFNR (21–326)-GFP; Fig. 3B], suggesting that some proteins also use internal targeting information.
To investigate whether the N-terminal peptides of AlFerredoxin, AlFrataxin, AlmtG3PDH, AlImp2, AlFNR, and EcISCU are sufficient for targeting to mitochondria, the N-terminal segments were fused to GFP and expressed in yeast. Of these, only the N terminus of Al-mtG3PDH (the first 31 residues) was sufficient to target GFP to mitochondria (Fig. 3C). This finding suggests that the N terminus is often necessary but, in targeted proteins with short extensions such as AlFerredoxin and EcISCU, additional internal signals are also required. Internal targeting signals in addition to N-terminal extensions are also used in yeast, for example in the ScISCU and ScGut2 proteins (Sc denotes a protein from S. cerevisiae), where the N terminus is sufficient for directing a reporter protein into the mitochondrion, but a N-terminally truncated protein is also targeted to the organelle (37).
Overall these data demonstrate that microsporidian mitosomal proteins contain targeting information that is functionally recognizable by canonical mitochondria. In most cases, the N terminus is necessary for targeting in yeast and in some cases also sufficient for targeting, but a combination of N-terminal and internal information is present in most examined proteins.
AlpremtG3PDH Is Processed by Imp1, Whereas EcpremtG3PDH Is Not Processed.
Of all of the mitosomal proteins identified in microsporidia, the mtG3PDH has the longest predicted presequence. In yeast, the mtG3PDH (Gut2) is targeted to the intermembrane space through a stop-transfer mechanism and processed by the IMP complex located in the mitochondrial inner membrane. Although a consensus for stop-transfer signals is not known, the general features include a segment rich in positive charges followed by a hydrophobic stretch and a cleavage site with characteristic P3 and P1′ residues for IMP cleavage (38). The AlG3PDH presequence resembles the ScmtG3PDH in that it consists of a region where a hydrophobic stretch preceded by positive charges can be identified (Fig. 2B).
The yeast IMP complex consists of two catalytic subunits with different substrate specificities, Imp1 and Imp2 (39, 40), and a noncatalytic subunit required for maturation of certain Imp1 substrates, Som1 (41, 42). In addition to Gut2, the IMP complex in yeast proteolytically processes several other proteins after their arrest in the TIM23 complex, thereby liberating these protein substrates for release into the intermembrane space. In A. locustae, a gene for Imp2 was recently identified (22). The presence of Imp2 in A. locustae, but not in E. cuniculi, and of a potential substrate in both organisms raises a number of questions about how the stop-transfer pathway and IMP processing work in microsporidia and how mtG3PDH may be differentially processed. These questions are all of the more interesting because both AlmtG3PDH and AlImp2 are successfully targeted to yeast mitochondria (Fig. 3A).
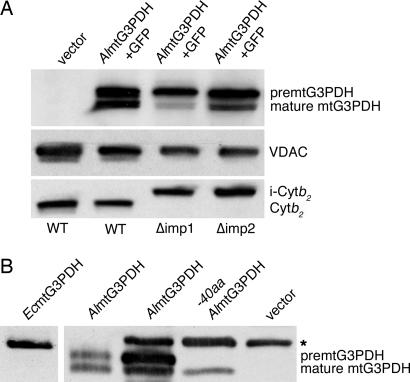
To determine whether mtG3PDH is a substrate of IMP, we expressed AlmtG3PDH-GFP in yeast strains lacking Imp1 and Imp2. Neither the Imp1 nor Imp2 subunits of the IMP complex are essential in S. cerevisiae, and mutants lacking either Imp1 or Imp2 are partially compromised in the processing of substrates like mtG3PDH (43). When expressed in yeast strains, both precursor and mature forms of AlmtG3PDH were detected in wild-type and Δimp2 mutants, but in Δimp1 mutants, processing was substantially attenuated (Fig. 4A). As a control we looked at processing of the intermembrane space cytochrome (Cyt) b2, which is first processed by MPP to generate an intermediate (i-Cytb2; Fig. 4A) and then by the IMP complex. In wild-type cells, processing to the mature form of Cytb2 occurred, whereas processing is completely blocked in Δimp1 cells (Fig. 4A). The Imp1 subunit is responsible for Cytb2 processing (40), but because yeast Δimp2 mutants express decreased levels of the Imp1 subunit (40), Cytb2 is also blocked in Δimp2 cells. In contrast, the outer membrane voltage-dependent anion channel (VDAC) is not processed and is expressed in all strains similarly (Fig. 4A). We conclude that AlmtG3PDH is a substrate of the yeast IMP complex, and in particular acted on by the Imp1 subunit. Preliminary experiments suggest this processing by the IMP complex would liberate AlmtG3PDH from the inner membrane to a soluble or membrane peripheral form (data not shown), but further work is required to know the suborganellar topology of the protein in microsporidians.
Fig. 4.
AlmtG3PDH is processed by the IMP complex, whereas EcmtG3PDH is not. (A) Mitochondria isolated from yeast transformed with an empty vector (lane 1) or preAlmtG3PDH fused to GFP (lanes 2–4) from either wild-type yeast cells (lane 1 and 2) or yeast mutants lacking Imp1 (Δimp1, lane 3) or Imp2 (Δimp2, lane 4) were compared for processing after SDS/PAGE separation of proteins. Proteins were analyzed by immunoblotting with antisera recognizing the GFP fused to AlmtG3PDH, the voltage-dependent anion channel (VDAC), and Cytb2. Positions of preprocessed and mature AlmtG3PDH-GFP and of i-Cytb2 and mature forms of Cytb2 (54, 57) are shown. (B) Western blot of total protein extracts from spores of either E. cuniculi (using an antibody raised against EcmtG3PDH, lane1) or A. locustae (using an AlmtG3PDH-specific antibody, lane 2). Mitochondria (100 μg of protein) were isolated from yeast expressing full-length AlmtG3PDH (lane 3) or a truncated AlmtG3PDH, where the first 40 aa have been deleted (lane 4). A background band seen in yeast is indicated with an asterisk.
To determine whether the mtG3PDH proteins are processed in microsporidia that either possess or lack IMP, we examined total protein preparations from purified A. locustae and E. cuniculi spores by Western blot analysis using antibodies raised against mtG3PDH-specific peptides from the two proteins. In E. cuniculi, a single band corresponding in size to the 68-kDa unprocessed form of EcmtG3PDH was observed (Fig. 4B, lane 1). In contrast, two distinct bands were visible in the A. locustae spores, corresponding closely to predicted size of unprocessed (66 kDa) and mature (61 kDa) forms of the enzyme (Fig. 4B, lane 2). AlmtG3PDH was expressed in yeast, and Western blots from purified mitochondria showed the same size protein species (Fig. 4B, lane 3). The predicted site of cleavage in AlmtG3PDH is between residues 40 and 41; as a marker for the size of the mature form we expressed an AlmtG3PDH mutant missing the first 40 codons in yeast, and Western blots of purified yeast mitochondria show the inferred mature protein band to be the same size as the construct (Fig. 4B, lane 4), confirming that the first 40 amino acids of AlmtG3PDH are processed. Thus, AlmtG3PDH is processed in both A. locustae and S. cerevisiae to species of similar size, and in S. cerevisiae, the processing must occur within mitochondria.
Overall, these data suggest the mtG3PDH is targeted in two different ways in the two microsporidia. In A. locustae the protein is processed by an IMP enzyme, and because no AlImp1 or AlSom1 could be identified so far, we suggest that the AlImp2 is not part of a complex but has simplified to operate alone. This is a plausible scenario, because the closely related bacterial signal peptidases often function as monomers (44). In contrast, the E. cuniculi system has been further simplified so that the IMP complex is lost altogether, and its substrates are apparently targeted by means of an N-terminal domain that is retained by the mature protein. The prediction of this, given the stop-transfer mechanism for targeting, is that the EcmtG3PDH would remain anchored to the inner mitosomal membrane in E. cuniculi. This result is direct evidence for mitochondrial presequence processing in microsporidia and a first glimpse at the functional implications of the degenerating complexity of the targeting system.
Reduced Complexity of Targeting Pathways in Microsporidian Mitosomes.
Microsporidia have one of the most reduced mitochondria both in terms of physical size and biochemical composition. Bioinformatic studies indicate that the mitochondrial import machinery is similarly pared down, with only five components identified to date, although there may be others that are difficult to recognize from sequence analysis. The missing components include some that are highly conserved in sequence across divergent species of eukaryotes, such as the MPP (39). Other organisms with relic mitochondria, such as Giardia and Trichomonas, also have targeting sequences of reduced size (45–49) and, at least in Giardia, there appears to be a similar combination of presequence-dependent and -independent protein import (47, 48). However, the short presequences on proteins targeted to these relic mitochondria are known to be processed by the MPP, which has been identified and studied in Giardia and Trichomonas (48). We have now shown that, even in the absence of MPP and with evidence for leaders in only very few proteins, microsporidian mitosomes have retained select elements of the import machinery found in canonical mitochondria.
In A. locustae, the stop-transfer pathway for import appears to have been retained with an IMP peptidase capable of liberating substrate proteins into the intermembrane space still present. The Imp subunit found in A. locustae is phylogenetically most similar to Imp2 (22), but it is possible that it has adapted to function in place of both proteins. Consistent with this, Imp2 is important for the stability and activity of Imp1 in the inner membrane but not vice versa (40, 50), and although the Som1 subunit in yeast is required for Imp1 function, it is dispensable for Imp2 function (42). In yeast, mtG3PDH is a substrate of Imp1; however, in mouse, it is processed by Imp2 (51), further supporting the suggestion that Imp2 could act alone to process diverse substrates for the stop-transfer pathway. In E. cuniculi, this reduction in processing has proceeded to the extreme, with the IMP complex lost altogether and mtG3PDH apparently unprocessed and retaining its N terminus.
With the mitosomal targeting system having degenerated to its present state in microsporidia, we suggest that the reduction of complexity of the mitosomal proteome has led to a targeting system with fewer constraints. With the N terminus of this dwindling number of proteins reduced in size, internal features of the proteins may have become more and more responsible for targeting to mitosomes. In the case where the N-terminal presequence is of minimal length, it might not be deleterious to the mature protein and would not need to be proteolytically removed. This scenario rationalizes the observation that microsporidia can make do without MPP and might also help explain the loss of other components of the protein import machinery.
Materials and Methods
DNA Extraction and PCR.
DNA was extracted from purified spores collected from E. cuniculi tissue culture cells (E. S. Didier, Tulane University, New Orleans, LA) using the Qiamp mini DNA extraction kit (QiAgen, Valencia, CA). DNA was extracted from A. locustae spores (M&R Durango, Bayfield, CO) by bead beating followed by phenol/chloroform extraction and ethanol precipitation. Gene sequences for putative mitochondrial proteins were downloaded from either the National Center for Biotechnology Information or the Antonospora locustae Genome Project (Marine Biological Laboratory, Woods Hole, MA) (funded by National Science Foundation Award 0135272). Where ORFs had not been verified and more than one possible starting methionine could be identified, 5′ RACE was used to eliminate some upstream methionines.
Cloning and Expression.
DNA fragments corresponding to whole ORFs, N termini, or ORFs lacking the N terminus were amplified by PCR by using primers that generated in-frame restriction sites. PCRs were carried out using Ready-to-Go PCR beads (Amersham, Piscataway, NJ). PCR products were cloned in front of GFP-S65T under the control of the MET25 promoter (52) for analysis by confocal or fluorescence microscopy. Constructs were then transformed into the diploid yeast strain JK9–3da/a (leu2-3,122/leu2-3,122 ura3-52/ura3-52 rme1/rme1 trp1/trp1 his4/his4 GAL+/GAL+ HMLa/HMLa) and plated on uracil and methionine deficient SD plates (2% [wt/vol] agar, 2% [wt/vol] glucose, and 0.67% [wt/vol] yeast nitrogen base supplemented with the relevant amino acids).
Positive colonies were grown overnight in SD medium lacking uracil and methonine and stained with MitoTracker (MitoTracker Red CM-H2XRos) according to the manufacturer's protocol (Molecular Probes, Eugene, OR). Yeast cells were visualized using the AxioplanII fluorescence microscope (Zeiss, Jena, Germany), C1 confocal microscope (Nikon, Tokyo, Japan), or Radiance confocal microscope (BioRad, Hercules, CA).
Preparation of Mitochondria, Spore Proteins, and Western Blot Analysis.
Crude mitochondria were isolated from S. cerevisiae strain W303 (Mata leu2-3,112 his3-11,15 ade2-2 ura3-1 trp1-1 cau1-100) as described (53). Samples of mitochondrial protein (100 mg) were separated by Tris-glycine SDS/PAGE, and Western blots were carried out according to published methods (54, 55). Spore proteins were obtained as described in ref. 56.
Acknowledgments
We thank Raheel Humayun and Irene Barrett for technical help. This work was supported by a grant (MOP-42517) from the Canadian Institutes for Health Research (CIHR) (to P.J.K.). P.J.K. is a Fellow of the Canadian Institute for Advanced Research and a new Investigator of the Michael Smith Foundation for Health Research (MSFHR) and CIHR. L.B. is supported by fellowships from MSFHR and CIHR. D.B. has an Australian Postgraduate Award, and T.L. is supported by a grant from the Australian Research Council.
Glossary
Abbreviations
- FNR
ferredoxin NADPH oxido-reductase
- Al
Antonospora locustae
- Ec
Encephalitozoon cuniculi
- Sc
Saccharomyces cerevisiae
- IMP
inner membrane peptidase
- ISC
iron-sulfur cluster
- MPP
mitochondrial processing peptidase
- mtG3PDH
mitochondrial glycerol-3-phosphate dehydrogenase
- Cyt
cytochrome
- TOM
translocase of the outer membrane
- TIM
translocase of the inner membrane.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Wittner M, Weiss LM. The Microsporidia and Microsporidiosis. Washington, DC: Am Soc Microbiol; 1999. [Google Scholar]
- 2.Franzen C, Muller A. Microbes Infect. 2001;3:389–400. doi: 10.1016/s1286-4579(01)01395-8. [DOI] [PubMed] [Google Scholar]
- 3.Edlind TD, Li J, Visvesvara GS, Vodkin MH, McLaughlin GL, Katiyar SK. Mol Phylogenet Evol. 1996;5:359–367. doi: 10.1006/mpev.1996.0031. [DOI] [PubMed] [Google Scholar]
- 4.Hirt RP, Logsdon JM, Jr, Healy B, Dorey MW, Doolittle WF, Embley TM. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keeling PJ. Fungal Genet Biol. 2003;38:298–309. doi: 10.1016/s1087-1845(02)00537-6. [DOI] [PubMed] [Google Scholar]
- 6.Keeling PJ, Doolittle WF. Mol Biol Evol. 1996;13:1297–1305. doi: 10.1093/oxfordjournals.molbev.a025576. [DOI] [PubMed] [Google Scholar]
- 7.Van de Peer Y, Ben Ali A, Meyer A. Gene. 2000;246:1–8. doi: 10.1016/s0378-1119(00)00063-9. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith T. In: Endocytobiology. Schwemmer W, Schenk HEA, editors. Vol 2. Berlin: de Gruyter; 1983. pp. 1027–1034. [Google Scholar]
- 9.Arisue N, Sanchez LB, Weiss LM, Muller M, Hashimoto T. Parasitol Int. 2002;51:9–16. doi: 10.1016/s1383-5769(01)00093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fast NM, Keeling PJ. Mol Biochem Parasitol. 2001;117:201–209. doi: 10.1016/s0166-6851(01)00356-5. [DOI] [PubMed] [Google Scholar]
- 11.Germot A, Philippe H, Le Guyader H. Mol Biochem Parasitol. 1997;87:159–168. doi: 10.1016/s0166-6851(97)00064-9. [DOI] [PubMed] [Google Scholar]
- 12.Hirt RP, Healy B, Vossbrinck CR, Canning EU, Embley TM. Curr Biol. 1997;7:995–998. doi: 10.1016/s0960-9822(06)00420-9. [DOI] [PubMed] [Google Scholar]
- 13.Peyretaillade E, Broussolle V, Peyret P, Metenier G, Gouy M, Vivares CP. Mol Biol Evol. 1998;15:683–689. doi: 10.1093/oxfordjournals.molbev.a025971. [DOI] [PubMed] [Google Scholar]
- 14.Williams BA, Hirt RP, Lucocq JM, Embley TM. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 15.Tovar J, Leon-Avila G, Sanchez LB, Sutak R, Tachezy J, van der Giezen M, Hernandez M, Muller M, Lucocq JM. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- 16.Embley TM, van der Giezen M, Horner DS, Dyal PL, Foster P. Philos Trans R Soc London B. 2003;358:191–201. doi: 10.1098/rstb.2002.1190. discussion 201–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tovar J, Fischer A, Clark CG. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- 18.Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Mol Cell Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truscott KN, Brandner K, Pfanner N. Curr Biol. 2003;13:R326–R337. doi: 10.1016/s0960-9822(03)00239-2. [DOI] [PubMed] [Google Scholar]
- 20.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al. Proc Natl Acad Sci USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 22.Williams BA, Keeling PJ. J Eukaryot Microbiol. 2005;52:271–276. doi: 10.1111/j.1550-7408.2005.05-00036.x. [DOI] [PubMed] [Google Scholar]
- 23.Claros MG, Vincens P. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 24.Claros MG, Brunak S, von Heijne G. Curr Opin Struct Biol. 1997;7:394–398. doi: 10.1016/s0959-440x(97)80057-7. [DOI] [PubMed] [Google Scholar]
- 25.Neupert W. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 26.Pfanner N, Geissler A. Nat Rev Mol Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 27.Wattenberg B, Lithgow T. Traffic. 2001;2:66–71. doi: 10.1034/j.1600-0854.2001.20108.x. [DOI] [PubMed] [Google Scholar]
- 28.Rapaport D. EMBO Rep. 2003;4:948–952. doi: 10.1038/sj.embor.embor937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawlitschek G, Schneider H, Schmidt B, Tropschug M, Hartl FU, Neupert W. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- 30.Arretz M, Schneider H, Guiard B, Brunner M, Neupert W. J Biol Chem. 1994;269:4959–4967. [PubMed] [Google Scholar]
- 31.von Heijne G, Steppuhn J, Herrmann RG. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- 32.Allison DS, Schatz G. Proc Natl Acad Sci USA. 1986;83:9011–9015. doi: 10.1073/pnas.83.23.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roise D, Schatz G. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- 34.Hendrick JP, Hodges PE, Rosenberg LE. Proc Natl Acad Sci USA. 1989;86:4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branda SS, Isaya G. J Biol Chem. 1995;270:27366–27373. doi: 10.1074/jbc.270.45.27366. [DOI] [PubMed] [Google Scholar]
- 36.Hermann GJ, Shaw JM. Annu Rev Cell Dev Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- 37.Gerber J, Neumann K, Prohl C, Muhlenhoff U, Lill R. Mol Cell Biol. 2004;24:4848–4857. doi: 10.1128/MCB.24.11.4848-4857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glick BS, Beasley EM, Schatz G. Trends Biochem Sci. 1992;17:453–459. doi: 10.1016/0968-0004(92)90487-t. [DOI] [PubMed] [Google Scholar]
- 39.Gakh O, Cavadini P, Isaya G. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 40.Nunnari J, Fox TD, Walter P. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- 41.Esser K, Pratje E, Michaelis G. Mol Gen Genet. 1996;252:437–445. doi: 10.1007/BF02173009. [DOI] [PubMed] [Google Scholar]
- 42.Jan PS, Esser K, Pratje E, Michaelis G. Mol Gen Genet. 2000;263:483–491. doi: 10.1007/s004380051192. [DOI] [PubMed] [Google Scholar]
- 43.Esser K, Jan PS, Pratje E, Michaelis G. Mol Genet Genomics. 2004;271:616–626. doi: 10.1007/s00438-004-1011-y. [DOI] [PubMed] [Google Scholar]
- 44.Tuteja R. Arch Biochem Biophys. 2005;441:107–111. doi: 10.1016/j.abb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Bradley PJ, Lahti CJ, Plumper E, Johnson PJ. EMBO J. 1997;16:3484–3493. doi: 10.1093/emboj/16.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hausler T, Stierhof YD, Blattner J, Clayton C. Eur J Cell Biol. 1997;73:240–251. [PubMed] [Google Scholar]
- 47.Regoes A, Zourmpanou D, Leon-Avila G, van der Giezen M, Tovar J, Hehl AB. J Biol Chem. 2005;280:30557–30563. doi: 10.1074/jbc.M500787200. [DOI] [PubMed] [Google Scholar]
- 48.Dolezal P, Smid O, Rada P, Zubacova Z, Bursac D, Sutak R, Nebesarova J, Lithgow T, Tachezy J. Proc Natl Acad Sci USA. 2005;102:10924–10929. doi: 10.1073/pnas.0500349102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Giezen M, Tovar J, Clark CG. Int Rev Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- 50.Schneider A, Oppliger W, Jeno P. J Biol Chem. 1994;269:8635–8638. [PubMed] [Google Scholar]
- 51.Luo W, Fang H, Green N. Mol Genet Genomics. 2006:1–6. doi: 10.1007/s00438-006-0099-7. [DOI] [PubMed] [Google Scholar]
- 52.George R, Beddoe T, Landl K, Lithgow T. Proc Natl Acad Sci USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daum G, Bohni PC, Schatz G. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 54.Lithgow T, Junne T, Wachter C, Schatz G. J Biol Chem. 1994;269:15325–15330. [PubMed] [Google Scholar]
- 55.Beilharz T, Suzuki CK, Lithgow T. J Biol Chem. 1998;273:35268–35272. doi: 10.1074/jbc.273.52.35268. [DOI] [PubMed] [Google Scholar]
- 56.Fast NM, Law JS, Williams BA, Keeling PJ. Eukaryot Cell. 2003;2:1069–1075. doi: 10.1128/EC.2.5.1069-1075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glick BS, Brandt A, Cunningham K, Muller S, Hallberg RL, Schatz G. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]