Abstract
B cells generate varied yet functional clones under high rates of mutation of their V genes. It has been proposed that as a result of the opposing demands of diversification and preservation of integrity, the V genes of heavy and light chains have evolved to overexpress codons prone to amino acid change in their complementarity determining regions (CDR) compared with the framework (FW) regions. We have analyzed the germ-line V genes of heavy and light chains (both κ and λ), comparing codons of CDR and FW of the germ-line V regions both to each other and to control regions. We found that in both germ-line heavy chains and λ chains, CDR codons are prone to replacement mutations, whereas in the FW, the opposite is true. Furthermore, the difference between CDR and FW in heavy chains and λ chains is based on codons that are prone to nonconservative changes of amino acid. In contrast, in germ-line κ chains, the codons in both CDR and FW are more prone to replacement mutations. We also demonstrated that negative selection during immune responses is more sensitive to nonconservative amino acid substitutions than overall amino acid change, demonstrating the applicability of our analysis to real-time process of selection in the immune system. The differences in germ-line κ and λ light chains' potential reaction to mutation suggests that via these two differently evolved light-chain types, the B cell repertoire encompasses two different strategies to balance diversity and stability in an immune response.
Keywords: affinity maturation, B cells, codon usage, selection, evolution
The immune system is capable of responding to nearly any pathogen it encounters. A large part of the system's flexibility and robustness stems from its ability to fine tune its diverse repertoire of antigen receptors through interaction (1). In the case of B cells, diversity is generated in two stages. First, a variety of B cell receptors are encoded in the germ line as gene segments, which undergo a process of random rearrangement to form the assembled receptor gene (2). Second, when a pathogen triggers an immune response, B cells undergo a process of proliferation, death, and mutation of their IgV regions. The mutation rate is high, 10−3 per bp·generation−1, leading to remarkable diversity of receptors among antigen-stimulated B cells during the acute phase of the immune response. During this response, there is selection of B cells with IgV region mutants that confer a high affinity for epitopes on the pathogen (3), a process commonly termed affinity maturation. Despite advances in identifying molecular mechanisms that generate diversity, the exact dynamic of the forces of selection that underlie this process of affinity maturation is not well understood.
Even before the initiation of the immune response, there is extraordinary genetic diversity in the B cell receptor repertoire, owing to somatic recombinations used to construct the B cell receptor. Each receptor is made of a heavy (H) chain and one of the two light (L) chain types, κ or λ. Each of these chains is itself the result of the imprecise and random combinatorial assembly of several gene fragments. Thus, every B cell has a different and, therefore, unpredictable receptor whose specific functional attributes could not be accounted for directly by their germ-line DNA. It is clear that B cell receptors follow certain canonical shapes (4) and that certain V regions have been selected because they are sufficient to confer binding to key recurrent pathogen antigenic motifs (5). Nevertheless, it is also clear that B cell receptors, as a whole, could not have evolved just to recognize specific ligands, because the immune system responds to a changing and unpredictable array of antigens (6). A further distinction between the V region of B cell receptors and other genes is that the DNA-encoding B cell receptors is expected to encounter and withstand high levels of mutation as part of its normal function. It must have evolved to both maximize the value of such point mutations in terms of total diversity and withstand their deleterious effects.
In considering how B cell receptors may have evolved to function in the face of high rates of somatic hypermutation along with their function in binding to a largely unpredictable array of epitopes, it is important to consider how change in genotype relates to a change in phenotype. For a given phenotype there is a neutral network of states in the genotype in which point mutations would not cause a functional change (7). Thus, two individuals with the same phenotype nonetheless can differ in the potential for change that the genotype encodes, i.e., what they can mutate to (8). This potential is determined by the DNA codons and the amino acids (AA) specified in their sequence. Most AAs are coded for by between 2 and 6 codons so that they have a neutral genotypic network of 2, 4, or 6 nodes. Thus, the genetic code shapes the way genotypic change effects AA change. In fact, the genetic code itself appears to have evolved to enhance stability in the face of random mutation (9–11).
In three mutations, a codon has the potential to change to a codon encoding any AA. During the process of B cell receptor affinity maturation, the number of cell divisions is low and, despite the high rate of mutation, it is highly unlikely that a codon will undergo more than one, or at most two, mutations (12). Thus, the precise germ-line DNA sequence encoding each AA in a B cell receptor also determines the phenotypes of its potential progeny after somatic hypermutation. The process of affinity maturation resembles evolution in the sense of recurrent cycles of mutation and selection (although over a very short time scale). However, the process of somatic hypermutation and affinity selection is itself an evolved part of the immune system, and one can assume it has been optimized by evolution for its selective value. Thus, we may expect the substrate on which the process operates, the germ-line sequences, to be optimized in a way that reflects how selection works in affinity maturation. In particular, we hypothesize that the B cell receptor sequences have evolved for viable change after mutation.
The V gene of both H and L chains is commonly divided into framework (FW) subregions that define the basic architecture of the receptor and the complementarity-determining regions (CDR) that encode the parts of the receptor that actually interact with antigen. The FW regions, which form the backbone of the receptor, are more constrained in terms of functionality, whereas the exact amino acid makeup of CDRs is not as important in overall protein folding (4). Indeed, it has been recognized that codons with a higher a priori tendency to mutate to encode a different amino acid [i.e., replacement (R) mutations] are enriched in the CDR compared with the FW (13–16). This predisposition is especially evident in the codon usage of serine (S). In the CDR, S is most often encoded by agt/c, the two codons of S that tend most to R mutations, whereas in the FW, the other codons for S (tcx) are more common (17).
Whereas this bias was shown to be the case for H chains, in L chains the results were unclear and at times contradictory. L chains, and especially κ, exhibited less distinctive differences between CDR and FW (13). Furthermore, κ was shown to have a less strict bias between FW and CDR in the use of agt/c (17). These studies were limited, as they did not sufficiently or equally sample the different L-chain types. Further, the study of S codon usage did not consider λ chains at all (17). Later studies, which looked at larger subsets of κ and λ chain types (15, 16), considered both nucleotide and codon composition to show the effect of the germ-line sequence on potential mutability (16). However, their results were contradictory as regards to κ chains, finding in one study that κ chains did not exhibit a significant difference between CDR and FW (15), but in another one, that both λ and κ showed contrast between CDR and FW similar to that in H chains (16). The ambiguous results regarding L chains call for a more rigorous measurement of the potential for change in the CDR and FW, because the contrasting of CDR and FW may not be the only way V genes have evolved under the opposing demands of variability and viability.
These previous studies used the FW as a control for results in the CDR. They could only test the possibility that these two regions have evolved differently from each other in terms of their reaction to mutation and ignored, therefore, the distinct possibility of selective forces operating on FWs. Two main elements were missing from previous research: first, a comparison of CDR and FW to other genes that were not subjected to somatic hypermutation, and second, a test of the tendency for different types of R mutation, both conservative and nonconservative.
Here, we have analyzed the codon usage of CDR and FW in H and L chains. Using a novel network view of the genetic code, we compared CDR and FW to each other and to CD8, a L-chain homolog that does not undergo somatic hypermutation. We also compared the codon usage in these different regions to the general codon usage in the human genome. For H chains, we corroborated the prior findings that the codon used in CDRs are more prone to change, whereas those used in the FW regions are more stable. We expected to find the same relationship of CDR and FW for L chains as in H chains. However, to our surprise, the two types of L chains (λ and κ) exhibit different relationships between codon usage in the CDR and FW. When compared with the general codon usage of the human genome, λ chains have significantly more changeable CDRs and less changeable FW regions, whereas κ chains are more changeable in both CDR and FW. Moreover, and as was hitherto unnoticed, the contrast between CDR and FW, which is found in H chains and in λ chains, does not correlate merely with codons more prone to R mutation but, in fact, with nonconservative R mutations. To more clearly identify the importance of different magnitudes of AA change, we analyzed the mutation profile in sequences taken from B cell mutants at days 10 and 16 of a λ-based immune reaction. As in our analysis of the germ line, we calculated the extent of nonconservative mutation separately from AA replacements in general. We found that the FWs of these mutants showed a marked selection against nonconservative mutations and not against R mutations in general.
Our results suggest that the immune system has two different strategies to balance generation of diversity with the maintenance of functionality during the process of somatic hypermutation and selection. Mutation in H and λ chains will tend to generate diverse mutants while maintaining their viability, whereas mutation in κ chains will lead to mutants with greater diversity but with less control of the magnitude of the effect of their mutations or their potential viability.
Results and Discussion
The Codon Network.
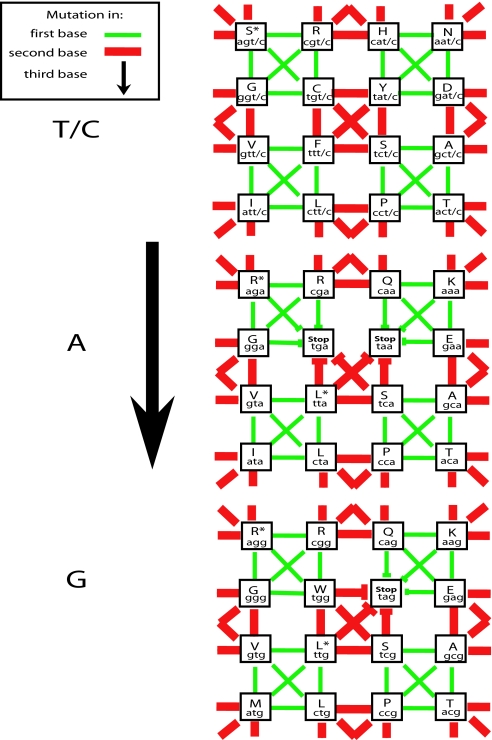
The nucleotide triplets that comprise the genetic code can be plotted on the corners of a hyper cube (18). Different regions of the cube tend to share traits relevant to the AA's function (10, 18, 19). Codons differing only in the third nucleotide usually encode the same AA, codons differing in the first nucleotide often encode AAs of similar traits (such as hydrophobicity), and codons differing in the second nucleotide share the same common precursor from which the AA is synthesized (10, 19). We consider this hypercube as a network, in which every codon is a node, and an edge is a mutation of one of its bases (Fig. 1).
Fig. 1.
The codon network. Represented here in three levels is the genetic code in the form of a network of codons. Such a view emphasizes the different potential for change that two codons encoding the same AA can have. Every node has three kinds of edges, those that are the result of a mutation in the first base (green), those that are the result of a mutation of the second base (red), and those that are the result of a mutation in the third or wobble base (up/down). For the encoding of AAs, the third nucleotide is completely redundant if it is a thymidine (t) or a cytosine (c). The network therefore contains only 48 nodes. Nodes that have AA denoted with an “∗” are codons encoding an AA with six codons that differ from the rest of the codons for that AA by the first nucleotide [arginine (R) and leucine (L)] or by both the first and second nucleotides (S). Nodes denoted with a “stop” are stop codons. The network is unbounded, movement off the left end will lead to the right end of a network, and movement off the top leads to the bottom. Each node is also connected to all of its equivalent nodes in the levels above and below it.
Not all nodes in the codon network are equally interconnected. First, not all nodes have the same number of edges, because some are adjacent to stop codons and, thus, have fewer viable mutational options. Second, because some AAs have more codons, movement between nodes is not equally likely to lead to changes in AAs. Furthermore, meaningful changes after mutation, in the AAs encoded, can be defined in many ways, because not all changes in AA are equal.
We considered two measures of potential for change in codons and the AAs they encode:
AA changeability: the chance that a mutation of a single nucleotide will change the encoded AA.
Trait changeability: the chance for a given codon that a mutation of a single nucleotide will be nonconservative and result in a codon encoding a different AA whose properties are radically different from the original AA. There are several ways to define the traits of AAs, including hydrophobicity, polarity, and size. Because we are studying IgV regions, we decided to use the trait definitions derived from the study of multiple variable domains of immunoglobulins (20). In this study, AAs were clustered by their hydrophobicity and their general tendency to be on the surface of the receptor. This resulted in a division of all AAs to three trait groups Hydrophobic/Buried, Neutral/Intermediate, and Hydrophilic/Surface (20). The distribution of AAs into these three groups is shown in Fig. 4, which is published as supporting information on the PNAS web site. For the purpose of our analysis, we consider all trait changes to be of equal size.
In calculating both changeability scores, we have taken into account that the likelihood of all mutations is not equal. There is a 2-to-1 transition bias (i.e., purine to purine and pyrimidine to pyrimidine) in somatic hypermutation (21).
Analyzing Germ-Line Sequences Through the Lens of the Codon Network.
We compared the AA changeability of CDR and FW in H chains, λ and κ L chains, and the L chain homolog CD8 to the expected changeability based on the codon usage of the entire human genome. As previous studies led us to predict (13–17), we found that the CDRs in H chains and in both types of L chain are significantly more changeable than expected from the human genome. We also found that the changeability of the CDRs of CD8 is not significantly different from the human genome as a whole, demonstrating that heightened changeability is not merely a property of CDR-like regions of an IgV-like domain (Table 1). In agreement with the conventional view of the relationship of CDR and FW, we found that the FW regions of H chains and λ chains are significantly less changeable than expected compared with the human genome. However, in κ chains, the FW regions are only slightly less changeable than the CDR and are significantly more changeable than expected compared with the human genome (Table 1). Thus, compared with λ chains, κ chains will generate mutant progeny during the antigen-driven immune response with greater variability in their receptors. However, because a large fraction of R mutations in FW will adversely affect V region folding and, thus, B cell receptor function (22), κ chains also will generate more nonfunctioning receptors.
Table 1.
Different regions in V genes exhibit significant differences from the expected changeability of the human genome
| Gene type | AA changeability |
Trait changeability |
|||||
|---|---|---|---|---|---|---|---|
| Region | erf(x)* | P† | Dir.‡ | erf(x) | P | Dir. | |
| λ chain | CDR | 0.999 | <10−3 | (+) | 0.998 | <10−3 | (+) |
| FW | −0.999 | <10−3 | (−) | −0.999 | <10−3 | (−) | |
| κ chain | CDR | 0.999 | <10−3 | (+) | 0.949 | 0.026 | (+) |
| FW | 0.905 | 0.050 | (+) | 0.950 | 0.025 | (+) | |
| H chain | CDR | 0.996 | 0.002 | (+) | 0.996 | 0.002 | (+) |
| FW | −0.997 | 0.001 | (−) | −0.641 | 0.179 | (NS) | |
| CD8 | CDR | 0.396 | 0.302 | (NS) | 0.169 | 0.415 | (NS) |
| FW | 0.468 | 0.266 | (NS) | 0.835 | 0.080 | (NS) | |
Human codon usage was calculated from a sample of 84,949 coding sequences (36,349,745 codons), www.kazusa.or.jp/codon.
*erf(x) is calculated as explained in Materials and Methods.
†P value is calculated as explained in Materials and Methods.
‡Direction (Dir.) of difference from expected from human genome: +, significantly more changeable; −, significantly less changeable; NS, not significantly different.
The results of the above analysis were the same when considering trait changeability. This result is not surprising because the comparison of the overall changeability to the expected changeability is influenced mostly by those AAs that have multiple codons with differing changeability scores, most prominently S, arginine, and leucine, which are each encoded by six different codons (23). This limitation does not create a problem in tracking changeability in the L chains, because S is plentiful. However, among the codons that encode S, those codons that are the most prone to AA change are also most prone to trait change, accounting for an inevitable concordance between the two measures in this instance.
Consistent with the above results we found that, although the bias exists toward encoding of S by the most changeable codons (agt/c) in the CDR and not in the FW, it is much less pronounced in FW of κ chains than in the FW of λ chains. Strikingly, whereas overall levels of S encoding codons are similar in both types of L chain, the prevalence of agt/c is 5-fold higher in the FW of κ chains compared with the FW of λ chains (χ2, 19.427; P = 1 × 10−5; see Table 4, which is published as supporting information on the PNAS web site).
To differentiate between the influence of trait and AA changeability described above, we calculated Pearson correlations between the AA and trait changeability scores and the frequencies of codons in CDR vs. FW of V regions and in CDR of V regions vs. CDR of CD8 (Table 2). In H chains and in λ chains, we found there was significant correlation between trait changeability and the relative frequency of codons. Such a correlation was not found with AA changeability. Therefore, we concluded that trait changeability is the distinguishing feature of CDR and FW in H chains and in λ chains.
Table 2.
Pearson correlation between changeability scores of codons and their relative frequency
| Gene type | Condition* | AA changeability correlation† | Trait changeability correlation |
|---|---|---|---|
| λ chain | CDR-FW | 0.175 | 0.283 |
| CDR-CDRCD8 | 0.104 | 0.315‡ | |
| λ-CD8 | 0.174 | 0.293 | |
| κ chain | CDR-FW | 0.281 | 0.240 |
| CDR-CDRCD8 | 0.238 | 0.279 | |
| κ-CD8 | 0.247 | 0.265 | |
| H chain | CDR-FW | 0.224 | 0.329 |
| CDR-CDRCD8 | 0.189 | 0.345 | |
| H-CD8 | 0.221 | 0.308 |
*Three relative frequencies of codons: CDR-FW, of the same gene type; CDR-CDRCD8, CDR of a given gene type: minus the CDR of CD8; λ/κ/H-CD8, the interaction of both differences between CDR and FW and the CDR of the V genes and that of CD8 [i.e. λ (cdr–fw)–CD8(cdr–fw), similarly for κ and H chains].
†For a two-tailed test Pearson correlation is significant when it is <0.288 (α< 0.05).
‡Results that are significant are in bold.
With regard to correlation between changeability and codon usage, we found that κ chains again differed from H and λ chains. κ chains did not exhibit significant correlations between the frequency of codon usage and the AA or trait changeability scores. Despite that the κ FW is markedly more changeable, it does not exhibit a tendency for trait change over general AA changeability (Table 2). The lack of difference could well be because in κ chains, CDR and FW are not as different in terms of changeability. It is interesting to note that in κ chains, although neither AA nor trait changeability is significantly correlated to relative codon frequency, both show similar levels of correlation just below significance. In λ chains, on the other hand, only correlation to trait changeability is significantly correlated to codon frequency, and this correlation is much stronger than that of AA changeability (Table 2). This result suggests that κ chains are primed for change, in general, after mutation but are not specially selected to maximize the impact of such change, as H chains and λ chains are.
Transition Neighborhoods.
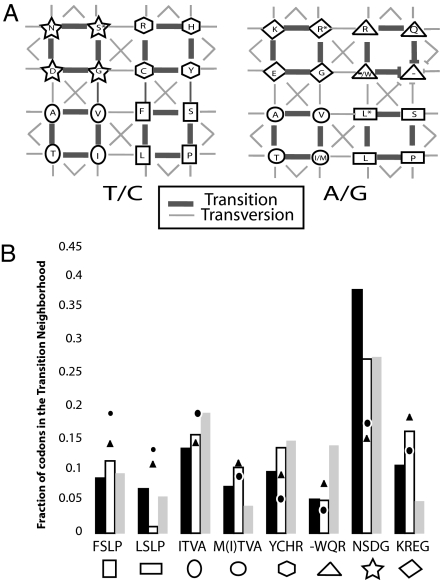
The bias of mutation toward transition mutations is also present in meiotic mutation (21), which has implications beyond the correct calculation of changeability scores. When the positive bias for transition mutations is taken into account, the general stabilizing tendencies of the genetic code are even more pronounced (10, 18). We divided the codon network into eight “transition neighborhoods.” Each neighborhood is comprised of eight codons that are connected by transition mutations. Because of the transition bias, codons in the same neighborhood will tend to mutate into each other more often than into codons outside their neighborhood, thus further segregating the types of mutants a given codon is likely to generate. We refer to specific neighborhoods of codons by the AAs they encode. For instance, the neighborhood that includes the codons that encode for asparagine (N), S, aspartic acid (D), and glycine (G) is called NSDG (Fig. 2A).
Fig. 2.
Transition neighborhoods. (A) In this image of the codon network, every node represents a pair of codons that differ only by a transition mutation in the third base. The codons in the left network end with t/c and in the right with a/g. Nodes show AAs because all such third base transition mutations are silent except M→I, whose representative node is shown as I/M. Stop codons are shown as “−,” and the fifth and sixth codon of an AA is represented by ∗. Transition mutations of either the first or second bases are represented by thick edges, and transversion mutations are represented by thin edges. The four codon couplets that are connected through transition mutations are considered to be in the same transition neighborhood. All nodes in the same neighborhood share the same enclosing symbol. (B) Distribution of codons from the CDR into the different transition neighborhoods. Each neighborhood is designated by the AAs it encodes and by its symbol as given in Fig. 2A. Results from the CDR of human L chains λ (black) and κ (gray) and H chains (white) are compared with those of CD8 (circle) and to overall levels in the human genome (triangle).
We compiled the distribution of codons across transition neighborhoods in the CDRs of H chains, L chains, and CD8. We found that codons from the NSDG neighborhood were overrepresented in the CDRs of H chains and λ chains compared with those of CD8 and overall levels in the human genome (Fig. 2B). NSDG codons also are overrepresented in κ chains, although to a lesser extent.
The overrepresentation of codons from a given transition neighborhood can make random mutations less disruptive and help preserve the overall antibody fold. During somatic hypermutation, codons are mutated usually once at most. Therefore, a mutated chain position generally will remain in its original transition neighborhood. This segregation to part of the genetic code is further stabilizing if we take into consideration that the genetic code puts amino acids that share certain properties in the same transition neighborhood. For instance, NSDG all favor non-α-helix and non-β-sheet secondary structure (24), which may serve to ensure that the mutated CDR still are able to fold into a stable structure.
At first glance, this segregation to a specific transition neighborhood may appear to contradict our conclusions regarding trait changeability being reinforced in H chains and λ chains. However, the specific neighborhood that is overrepresented, NSDG, is the most prone to AA changeability and the second-most prone-to-trait change. On the other hand, NSDG is at least two mutations away from any of the stop codons. In addition arginine, which is involved in many autoreactive receptors (14), is not reachable by a transition mutation. Furthermore, the trait changes that can happen after transition mutations in NSDG are not from hydrophilic to hydrophobic but only to intermediate AAs. The overrepresentation of the NSDG transition neighborhood suggests that H chains and λ chains strike a balance in generating meaningfully different receptors after mutation, ensuring that receptors that have not changed enough or have become nonfunctional will be less likely to be generated by mutation.
We have so far focused on how the FW of κ chains is different from that of λ and H chains. With regard to transition neighborhoods, the CDR is also different between the two L-chain types. In addition to the overrepresentation of NSDG, κ chains also exhibit a higher fraction of codons from the WQR neighborhood compared with both λ chains and CD8s (see Fig. 2B). Having codons in different transition neighborhoods will lead to a greater variety in κ-chain mutants. However, WQR includes codons for arginine, and all but one of its codons are a mutation away from stop codons; therefore, κ chains are more likely to have lethal or ineffectual mutations than λ chains, in which only NSDG is overexpressed. In this context, it is important to consider the relationship between glutamine (Q) and N. Previous studies have noted that the CDRs of H chains exhibit a strong bias toward N (25). Q and N, although distinct, often are found to be substituted when comparing aligned sequences of proteins with known homology. They have related chemical properties, and a priori there is no reason why one should be preferred over another in a random repertoire of V regions that in general is not selected for particular binding specificities (25). However, from the perspective of viability after mutation, Q is a transition mutation away from stop codons and N is two mutations away. As expected from the distribution of codons to transition neighborhoods, H and λ chains exhibit a strong bias toward N (14 to 1 and 3 to 1, respectively), whereas in κ chains, the CDR shows the opposite trend (3 to 5, χ2, 7.984; P = 0.005; see Table 4). Once more, we find that κ generates greater diversity but is more likely to have a lethal mutation, this time in the CDR.
Our results indicate that, after mutation, B cell receptors with κ chains will generate greater variability than those with λ chains. However, much of the variability of the repertoire of κ chain mutants will consist of cells that do not express receptors that are meaningfully different from the original clone. This limited functional variability could happen either because the κ chain mutants are not significantly different from their progenitors because of a lower frequency of R mutations being nonconservative or because they are nonfunctional. Nonfunctional mutants can arise either because of mutations to stop codons in the CDR or mutations in the FW region that result in an unfoldable receptor.
We speculate that the B cell repertoire can withstand the lower viability of κ-chain mutants because of the differences in gene organization between κ chains and λ chains (26). In λ chains, the different families of the germ-line genes are clearly separated (27). In κ chains, the different families are closely related with specific members of different families, typically showing a similarity of >80% (26, 28). This difference between κ and λ predates the separation of the orders to which mice and men belong (27–29). Thus, the repertoire of responding B cells is more resilient to the loss of a specific κ-chain mutant because it is likely to have one with a similar V region on hand.
Trait Change Analysis During the Process of Selection.
We tested whether measures of changeability could provide insight into real-time selection of V region mutants in the germinal center (GC). We have been studying an immune response to the (4-hydroxy-3-nitrophenyl)acetyl (NP) hapten in two types of B cell-receptor transgenic mice, V23 and B1-8, in which fixed, nonmutable H chains are expressed in every B cell. In combination with an endogenous λ chain, found in 3% of B cells, the V23 H chain has a low affinity for NP, whereas the B1-8 H chain has a high innate affinity (30, 31). These mice are an ideal substrate for testing our measures of selection because all affinity-based selection occurs in λ chains, canceling the need to analyze the effects on separate H+L chains simultaneously, giving a clearer signal on which to test our measures of selection on somatic hypermutation.
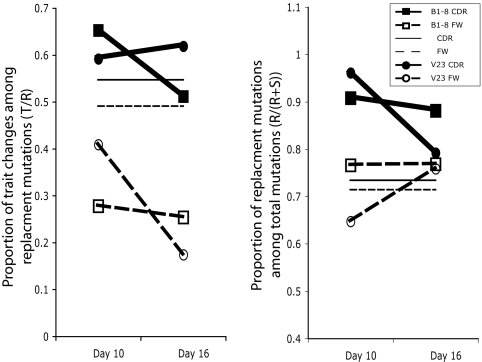
We analyzed the CDR and FW of these L chains at days 10 and 16 after infection and compared the number of mutations of different types to the expected distribution of mutations of these types. In the CDR, we found that the fraction of R mutations out of total mutations was significantly above the random expected value. We only see purification of trait changes over time in low-affinity V23 mice. It appears that only where very low affinity pushes selection to the extreme, positive selection for trait change beyond overall change is evident even over a 16-day span. In the FW region, we found that trait changes were actively being selected against, whereas R mutations in general were not being selected against (Fig. 3 and Table 3).
Fig. 3.
Fraction of AA and trait changes in mouse λ chains 10 and 16 days after immunization. (Left) Fraction of nonconservative trait changing mutations out of R mutations. (Right) Fraction of R mutations out of total. The lines marked CDR and FW represent the level of mutations expected.
Table 3.
Selection for trait change mutations compared with other replacement mutations
| Mouse type | Day | Region | T/R* |
R/(R + S)† |
||
|---|---|---|---|---|---|---|
| Frq.‡ | P§ | Frq.¶ | P | |||
| V23 | 10 | CRD | 0.59 | 0.171 | 0.96 | <10−3 |
| FW | 0.41 | 0.356 | 0.65 | 0.369 | ||
| 16 | CDR | 0.62 | 0.036‖ | 0.79 | 0.031 | |
| FW | 0.17 | <10−3 | 0.76 | 0.115 | ||
| B1-8 | 10 | CDR | 0.65 | 0.018 | 0.91 | <10−3 |
| FW | 0.28 | 0.01 | 0.77 | 0.122 | ||
| 16 | CDR | 0.51 | 0.742 | 0.88 | <10−3 | |
| FW | 0.25 | <10−3 | 0.77 | 0.111 | ||
*Numberof nonconservative trait change mutations out of total replacement.
†Number of replacement mutations out of total mutations.
‡The expected frequency (Frq.) T/R, based on the mouse λ V1J1 germ line, is 0.55 in the CDR and 0.49 in the FW.
§In both cases P is the result of a one-tailed binomial test (α< 0.05).
¶The expected frequency R/(R + S) is 0.73 in the CDR and 0.71 in the FW.
‖Results that are significant are in bold.
These unexpected divergences of selection, for or against trait but not AA change, indicated that trait differences could be a meaningful signal for the process of selection in the germinal center. These results suggest that trait changeability measures could be an important tool for describing the levels of negative and positive selection in affinity maturation. They also corroborate our findings, showing that the germ-line sequences indeed have evolved to reflect changes in AA that are relevant to the actual process of somatic selection. Finally our results show that differentiating between AA by their traits adds to our understanding of the phenotype being selected for. It is important to note that we do not suggest that the traits we have distinguished in V regions are applicable universally. The relevant traits of AAs depend on the phenotype being selected and are specific to the kinds of AA interactions relevant to the specific biological process under study. This relationship must be considered to incorporate the traits of AAs into future research.
The Importance of the Codon View.
Based on the codon view, our results show how V regions have evolved to balance the immune system's requirement for variable B cell receptors with the need to have enough viable mutants under high levels of mutation. The full complexity of this balance in both CDR and FW cannot be understood by nucleotide analysis alone. Recent studies of H chains have suggested that in the CDR, c nucleotides are positioned preferentially so that they will result in silent mutations (25). In effect, this positioning implies the use of codons ending in t/c, because such nucleotides will cause only R mutations at this position in the less likely case of a transversion mutation. It was reasoned that such a bias will downplay the greater tendency of somatic mutation to occur in c compared with a nucleotides (25). However, this overexpression of codons with t/c in the third position also fits with the overexpression of codons from the NSDG neighborhood that we describe. By taking into consideration the codon usage of the different V genes, we find that the c nucleotide bias in L and H chains goes hand in hand with the usage of codons that are in general more prone to R mutations. Thus, the overexpression of codons with silent mutations in c/t is not merely a mechanism to reduce the propensity for change in c but, in addition, is balanced by the use of more changeable codons and AAs.
A Functional Explanation for the Differences Between λ and κ Chains.
The differences we have described between λ chains and κ chains suggest that the balancing of variability and viability of V-region mutants has evolved in more than one way. The fact that both L-chain families are found in many different immune systems (32) implies that they may have been selected to suit specific needs of the immune system. This multiplicity is possible, because the immune reaction is the result of the selection of a few clones from many participants. Affinity maturation is not at the level of the single clone but rather reflects changes in the entire B cell repertoire, which includes clones with either type of L chain.
Models of affinity maturation have represented the movement toward maximal affinity of interaction between receptor and antigen as movement on an affinity landscape that occurs on a rough landscape, with many local minima and maxima of affinity, or a smooth landscape, rising to a clear maxima (33). The differences in codon composition of κ and λ and, thus, their responses to mutation, imply that during a typical response, evolution of receptors is occurring on two generally different types of landscapes. λ chains mutate in measured steps of a certain size and mediate a search in a smooth landscape. κ chains take more uneven steps. Some κ chains mutate to radically different shapes but many mutants remain practically unchanged, close to the original germ line in affinity. These kinds of changes describe a search in a rougher landscape in which there is a need to escape local minima. Which strategy would be more effective cannot be predicted a priori and would depend on the initial B cell and the Ag. Using a repertoire that includes both κ or λ chains in tandem, the immune response can search simultaneously the shape space of antigens at both a rough and a smooth level of acuity.
Methods
Source of Genomic Data.
All germ-line sequences were taken from the IMGT database (27; http://imgt.cines.fr). To eliminate bias from subgroup family size differences, we selected one sequence at random from each of the subgroups of the H- or L-chain families. These sequences represented 7 subgroups for H chains, 10 subgroups for λ, and 5 subgroups for κ (Data Set 1, which is published as supporting information on the PNAS web site). We based our division of the L chains into CDR and FW on Kabat et al. (34). The sequence data from actual stages of the process of selection is from unpublished sequences derived from microdissection of splenic germinal centers in B1-8 and V23 Ig transgenic mice engineered in the Shlomchik laboratory (30, 31).
We have repeated the analyses described above on similarly selected representative sequences of murine λ and κ V genes and on the entire repertoire of human κ and λ V genes (sequences in FASTA format in Data Sets 2 and 3, which are published as supporting information on the PNAS web site). The results of these analyses fit well with our results presented here (Fig. 5 and Tables 5–7, which are published as supporting information on the PNAS web site).
Statistical Methods.
Comparing overall changeability.
By following Plotkin et al. (35), we defined the changeability of CDR or FW [C(g)] as the summed changeability of their codons [C(i)]. We then adapted their statistical test (as described in Supporting Methods, which is published as supporting information on the PNAS web site) to determine whether the overall changeability of CDR and FW regions was significantly elevated or depressed compared with the rest of the human genome while controlling for the length and AA composition.
Pearson correlation.
To measure how codon usage is related to codon changeability, we calculated whether there was a significant Pearson correlation between the changeability scores (AA or trait) of a codon and the frequency of that codon's usage. To evaluate the differences between CDR and FW and between CDR in L chains and in CD8, we calculated Pearson correlations with the following three relationships of codon usage.
The relative frequency of a codon in CDR vs. FW.
The relative frequency of a codon in the CDR of an H or L chain vs. the CDR of CD8.
The interaction of these two frequencies [(CDR–L chain FW) – (CD8 CDR–CD8 FW)].
Analysis of Numbers of Mutations After Somatic Hypermutation.
We analyzed the CDR and FW of the λ L chains counting the number of independent mutations of a given type (R or T), i.e., in how many different germinal centers a certain mutation occurred. We thus could treat mutations in CDR and FW independently from each other and test each region with a separate binomial test (36), which compared the number of independent mutations to the expected levels, based on the germ line of mouse λ chains.
Supplementary Material
Acknowledgments
We thank Gunter Wagner, Michael Levitt, Anat Ninio, Martin Weigert, and John Jungk for insightful comments given during the process of writing this manuscript. U.H. is a recipient of the Pharmaceutical Research and Manufacturers of America Foundation postdoctoral fellowship in informatics, and M.J.S. is funded in part by National Institutes of Health Grant R01 AI43603.
Abbreviations
- AA
amino acid
- CDR
complementarity determining regions
- FW
framework
- H
heavy
- L
light
- N
asparagine
- NSDG
asparagine, serine, aspartic acid, and glycine
- R
replacement
- S
serine.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hershberg U, Efroni S. Complexity. 2001;6:14–21. [Google Scholar]
- 2.Weigert M, Gatmaitan L, Loh E, Schilling J, Hood L. Nature. 1978;276:785–790. doi: 10.1038/276785a0. [DOI] [PubMed] [Google Scholar]
- 3.Siskind GW, Benacerraf B. Adv Immunol. 1969;10:1–50. doi: 10.1016/s0065-2776(08)60414-9. [DOI] [PubMed] [Google Scholar]
- 4.Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, Sheriff S, Padlan EA, Davies D, Tulip WR, et al. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 5.Conger JD, Sage HJ, Kawaguchi S, Corley RB. J Immunol. 1991;146:1216–1219. [PubMed] [Google Scholar]
- 6.Burrnet FM. Aust J Sci. 1957:67–69. [Google Scholar]
- 7.Smith JM. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 8.Stadler BM, Stadler PF, Wagner GP, Fontana W. J Theor Biol. 2001;213:241–274. doi: 10.1006/jtbi.2001.2423. [DOI] [PubMed] [Google Scholar]
- 9.Knight RD, Freeland SJ, Landweber LF. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 10.Aita T, Urata S, Husimi Y. J Mol Evol. 2000;50:313–323. doi: 10.1007/pl00020999. [DOI] [PubMed] [Google Scholar]
- 11.Freeland SJ, Knight RD, Landweber LF, Hurst LD. Mol Biol Evol. 2000;17:511–518. doi: 10.1093/oxfordjournals.molbev.a026331. [DOI] [PubMed] [Google Scholar]
- 12.Kleinstein SH, Louzoun Y, Shlomchik MJ. J Immunol. 2003;171:4639–4649. doi: 10.4049/jimmunol.171.9.4639. [DOI] [PubMed] [Google Scholar]
- 13.Chang B, Casali P. Immunol Today. 1994;15:367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shlomchik MJ, Aucoin AH, Pietsky DS, Weigert MG. Proc Natl Acad Sci USA. 1987;84:9150–9154. doi: 10.1073/pnas.84.24.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kepler TB. Mol Biol Evol. 1997;14:637–643. doi: 10.1093/oxfordjournals.molbev.a025803. [DOI] [PubMed] [Google Scholar]
- 16.Oprea M, Kepler TB. Genome Res. 1999;9:1294–1304. doi: 10.1101/gr.9.12.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner SD, Milstein C, Neuberger MS. Nature. 1995;376:732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez-Montano MA, de la Mora-Basanez CR, Poschel T. BioSystems. 1996;39:117–125. doi: 10.1016/0303-2647(96)01605-x. [DOI] [PubMed] [Google Scholar]
- 19.Copley SD, Smith E, Morowitz HJ. Proc Natl Acad Sci USA. 2005;102:4442–4447. doi: 10.1073/pnas.0501049102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chothia C, Gelfand I, Kister A. J Mol Biol. 1998;278:457–479. doi: 10.1006/jmbi.1998.1653. [DOI] [PubMed] [Google Scholar]
- 21.Oprea M, Cowell LG, Kepler TB. J Immunol. 2001;166:892–899. doi: 10.4049/jimmunol.166.2.892. [DOI] [PubMed] [Google Scholar]
- 22.Davies DR, Metzger H. Annu Rev Immunol. 1983;1:87–117. doi: 10.1146/annurev.iy.01.040183.000511. [DOI] [PubMed] [Google Scholar]
- 23.Hahn MW, Mezey JG, Begun DJ, Gillespie JH, Kern AD, Langley CH, Moyle LC. Nature. 2005;433:E5–E6. doi: 10.1038/nature03221. discussion E7–E8. [DOI] [PubMed] [Google Scholar]
- 24.Levitt M. Biochemistry. 1978:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 25.Zheng NY, Wilson K, Jared M, Wilson PC. J Exp Med. 2005;201:1467–1478. doi: 10.1084/jem.20042483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strohal R, Helmberg A, Kroemer G, Kofler R. Immunogenetics. 1989;30:475–493. doi: 10.1007/BF02421180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, Ginestoux C, Clement O, Chaume D, Lefranc G. Nucleic Acids Res. 2005;33:593–597. doi: 10.1093/nar/gki065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirschbaum T, Röschenthaler F, Bensch A, Hölscher B, Lautner-Rieske A, Ohnrich M, Pourrajabi S, Schwendinger J, Zocher I, Zachau HG. Eur J Immunol. 1999;29:2057–2064. doi: 10.1002/(SICI)1521-4141(199907)29:07<2057::AID-IMMU2057>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 29.Klein R, Jaenichen R, Zachau HG. Eur J Immunol. 1993;23:3248–3271. doi: 10.1002/eji.1830231231. [DOI] [PubMed] [Google Scholar]
- 30.Hannum LG, Haberman AM, Anderson SM, Shlomchik MJ. J Exp Med. 2000:931–942. doi: 10.1084/jem.192.7.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dal Porto JM, Haberman AM, Kelsoe G, Shlomchik MJ. J Exp Med. 2002;195:1215–1221. doi: 10.1084/jem.20011550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hood L, Gray WR, Sanders BG, Dreyer WJ. Cold Spring Harbor Symp Quant Biol. 1967;32:133–146. [Google Scholar]
- 33.Perelson AS, Weisbuch G. Rev Mod Phys. 1997;69:1219–1267. [Google Scholar]
- 34.Kabat EA, Wu TT, Reid-Miller M, Perry H, Gottesman K. Sequences of Proteins of Immunological Interest. Washington, DC: US Govt Printing Off; 1987. pp. 165–492. [Google Scholar]
- 35.Plotkin JB, Dushoff J, Fraser HB. Nature. 2004;428:942–945. doi: 10.1038/nature02458. [DOI] [PubMed] [Google Scholar]
- 36.Hogg RV, McKean JW, Craig AT. Introduction to Mathematical Statistics. Edgewood Cliffs, NJ: Prentice–Hall; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.