Abstract
Host-defense cationic antimicrobial peptides (≈12–50 aa long) play an essential protective role in the innate immune system of all organisms. Lipopeptides, however, are produced only in bacteria and fungi during cultivation, and they are composed of specific lipophilic moieties attached to anionic peptides (six to seven amino acids). Here we report the following. (i) The attachment of an aliphatic chain to otherwise inert, cationic D,L tetrapeptides endows them with potent activity against various microorganisms including antibiotic resistance strains. (ii) Cell specificity is determined by the sequence of the short peptidic chain and the length of the aliphatic moiety. (iii) Despite the fact that the peptidic chains are very short, their mode of action involves permeation and disintegration of membranes, similar to that of many long antimicrobial peptides. Besides adding important information on the parameters necessary for host-defense lipopeptides to kill microorganisms, the simple composition of these lipopeptides and their diverse specificities should make them economically available, innate immunity-mimicking antimicrobial and antifungal compounds for various applications.
Keywords: antimicrobial peptides, innate immunity, peptide–membrane interaction, carpet model, lytic peptides
The increasing resistance of bacteria and fungi to available antibiotics is a major concern worldwide, leading to enormous efforts to develop new antibiotics with new modes of action. Two promising families of drugs that meet these criteria are host-defense cationic antimicrobial peptides (AMPs) and lipopeptides. AMPs are produced by all species of life and represent key components of the innate immune system, providing a fast acting weapon against invading pathogens including bacteria, fungi, and yeast (1–4). Importantly, many AMPs physically and rapidly permeate and destroy the cell membrane, causing damage hard to fix, in contrast to conventional antibiotics, which act on specific targets such as enzymes or DNA. Therefore, microbial resistance may occur with lower probability than that observed with available antibiotics (2). Structurally, AMPs are ≈12–50 aa long, carry a net positive charge of >2, and are composed of ≈50% hydrophobic amino acids. Many of them adopt an amphipathic structure only in membrane environments, which is considered to be a prerequisite for their lytic activity (5, 6). The diversity in AMPs' lengths, sequences, structures, and spectra of activities is matched by a number of models attempting to describe and explain their modes of action (7–10). There is a consensus, however, that one major step in the activity of cationic AMPs is their initial binding to the negatively charged lipopolysaccharide (LPS) of Gram-negative bacteria or to the lipoteichoic acid of Gram-positive bacteria. AMPs then traverse into the inner phospholipid membrane [highly enriched in phosphatidylglycine (PG)] and permeate it. In fungi, the peptides bind to the negatively charged membrane phosphatidylinositol (PI) and/or to the cell wall, which is composed of polybranched (1,3)-β-d-glucan (11–14). Many AMPs lyse membranes via the “carpet” mechanism, in which the peptide accumulates on the surface of the membrane until a threshold concentration has been reached; membrane permeation and/or disintegration in a detergent-like manner (10, 15) follow. If long enough, AMPs can form toroidal-like pores (16).
Lipopeptides differ from AMPs in that they are produced only in bacteria and fungi during cultivation on various carbon sources (11, 12). They are composed of a specific lipophilic moiety attached to an anionic peptide (six to seven amino acids). Some of them destabilize the membrane by unknown mechanisms (17–19). Unfortunately, native lipopeptides belonging to this group are non-cell-selective and therefore toxic to mammalian cells, too. Despite this toxicity, a member of this family, daptomycin, which is active only toward Gram-positive bacteria, was recently approved by the Food and Drug Administration (FDA) for the treatment of complicated skin and skin structure infections (20).
Recently, we reported that the conjugation of aliphatic acids to the N terminus of native or designed AMPs endowed them with selective antimicrobial, antifungal, or both antimicrobial and antifungal activities (14, 21–23). These studies revealed that a threshold of hydrophobicity and a defined structure of peptidic moiety are required for antimicrobial activity. Intriguingly, here we report on a family of ultrashort lipopeptides with potent antimicrobial activity against a variety of microorganisms. They are composed of only four D,L amino acids conjugated to aliphatic acids with different chain lengths. The sequence of the peptidic moiety and the length of the aliphatic acid determine their spectra of antimicrobial activity. Importantly, despite their short peptidic chain length, their mode of action involves the permeation and disintegration of the membrane, similar to that of many classical AMPs.
Results
We synthesized a series of lipopeptides composed of four L and D amino acids linked to aliphatic acids with different lengths. The sequence of the peptidic moiety was KXXK (X here represents L, A, G, K, or E). A peptide comprising the four N-terminal amino acids of magainin served as a negative control. All of the peptides were amidated at their C terminus, and one of their amino acids was replaced with the d-enantiomer. Table 1 lists the lipopeptides investigated.
Table 1.
Antimicrobial MICs of the ultrashort lipopeptides (μM) and their hemolytic activity
| Peptide designation |
E.c. |
P.a. 27853 | S.a. 6538P | A.b. 19606* | A.fu. 26430 | A.fl. 9643 | C.a. 10231 | C.n. MYA-422 | Hemolysis, % |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D21 | 25922 | At 20 μM | At 50 μM | ||||||||
| C16-KLLK | 100 | 100 | 100 | 3.12 | 50 | 6.25 | 12.5 | 6.25 | 1.5 | 9 | 45 |
| C14-KLLK | 25 | 25 | 12.5 | 3.12 | >100 | 6.25 | 12.5 | 6.25 | 2.3 | 13 | 50 |
| C12-KLLK | 50 | 50 | 25 | 12.5 | >100 | 25 | 100 | 12.5 | 3 | 0 | 0 |
| C16-KAAK | 6.25 | 6.25 | 12.5 | 6.25 | 12.5 | 6.25 | 100 | 12.5 | 3 | 0 | 3 |
| C14-KAAK | 50 | 50 | 50 | 12.5 | 100 | 25 | 100 | 50 | 6.25 | 0 | 0 |
| C12-KAAK | >100 | 100 | >100 | 100 | >100 | >100 | >100 | >100 | 50 | 0 | 0 |
| C16-KGGK | 3 | 3 | 6.25 | 6.25 | 12.5 | 12.5 | 12.5 | 12.5 | 1.5 | 8 | 75 |
| C16-KKKK | 6.25 | 6.25 | 3 | 6.25 | 12.5 | 25 | 100 | 25 | 1.5 | 0 | 6 |
| C16-KKEK | 12.5 | 12.5 | 50 | 12.5 | 37 | 25 | 100 | 100 | 6.25 | 0 | 0 |
| C16-GIGK | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 100 | 6 | 10 |
| Gentamicin | 3.12 | 6.25 | 1.56 | 6.25 | >100 | — | — | — | — | — | — |
| Ampotericin B | — | — | — | — | — | 1.6 | 3.2 | 1.5 | 1.5 | — | — |
The abbreviations used for genus and species names are as follows: E.c., Escherichia coli; P.a., Pseudomonas aeruginosa; S.a., Staphylococcus aureus II; A.b., Acinetobacter baumannii; A.fu., Aspergillus fumigatus; A.fl., Aspergillus flavus; C.a., Candida albicans; C.n., Cryptococcus neoformans. The numbers that follow the abbreviated genus and species names correspond to American Type Culture Collection (ATCC) numbers. Amino acids in italics are the d-enantiomers.
*Gentamicin-resistant.
Biological Activity of the Lipopeptides.
The lipopeptides were assayed against representative Gram-positive and Gram-negative bacteria, yeast, and fungi, including gentamicin-resistant Acinetobacter baumannii, which are common in human infections, as well as against a highly diluted solution [4% (vol/vol)] of human erythrocytes. The antibiotics gentamicin and amphotericin B served as controls for bacteria and fungi, respectively, and gave the reported minimal inhibitory concentrations (MICs). Importantly, the data shown in Table 1 indicate a repertoire of very short lipopeptides with potent antibacterial and antifungal activities. Interestingly, in contrast to many AMPs, in which increasing the hydrophobicity of the amino acids increases biological activity, our lipopeptides preserved potent biological activity independently of the hydrophobicity of the uncharged amino acids (L, A, or G). However, the type of the hydrophobic amino acid determined the microorganism's specificity. For example, C16-KLLK was active only toward the Gram-positive bacteria and fungi tested, whereas C16-KGGK was active toward all types of cells tested. Furthermore, specificity was also dictated by the length of the aliphatic acid, as can be seen with KLLK and KAAK, each of which is attached to hexadecanoic, tetradecanoic, and dodecanoic acid.
The hemolytic activity of the lipopeptides against the highly diluted solution of human erythrocytes is also shown in Table 1. Unexpectedly, the hexadecanoyl-linked peptide of the less hydrophobic amino acid glycine was hemolytic, whereas the alanine-containing lipopeptide was practically nonhemolytic. Note, however, that even the hemolytic lipopeptides were practically inactive up to ≈20 μM.
Membrane Disruption Induced by the Lipopeptides.
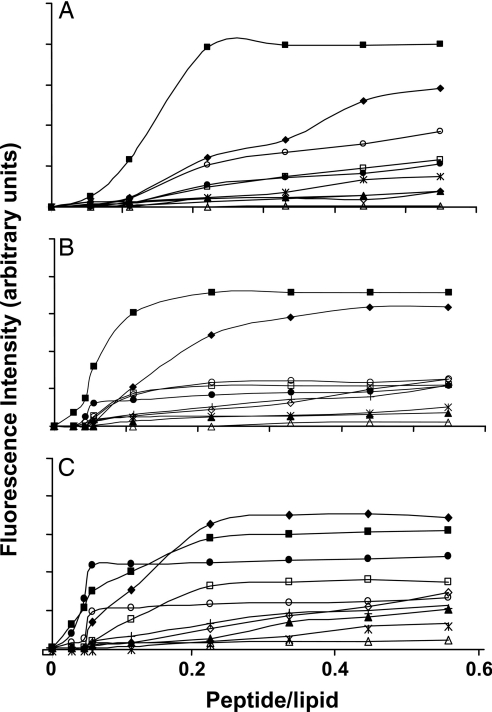
The lipopeptides were investigated for their ability to induce calcein release from small unilamellar vesicles (SUVs) composed of three types of phospholipids, which mimic the membrane composition of fungi, bacteria, and mammalian cells. Lipopeptides at increasing concentrations were added to a suspension of SUVs encapsulated with calcein, and membrane permeability was followed by monitoring the fluorescence recovery. The level of the maximum leakage reached as a function of peptide-to-lipid molar ratio is shown in Fig. 1 A, B, and C for phosphatidylcholine (PC)/phosphatidylethanolamine (PE)/PI/ergosterol (5:4:1:2, wt/wt/wt/wt), PE/PG (7:3, wt/wt), and PC/cholesterol (10:1 wt/wt), respectively. The data reveal a direct correlation between the activity of the lipopeptides on these model membranes and the corresponding microorganisms. The only exception is C16-KLLK, which was not active on Escherichia coli but strongly permeated bacterial model membranes, the reason for which will be discussed later.
Fig. 1.
Calcein release induced by the lipopeptides. The lipopeptides were added to SUVs with different phospholipid compositions (final concentration of 2.5 μM) and encapsulated with calcein. (A) PC/cholesterol (10:1). (B) PC/PE/PI/ergosterol. (C) PE/PG. Designations are as follows: □, C16-KAAK; ◇, C14-KAAK; ▵, C12-KAAK; ■, C16-KLLK; ♦, C14-KLLK; ▴, C12-KLLK; ●, C16-KKKK; ○, C16-KGGK; ∗, C16-GIGK; +, C16-KKEK.
Permeation of Bacterial and Fungal Cell Membranes.
The extent of the membrane damage caused by the lipopeptides was investigated by using two independent methods, described below.
Measuring the entrance of SYTOX green into the microorganism's cells.
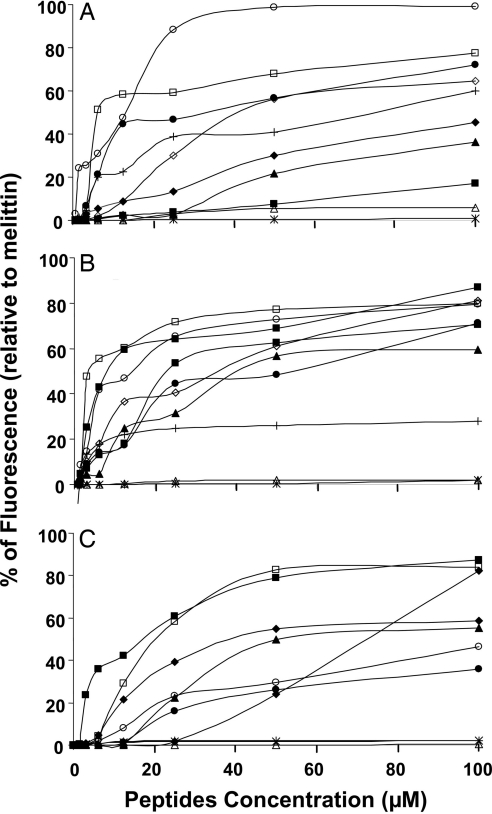
The cationic dye SYTOX green (molecular mass of 900 Da) cannot enter into an intact cell unless its membrane is disrupted by external compounds. The fluorescence of SYTOX green increases drastically when bound to intracellular nucleic acids. We found a marked enhancement in the fluorescence of E. coli, Staphylococcus aureus, and Candida albicans that were treated with the biologically active lipopeptides (e.g., C16-KGGK and C16-KAAK), whereas no activity was observed with these cell types when treated with lipopeptides that were not bioactive on these microorganisms (e.g., C12-KAAK and C16-GIGK) (Fig. 2). Overall, there was a direct correlation between the potency of the different lipopeptides in inducing the influx of SYTOX green into the cells and their corresponding biological activities on those cells.
Fig. 2.
Influx of the vital dye SYTOX green into E. coli (A), S. aureus (B), and C. albicans (C) after the addition of the lipopeptides. The increase in fluorescence was monitored with excitation set at 485 nm and emission at 520 nm. The fluorescence increase obtained by using 50 μM melittin was taken as 100%. All readings were normalized by subtracting parasite scattering and the basal fluorescence of the dye. Designations are as follows: □, C16-KAAK; ◇, C14-KAAK; ▵, C12-KAAK; ■, C16-KLLK; ♦, C14-KLLK; ▴, C12-KLLK; ●, C16-KKKK; ○, C16-KGGK; ∗, C16-GIGK; +, C16-KKEK.
Measuring the entrance of FITC.
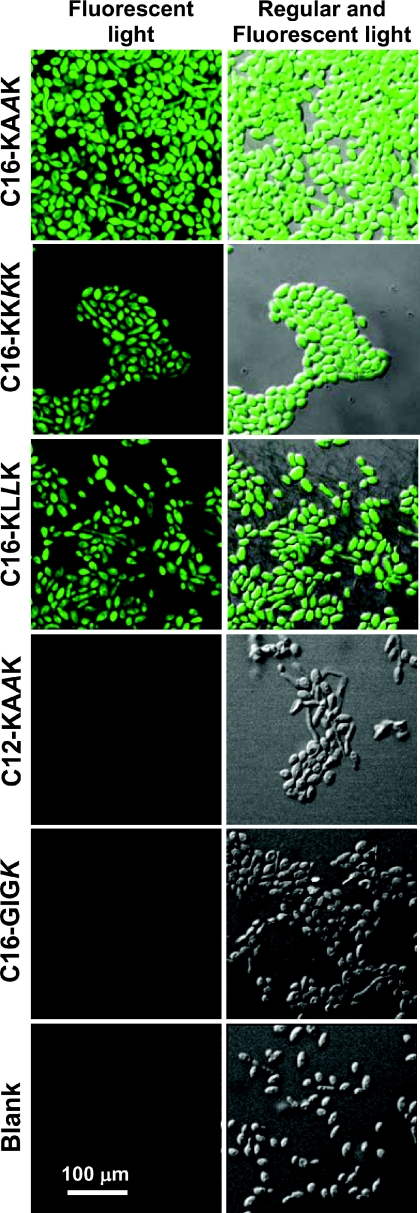
FITC (molecular mass of 389.4 Da), a green fluorescent probe, can also traverse only through the damaged cytoplasmic membrane of a cell. When we treated C. albicans and E. coli ATCC 25922 with the different lipopeptides in the presence of FITC, we observed that FITC could penetrate only into cells that were treated with the lipopeptides that were active on them. More specifically, there was a marked fluorescence signal within C. albicans when treated with the three active lipopeptides C16-KAAK, C16-KKKK, and C16-KLLK (Fig. 3 Left), but not with the untreated control and the inactive C12-KAAK and C16-GIGK (Fig. 3 Right). Similar results were obtained with E. coli ATCC 25922 (data not shown). These results further support a membranolytic mechanism of killing.
Fig. 3.
Lipopeptide-induced influx of FITC into C. albicans. Cells (2 × 107) were incubated with 50 μM lipopeptides at 30°C for 60 min and immobilized on poly(l-lysine)-coated glass slides. FITC (500 μl of a diluted solution in 10 mM sodium phosphate buffer) was spread on the slides, which were then incubated at 30°C for 30 min. After washing, the slides were examined by confocal laser scanning microscopy to assess the affect of the lipopeptides on FITC influx inside the bacterial and yeast cells. Blank, cells with FITC.
Visualization of Cell Damage by Using Transmission EM.
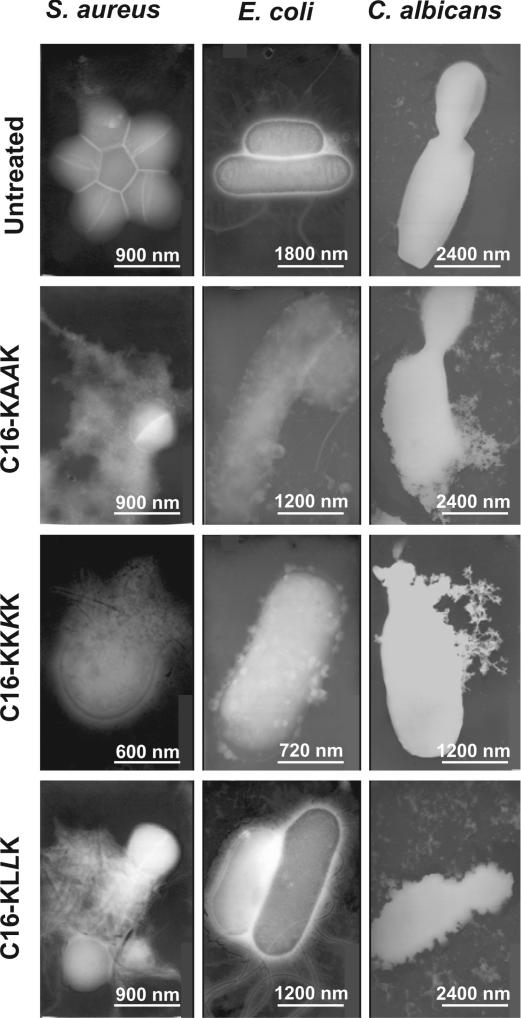
C. albicans, S. aureus, and E. coli were treated with the different lipopeptides for 15 min at their MICs and visualized by using transmission EM (Fig. 4). The images reveal a direct correlation between the biological function of the lipopeptides (Table 1) and their ability to disrupt the cell membrane (Fig. 4). For example, C16-KAAK, C16-KLLK, and C16-KKKK lyse C. albicans, whereas C16-GIGK is not active. Similar patterns were observed with the two bacteria tested (data not shown). Note that C16-KLLK, which is practically inactive toward E. coli, does not affect most of the bacteria, but some of the population has a “ghost-like” shape.
Fig. 4.
Electron micrographs of negatively stained C. albicans, E. coli, and S. aureus untreated or treated with the lipopeptides. The lipopeptide concentrations used were their MICs.
Discussion
The repertoire of AMPs has dramatically increased during the past decade (2). The list includes >800 natural peptides with a length ranging mostly from ≈12 to 100 aa (an online list can be found at www.bbcm.univ.trieste.it/∼tossi/antimic.html). Their amino acid composition, amphipathic structure, net positive charge, and size were found to be important properties that allow them to bind, insert, and destabilize the membrane of the pathogen via various mechanisms (2, 8, 10). Other properties of AMPs, such as the ability to assemble in solution and in membranes, also affect their propensity to selectively kill a particular family, or several families, of microorganisms.
Our study demonstrates three intriguing findings. First, the attachment of an aliphatic chain to otherwise very short, inert cationic peptides can compensate for the length and hydrophobicity of the peptidic chain and endow the resulting lipopeptides with potent antimicrobial activity. Importantly, the MICs of these lipopeptides are similar or better than the values reported for many native AMPs, particularly while taking into account their relatively low molecular mass (≈600 Da). Second, the sequence of a peptidic chain as short as four amino acids alone, or the size of the attached aliphatic chain, is sufficient to control the specificity of the lipopeptides toward the different types of cells. Importantly, most of the lipopeptides are practically not hemolytic against a highly diluted solution of human RBCs (hRBCs) at their MICs or at higher concentrations. Third, in contrast to most known AMPs or natural lipopeptides that are active either on bacteria alone or fungi alone, several members of this family of lipopeptides are highly potent against both bacteria and fungi.
A major question asks how these lipopeptides exert their activity. Most AMPs studied so far adopt α-helical and/or β-sheet amphipathic structures that have been shown to be important for their function (5, 6, 9). Here, all of the peptides contain only four amino acids, which makes it difficult for the peptides to create a defined and stable amphipathic structure. Note also that neither the hydrophobicity of the peptidic chain nor the length of the aliphatic moiety directly correlates with the biological function of the peptides. For example, the less hydrophobic lipopeptide C16-KGGK is the most potent compound. A second example is C14-KLLK, which is active against Gram-negative bacteria, whereas the more hydrophobic analog C16-KLLK is not active.
Most natural antifungal lipopeptides are composed of six to seven amino acids, are cyclic (12), relatively hydrophobic, and carry a net negative charge. They act via two major mechanisms: (i) inhibiting the synthesis of cell wall components such as (1,3)-β-d-glucan or chitin (11, 24) and (ii) lysing membranes, although the details in most cases are unknown (e.g., iturins, bacillomycin, and surfactin) (17–19). Daptomycin, for example, a lipopeptide recently approved by the Food and Drug Administration, can depolarize the membrane of only Gram-positive bacteria and only in the presence of calcium ions (20, 25).
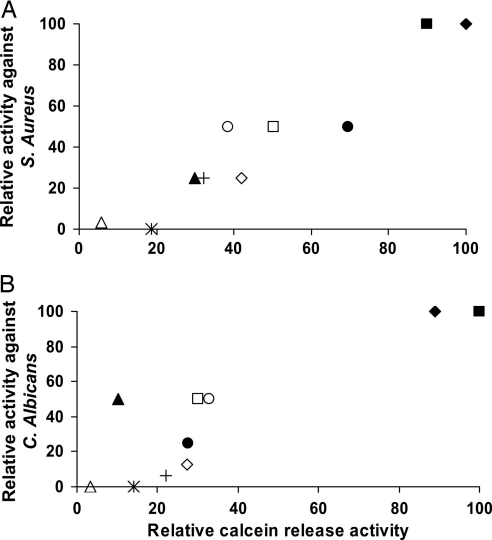
The following data suggest that, similar to those of most native AMPs, a major target of our lipopeptides is the membrane of the pathogen. Importantly, we found a direct correlation among the MICs of the lipopeptides on the different microorganisms (Table 1), their ability to disrupt and increase the permeability of the cytoplasmic membrane of these microorganisms (Figs. 2 and 3), as well as the corresponding model vesicles (Fig. 1), and their ability to damage the structures of the pathogens, as revealed by the EM studies (Fig. 4). This is demonstrated in Fig. 5, which shows in general a direct correlation between the activities of the lipopeptides on the selected microorganisms (Table 1) and the corresponding model membranes (Fig. 1). A similar relationship (data not shown) was found between the activities shown in Table 1 and the data presented in Fig. 2. C12-KLLK (Fig. 5B) seems to be an exception because it is highly active on C. albicans but could disrupt the fungal membrane only slightly. A second exception is C16-KLLK, which is highly potent against all microorganisms except E. coli, although it strongly disrupts E. coli-mimicking model membranes (Fig. 1C). A possible explanation is that C16-KLLK, being the most hydrophobic lipopeptide, forms aggregates that hardly traverse the LPS outer layer of E. coli, similar to what has been shown with a 12-mer lipopeptide (26).
Fig. 5.
Relationship between the relative activities of the different lipopeptides against S. aureus (A) and C. albicans (B) and their ability to induce calcein release from the corresponding model membranes. The most active lipopeptide in each assay was scored as 100. Designations are as follows: □, C16-KAAK; ◇, C14-KAAK; ▵, C12-KAAK; ■, C16-KLLK; ♦, C14-KLLK; ▴, C12-KLLK; ●, C16-KKKK; ○, C16-KGGK; ∗, C16-GIGK; +, C16-KKEK.
Note that the ultrashort lipopeptides comprise fatty acids attached to short cationic peptides and therefore could mimic partially alkylated ammonium salts, which constitute a broad class of metabolites commonly occurring in nature. However, whereas the lipopeptides can be modified easily to acquire different spectra of biological activities with no hemolytic activity against a highly diluted solution of hRBCs [4% (vol/vol)], alkylated “ammonium” salts are toxic to all types of cells (27). Furthermore, ammonium salts with a short alkyl chain, C12, are practically inactive (minimal bactericidal activity at the millimolar range) (28), compared with our C12 lipopeptides, which are highly active at the low micromolar range, depending on the sequence of the peptide (e.g., C12-KLLK).
It has been shown in the case of chemical surfactants that the longer the lipophilic acyl chain, the more effective the micellization, because of an increase in intermolecular hydrophobic interactions (29). For example, the micellar concentrations of quaternary ammonium salts conjugated to alkyl groups are 12,700, 1,790, and 210 μM for C12, C14, and C16 alkyl groups, respectively (30). Furthermore, previous studies (38) have also shown that the micellar concentration of hexadecanoic acid conjugated to an amino acid is in a micromolar range and should significantly decrease by increasing the peptidic chain. This suggests that the lipopeptides with long fatty acids exist, at least partially, as micelles at the concentrations at which they exert biological function.
In summary, besides adding important information about the parameters required for host-defense-like lipopeptides to kill microorganisms, we present a strategy that allows the creation of a repertoire of a promising family of very short cationic peptides: although they cannot perturb the integrity of phospholipid membranes alone, they are highly potent toward various microorganisms when conjugated to aliphatic chains with different lengths. Studies on their plausible mode of action support a membranolytic or detergent-like effect [probably via the carpet mechanism (10)] that, similar to many membrane-active AMPs, should make it difficult for the microorganisms to develop resistance. The detergent-like properties of AMPs have been discussed (31–34). Furthermore, the incorporation of D amino acids should give these lipopeptides several advantages compared with their all L amino acid parental lipopeptides, such as controlled enzymatic degradation, which has been recently shown for 15-mer diastereomeric antibacterial peptides (35). Finally, their simple composition and diverse specificities should make them economically viable antimicrobial and antifungal compounds for many applications.
Materials and Methods
Materials.
Rink amide-4-methylbenzhydrylamine hydrochloride salt (MBHA) resin, 4-methylbenzhydrylamine (BHA) resin, and 9-fluorenylmethoxycarbonyl (Fmoc) amino acids were obtained from Calbiochem Novabiochem (Darmstadt, Germany). Lauric acid (dodecanoic acid) was purchased from Sigma (St. Louis, MO). Other reagents used for peptide synthesis included trifluoroacetic acid (TFA, Sigma), piperidine (Merck, Darmstadt, Germany), N,N-diisopropylethylamine (DIEA; Sigma), N-methylmorpholine (NMM; Fluka, Seelze, Germany), N-hydroxybenzotriazole hydrate (HOBT, Aldrich, St. Louis, MO), and 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and dimethylformamide (DMF, peptide synthesis grade; Bio-Lab, St. Paul, MN). PC (from egg yolk), PE (from E. coli), PI (from bovine liver), egg PG, and ergosterol were purchased from Sigma. Cholesterol (extra pure) was supplied by Merck. Calcein was purchased from Molecular Probes (Junction City, OR). All other reagents were of analytical grade. Buffers were prepared in double-distilled water. Amphotericin B and gentamicin were purchased from Sigma. RPMI medium 1640 was purchased from Biological Industries (Beit Haemek, Israel).
Peptide Synthesis, Acylation, and Purification.
Peptides were synthesized by an Fmoc solid-phase method on Rink amide-4-methylbenzhydrylamine hydrochloride salt (MBHA) resin, by using an Applied Biosystems (Foster City, CA) 433A automatic peptide synthesizer. The lipophilic acid was attached to the N terminus of a resin-bound peptide by standard Fmoc chemistry followed by peptide cleavage from the resin and purification by RP-HPLC (>98%) (21). Lipopeptide composition was confirmed by electrospray mass spectroscopy and amino acid analysis.
Antifungal Activity.
The antifungal activity of the lipopeptides was measured according to the conditions of National Committee for Clinical Laboratory Standards document M27-A. The peptides were examined in sterile 96-well plates [Nunc (Roskilde, Denmark) F96 microtiter plates] in a final volume of 200 μl as follows: 100 μl of a suspension containing fungi at a concentration of 2 × 103 cfu/ml in culture medium (RPMI medium 1640 and 0.165 M Mops, pH 7.4, withl-glutamine, without NaHCO3 medium) was added to 100 μl of water containing the peptide in serial 2-fold dilutions. The fungi were incubated for 24 h for Aspergillus fumigatus (ATCC 26430) and Aspergillus flavus (ATCC 9643) and 48–72 h for C. albicans (ATCC 10231) and Cryptococcus neoformans (ATCC MYA-422) using a Binder (Tuttingen, Germany) KB115 incubator. Growth inhibition was determined by measuring the absorbance at 620 nm in an El309 microplate autoreader (Biotek Instruments, Luton, U.K.). Antifungal activities are expressed as the MIC, the concentration at which no growth was observed.
Antibacterial Activity.
The antibacterial activity of the lipopeptides was examined in sterile 96-well plates (Nunc F96 microtiter plates) in a final volume of 100 μl as follows: aliquots (50 μl) of a suspension containing bacteria at a concentration of 106 cfu/ml in culture medium were added to 50 μl of water containing the peptide (prepared from a stock solution of 1 mg/ml peptide in water) in serial 2-fold dilutions in LB. Inhibition of growth was determined by measuring the absorbance at 492 nm with an El309 microplate autoreader (Biotek Instruments) after an incubation of 18–20 h at 37°C. Antibacterial activities were expressed as the MIC, the concentration at which no growth was observed after 18–20 h of incubation. The bacteria used were E. coli ATCC 25922, E. coli ATCC D21, Pseudomonas aeruginosa ATCC 27853, S. aureus ATCC 6538P, and gentamicin-resistant A. baumannii.
Hemolysis of hRBCs.
The assay was done by using a protocol described previously (36), with a final volume of 100 μl of PBS solution containing the lipopeptides and hRBCs [final concentration of 4% (vol/vol)]. The release of hemoglobin was monitored by measuring the absorbance of the supernatant at 540 nm. Controls for 0% hemolysis (blank) and 100% hemolysis consisted of hRBCs suspended in PBS and 1% Triton X-100, respectively.
Preparation of Liposomes.
SUVs were prepared by dissolving dry lipids in chloroform/MeOH (2:1, vol/vol), evaporating the solvent under a stream of nitrogen, lyophilizing overnight, resuspending in buffer (10 mg/ml), vortex mixing, and sonicating. Lipid films were prepared from PC/PE/PI/ergosterol (5:4:1:2, wt/wt/wt/wt), PE/PG (7:3, wt/wt), and PC/cholesterol (10:1, wt/wt), which mimic the outer leaflets of the plasma membranes of C. albicans (32), E. coli (33), and hRBCs (34), respectively.
Membrane Permeability Studies.
Calcein release from vesicles induced by the lipopeptides.
Calcein (60 mM, self-quenching concentration) was entrapped in self-quenched concentrations in SUVs composed of PC/cholesterol (10:1, wt/wt), PE/PG (7:3, wt/wt), or PE/PC/PI/ergosterol (5:4:1:2, wt/wt/wt/wt) as described previously (21). The buffer was 10 mM Hepes and 150 mM NaCl (pH 7.4). The nonencapsulated calcein was removed from the liposome suspension by gel filtration (35). Calcein release due to membrane permeation induced by the lipopeptides was monitored by the fluorescence increase at room temperature (excitation wavelength of 485 nm and emission wavelength of 515 nm). Complete dye release was obtained by using 0.1% Triton X-100.
SYTOX green uptake assay.
C. albicans was grown in potato dextrose broth, whereas E. coli and S. aureus were grown in LB at 37°C, washed, and suspended in PBS. Cells were suspended (2 × 107 cells per ml) in 10 mM sodium phosphate buffer (pH 7.4) and were incubated with 1 μM SYTOX green for 15 min with agitation in the dark (37). After the addition of the peptides, the increase in fluorescence, due to the binding of the dye to intracellular DNA, was monitored (excitation wavelength of 485 nm and emission wavelength of 520 nm).
FITC uptake assay.
The assay was done as previously described (37). E. coli and S. aureus were grown in LB at 37°C, and C. albicans was grown in potato dextrose broth. Cells were suspended (2 × 107 cells per ml) in 10 mM sodium phosphate buffer (pH 7.4) and incubated with the lipopeptides for 60 min at 30°C. The microorganisms were immobilized on poly(l-lysine)-coated glass slides by incubation at 30°C for 45 min. The slides were washed with 10 mM sodium phosphate buffer, 6 μg/ml FITC (stock solution is 10 mg/ml in acetone) in 10 mM sodium phosphate buffer was spread on them, and they were incubated at 30°C for 30 min. The slides were then washed with 10 mM sodium phosphate buffer. Controls were run in the presence of peptide solvents. After washes with sodium phosphate buffer (pH 7.4), the slides were examined by confocal microscopy with an excitation wavelength of 488 nm.
EM.
Samples containing C. albicans ATCC 10231, E. coli ATCC 25922, and S. aureus ATCC 6538P (3 × 107 cfu/ml) were incubated with 50 μM lipopeptides dissolved in PBS at their MICs for 15 min and were centrifuged at 420 × g. Controls were made in the presence of lipopeptides. The yeasts were fixed by incubation with 1% glutaraldehyde in PBS for 20 min. The pellets were resuspended; a drop containing the fungi was deposited onto a carbon-coated grid and negatively stained with 1% (wt/vol) uranyl acetate, and the bacteria were negatively stained with 2% (wt/vol) phosphotungstic acid. The grids were examined by using a JEOL (Tokyo, Japan) JEM 100B electron microscope.
Acknowledgments
We thank Vladimir Kiss for help with the fluorescence microscopy, and Batya Zarmi for technical assistance, and Yehuda Marikovsky for EM studies. This work was supported in part by the Dr. Joseph Cohn Minerva Center for Biomembrane Research. Y.S. is the incumbent of the Harold S. and Harriet B. Brady Professorial Chair in Cancer Research.
Abbreviations
- AMP
antimicrobial peptide
- PG
phosphatidylglycine
- PI
phosphatidylinositol
- MIC
minimal inhibitory concentration
- SUV
small unilamellar vesicles
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- hRBC
human RBC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Boman HG. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Selsted ME, Ouellette AJ. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 4.Hancock RE, Brown KL, Mookherjee N. Immunobiology. 2006;211:315–322. doi: 10.1016/j.imbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Shai Y, Oren Z. Peptides. 2001;22:1629–1641. doi: 10.1016/s0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- 6.Tossi A, Sandri L. Curr Pharm Des. 2002;8:743–761. doi: 10.2174/1381612023395475. [DOI] [PubMed] [Google Scholar]
- 7.Gazit E, Boman A, Boman HG, Shai Y. Biochemistry. 1995;34:11479–11488. doi: 10.1021/bi00036a021. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki K. Biochim Biophys Acta. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 9.Hancock RE, Rozek A. FEMS Microbiol Lett. 2002;206:143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x. [DOI] [PubMed] [Google Scholar]
- 10.Shai Y. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 11.Balkovec J. Exp Opin Investig Drugs. 1994;3:65–82. [Google Scholar]
- 12.De Lucca AJ, Walsh TJ. Antimicrob Agents Chemother. 1999;43:1–11. doi: 10.1128/aac.43.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thevissen K, Terras FR, Broekaert WF. Appl Environ Microbiol. 1999;65:5451–5458. doi: 10.1128/aem.65.12.5451-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avrahami D, Shai Y. J Biol Chem. 2004;279:12277–12285. doi: 10.1074/jbc.M312260200. [DOI] [PubMed] [Google Scholar]
- 15.Pouny Y, Rapaport D, Mor A, Nicolas P, Shai Y. Biochemistry. 1992;31:12416–12423. doi: 10.1021/bi00164a017. [DOI] [PubMed] [Google Scholar]
- 16.Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 17.Maget-Dana R, Peypoux F. Toxicology. 1994;87:151–174. doi: 10.1016/0300-483x(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 18.Maget-Dana R, Ptak M. Biophys J. 1995;68:1937–1943. doi: 10.1016/S0006-3495(95)80370-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peypoux F, Bonmatin JM, Wallach J. Appl Microbiol Biotechnol. 1999;51:553–563. doi: 10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- 20.Steenbergen JN, Alder J, Thorne GM, Tally FP. J Antimicrob Chemother. 2005;55:283–288. doi: 10.1093/jac/dkh546. [DOI] [PubMed] [Google Scholar]
- 21.Avrahami D, Shai Y. Biochemistry. 2002;41:2254–2263. doi: 10.1021/bi011549t. [DOI] [PubMed] [Google Scholar]
- 22.Avrahami D, Shai Y. Biochemistry. 2003;42:14946–14956. doi: 10.1021/bi035142v. [DOI] [PubMed] [Google Scholar]
- 23.Makovitzki A, Shai Y. Biochemistry. 2005;44:9775–9784. doi: 10.1021/bi0502386. [DOI] [PubMed] [Google Scholar]
- 24.Debono M, Gordee RS. Annu Rev Microbiol. 1994;48:471–497. doi: 10.1146/annurev.mi.48.100194.002351. [DOI] [PubMed] [Google Scholar]
- 25.Hancock RE. Lancet Infect Dis. 2005;5:209–218. doi: 10.1016/S1473-3099(05)70051-7. [DOI] [PubMed] [Google Scholar]
- 26.Papo N, Shai Y. J Biol Chem. 2005;280:10378–10387. doi: 10.1074/jbc.M412865200. [DOI] [PubMed] [Google Scholar]
- 27.Podolak M. Curr Top Biophys. 2002;26:211–216. [Google Scholar]
- 28.Ahlstrom B, Chelminska-Bertilsson M, Thompson RA, Edebo L. Antimicrob Agents Chemother. 1995;39:50–55. doi: 10.1128/aac.39.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosen MJE. Surfactants and Interfacial Phenomena. New York: Wiley; 1989. [Google Scholar]
- 30.Ahlstrom B, Chelminska-Bertilsson M, Thompson RA, Edebo L. Antimicrob Agents Chemother. 1997;41:544–550. doi: 10.1128/aac.41.3.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bechinger B. Biochim Biophys Acta. 1999;1462:157–183. doi: 10.1016/s0005-2736(99)00205-9. [DOI] [PubMed] [Google Scholar]
- 32.Shai Y. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 33.Papo N, Shai Y. Peptides. 2003;24:1693–1703. doi: 10.1016/j.peptides.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Bechinger B, Lohner K. Biochim Biophys Acta. 2006 Jul 13; doi: 10.1016/j.bbamem.2006.07.001. 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Papo N, Oren Z, Pag U, Sahl HG, Shai Y. J Biol Chem. 2002;277:33913–33921. doi: 10.1074/jbc.M204928200. [DOI] [PubMed] [Google Scholar]
- 36.Oren Z, Shai Y. Biochemistry. 1997;36:1826–1835. doi: 10.1021/bi962507l. [DOI] [PubMed] [Google Scholar]
- 37.Mangoni ML, Papo N, Barra D, Simmaco M, Bozzi A, Di Giulio A, Rinaldi AC. Biochem J. 2004;380:859–865. doi: 10.1042/BJ20031975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinitzky M, Haimovitz R. J Am Chem Soc. 1993;115:12545–12549. [Google Scholar]