Abstract
The import of nuclear-encoded proteins into chloroplasts is tightly controlled on both sides of the envelope membranes. Regulatory circuits include redox-control as well as calcium-regulation, with calmodulin being the likely mediator of the latter. Using affinity-chromatography on calmodulin-agarose, we could identify the inner envelope translocon component Tic32 as the predominant calmodulin-binding protein of this membrane. Calmodulin-binding assays corroborate the interaction for heterologously expressed as well as native Tic32. The interaction is calcium-dependent and is mediated by a calmodulin-binding domain between Leu-296 and Leu-314 close to the C-proximal end of the pea Tic32. We furthermore could establish Tic32 as a bona fide NADPH-dependent dehydrogenase. NADPH but not NADH or NADP+ affects the interaction of Tic110 with Tic32 as well as Tic62. At the same time, dehydrogenase activity of Tic32 is affected by calmodulin. In particular, binding of NADPH and calmodulin to Tic32 appear to be mutually exclusive. These results suggest that redox modulation and calcium regulation of chloroplast protein import convene at the Tic translocon and that both could be mediated by Tic32.
Keywords: calcium signaling, protein import, Tic translocon, redox-regulation, short-chain dehydrogenases
Chloroplast development and differentiation requires a massive import of nuclear-encoded proteins to build up the photosynthetic machinery and establish the functionality of the organelle (1). Maintenance of the photosynthetic capacity, which is highly prone to oxidative damage, requires an unceasing but highly regulated replacement of proteins. To ensure a regulated supply of the organelle with polypeptides, a tight regulatory network exists between the chloroplasts and the surrounding cell to coordinate gene expression, translation, and import of chloroplast-destined proteins. Chloroplasts import ≈3,000 different proteins from the cytosol in a posttranslational process (1). Most, but not all, precursor proteins use a standard import pathway that requires an N-terminal, cleavable transit sequence and involves two proteinaceous multisubunit complexes in the outer and inner envelope membrane, the Toc and the Tic translocons (2). Translocation of precursor proteins is highly regimented, including GTP-regulation by two prominent Toc components, Toc34 and Toc159, at the state of precursor recognition and translocation initiation on the outer envelope (3). In contrast, Tic translocation appears to be regulated by the redox status of the chloroplast as imparted to the translocon by the ratio of NADPH/NADP in the stroma (4–6). In particular, Tic32 displays sequence homology to a class of short-chain dehydrogenases (SDRs) (5). The family of SDRs comprises a wide range of enzymes with a great variety of functions (7). Common to all SDRs is the presence of a NAD(P)H-binding domain close to the N terminus as well as several conserved domains within the central part of the protein required for catalytic activity. The characteristics of Tic32 might indicate that the protein is part of a redox-sensing circuit that regulates the import of proteins into the organelle (5).
We have shown recently that chloroplast protein import is influenced by calcium, a regulation that most likely takes place at the inner envelope membrane (8). Calcium controls many biochemical processes in the cell by acting as a secondary messenger that relays primary, e.g., environmental, signals into cellular response pathways (9, 10). Calcium control is exerted by its interaction with a set of calcium-binding proteins, which often undergo conformational changes upon calcium binding. One such calcium sensor is calmodulin, a protein found ubiquitously in eukaryotic organisms (10). Dependent on its calcium-loaded state, calmodulin displays an altered affinity for a number of downstream target proteins. Calmodulin was identified as the most likely candidate to impart the calcium regulation of the chloroplast translocation process (8). This finding indicates that chloroplast protein import is integrated into the calcium-signaling network of the cell, thereby adding a further layer of regulation by which the chloroplast can fine-tune its protein content in correlation to the metabolic needs of the surrounding cell.
In this work, we have identified one of the translocon components of the inner envelope of chloroplasts, Tic32, as the predominant calmodulin-binding protein in this membrane. Calmodulin binding to Tic32 is calcium-dependent and affects NADPH-dependent dehydrogenase activity of Tic32. Besides, NADPH affects the interaction of Tic32 with other Tic components, such as Tic110 and Tic62. Our results indicate that calmodulin regulation of chloroplast protein import is conveyed by interaction with Tic32, making this protein the potential transducer of redox as well as calcium regulation of protein import.
Results
Tic32 Is the Predominant Calmodulin-Binding Protein in the Inner Envelope of Chloroplasts.
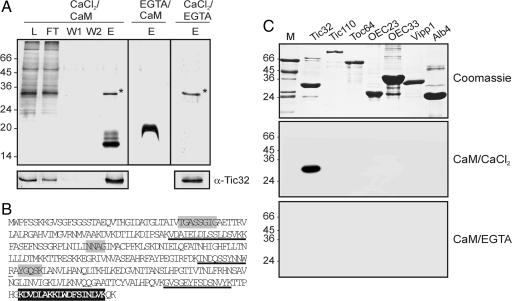
Previous work of our group has shown that the import of nuclear-encoded chloroplast proteins is regulated by calcium (8), which most likely is mediated by calmodulin and occurs at the stage of Tic translocation. We therefore attempted to isolate potential calmodulin-binding proteins from the inner envelope of pea chloroplasts by affinity chromatography on calmodulin-agarose (Fig. 1A). Detergent-solubilized inner envelope membranes were incubated with calmodulin-agarose in the presence of either 0.1 mM calcium or 5 mM EGTA, and, after extensive washing, bound proteins were eluted from the matrix by an excess of calmodulin (Fig. 1A Upper, CaCl2/CaM and EGTA/CaM). Protein analysis from all fractions by SDS/PAGE and silver staining revealed the binding of a single protein of ≈32 kDa to the matrix solely in the presence of calcium, indicating that the interaction of this protein with calmodulin-agarose is calcium-dependent. The protein bands visible at ≈16 kDa in the eluate fractions represent the calmodulin used to elute the matrix. The difference in size of the calmodulin between the calcium and EGTA experiment is attributable to an increased mobility of calmodulin in the presence of calcium, a specific feature of this protein (11). Once bound to the matrix in the presence of calcium, the 32-kDa protein also could be eluted by exchanging calcium with EGTA, thus supporting further the calcium dependency of its binding to calmodulin (Fig. 1A Upper, CaCl2/EGTA). Mass spectrometry of the 32-kDa protein resulted in the identification of three peptides that matched to Tic32 from pea (Fig. 1B), and the identification was verified by using an antiserum specific to Tic32 (Fig. 1A Lower).
Fig. 1.
Identification of Tic32 as a calmodulin-binding protein. (A) Solubilized inner envelope membranes from pea chloroplasts were incubated with calmodulin-agarose in the presence (CaCl2/CaM and CaCl2/EGTA) and absence (EGTA/CaM) of calcium. L, load; FT, flow through; W, wash; E, eluate. With calcium, a single protein bound to the matrix that could be eluted either with calmodulin in the presence of CaCl2 (∗) or by EGTA alone (∗). The protein did not bind to the matrix in the presence of EGTA. The protein was identified as Tic32 by Western blot analysis (Lower) and by mass spectrometry (B). (B) Deduced amino acid sequence of Tic32 from Pisum sativum (AAS38575). Peptides obtained by mass spectroscopic analysis from the protein isolated in A are underlined. Black boxes indicate the calmodulin-binding site of Tic32. Gray boxes indicate conserved active side residues of SDRs. (C) A calmodulin-binding assay was performed by using a number of different heterologously expressed proteins with biotinylated calmodulin (Bottom). Interaction of Tic32 with calmodulin occurred in the presence of calcium but not with EGTA. M, protein marker; Tic32 and Tic110, components of the Tic translocon; Toc64, component of the Toc translocon; OEC23 and OEC33, components of the oxygen-evolving system; Vipp1, protein associated with the inner envelope; Alb4, integral thylakoid protein.
To confirm that the binding of Tic32 to calmodulin-agarose is in fact caused by an interaction of the protein with calmodulin, we performed a calmodulin-binding assay with Tic32 heterologously expressed in Escherichia coli. This assay has been widely used to identify calmodulin-binding proteins from a variety of organisms (12). Tic32 and several control proteins were separated on SDS/PAGE (Fig. 1C Top) and subsequently transferred onto PDVF membrane. Binding of calmodulin was elucidated by incubation with biotinylated calmodulin in the presence of either calcium or EGTA. In the presence of calcium, the assay revealed binding of biotinylated calmodulin exclusively to Tic32 (Fig. 1C Middle). When the calmodulin-binding assay was performed in the presence of EGTA instead of calcium, no binding to Tic32 was observed (Fig. 1C Bottom). The results of this experiment substantiate that the interaction of calmodulin with Tic32 is specific as well as calcium-dependent.
Identification of the Calmodulin-Binding Domain of Tic32.
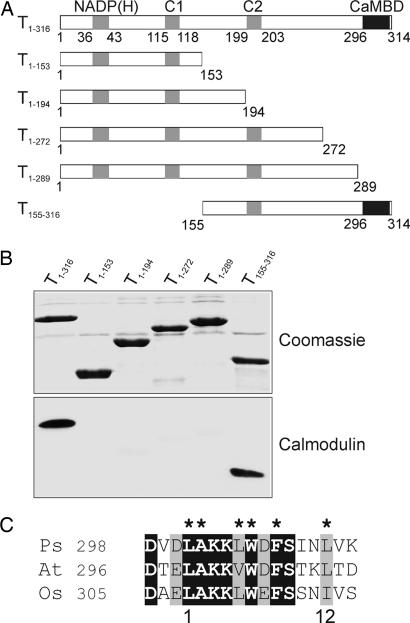
To determine the calmodulin-binding domain, we prepared a number of N- and C-terminal truncated constructs of Tic32 (Fig. 2A), and binding of calmodulin to the truncated proteins was analyzed by calmodulin-binding assays (Fig. 2B). As seen before, the full-length Tic32 displayed a strong calcium-dependent binding to calmodulin, but no interaction could be observed for any of the C-terminally truncated proteins, even though the longest construct lacked only the 26 most C-proximal amino acids. In contrast, removal of any N-terminal parts of the protein did not affect calmodulin interaction (Fig. 2B and data not shown). Thus, we conclude that the very C-proximal region from amino acids 290 to 316 contains the calmodulin-binding domain of Tic32.
Fig. 2.
Identification of the calmodulin-binding domain of Tic32. (A) Schematic representation of Tic32 and various N- and C-terminal deletion constructs. The NADP(H) binding site as well as two regions identified in other SDRs as part of the active site (C1 and C2) are marked by gray boxes. The black box indicates the calmodulin-binding domain (CaMBD) identified in the experiment. Numbers indicate amino acid residues in correlation to the wild-type Tic32. (B) Equal amounts of heterologously expressed and purified proteins were analyzed for calmodulin binding by using biotinylated calmodulin (Lower). Removal of the 26 most C-proximal amino acids (T1–289) resulted in a loss of interaction between calmodulin and Tic32. (C) Alignment of the putative calmodulin-binding domain of Tic32 from pea (Ps; AAS38575) with the corresponding sequences of Tic32 from Arabidopsis thaliana (At; NP_849428), and Oryza sativa (Os; AAX96188). Identical residues in all sequences are boxed in black, whereas conserved residues are boxed in gray. Conserved hydrophobic residues within the sequence are marked by asterisks. A potential 1–12 motif for calmodulin binding is indicated below the sequences.
Based on these results, we performed a closer investigation of this region. A characteristic of calmodulin-binding domains is the formation of a basic amphipathic helical structure. Such a structure can be predicted for the amino acids Leu-296 to Leu-314 of Tic32, forming a hydrophobic side and a hydrophilic side with an overall positive charge (Fig. 6, which is published as supporting information on the PNAS web site). A sequence alignment of the calmodulin-binding domain of Tic32 from pea with the corresponding region of Tic32 from Arabidopsis thaliana (araTic32) and Oryza sativa (oryzaTic32) reveals a high degree of homology within this region (Fig. 2C). Furthermore, at least three of the established calcium-dependent calmodulin-binding motifs can be found within the region spanning amino acids Leu-296 to Leu-314 of Tic32 from pea (http://calcium.uhnres.utoronto.ca/ctdb/pub_pages/search/index.htm). At least one of the motifs, called 1–12, can be found in all three Tic32 homologues (Fig. 2C). Thus, all three proteins contain the potential for calmodulin binding, indicating that this type of regulation might be conserved within higher plants.
Calmodulin Interacts with Tic32 at the Inner Envelope Membrane.
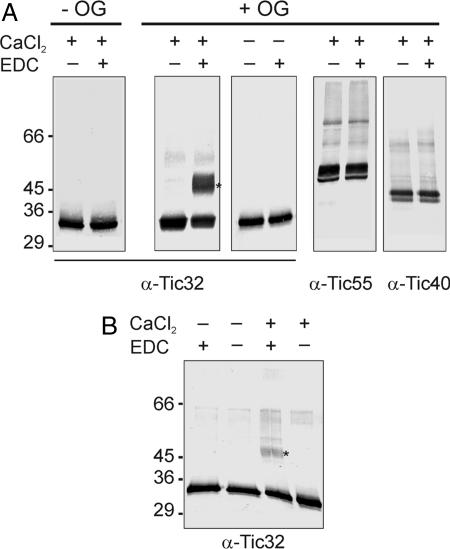
In order for calmodulin to regulate protein import by binding to Tic32, the reaction would have to occur at the inner envelope of chloroplasts. To elucidate this assumption, we cross-linked isolated inner envelope vesicles with calmodulin in the presence and absence of n-octylglycoside (OG). Because purified inner envelope vesicles are believed to be right-side-out, solubilization of the membrane is required to allow calmodulin to get into close proximity to Tic32. To test for interaction, we used the 0-Å cross-linker 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (13). In untreated inner envelope vesicles, a single immunoreactive band was observed slightly above 30 kDa, which represents Tic32 itself (Fig. 3A, first set of gels). In contrast, an additional immunoreactive band occurs in the presence of calmodulin after treatment of the inner envelope with OG. This band occurs at ≈48 kDa, matching the size of a cross-linking product between Tic32 and calmodulin (Fig. 3A, second set of gels). No cross-linking product at 48 kDa appeared in the absence of CaCl2 (Fig. 3A, third set of gels). As a control, the cross-linking reaction was analyzed with antisera against Tic55 and Tic40 (Fig. 3A, fourth and fifth sets of gels). No additional immunoreactive bands were observed for these proteins, supporting the specificity of the interaction between calmodulin and Tic32. When the experiment was performed without the addition of exogenous calmodulin, a weak immunoreactive band at 48 kDa could be observed in the presence of both cross-linker and calcium (Fig. 3B). This band was not observed in the absence of calcium. This result indicates that there might be some indigenous calmodulin present, in the purified inner envelope vesicles, that is associated with Tic32.
Fig. 3.
Interaction of Tic32 with calmodulin occurs at the inner envelope. (A) Chloroplast inner envelope membranes were treated with the 0-Å cross-linker EDC in the presence of 5 μM exogenous calmodulin (CaM) with (+OG) or without (−OG) a pretreatment with OG. Aliquots of the samples were analyzed by using antibodies against Tic32, Tic40, and Tic55. The appearance of a new 48-kDa cross-linking product was observed exclusively with α-Tic32 in solubilized vesicles (+OG) and only in the presence of CaCl2. The position of the calmodulin/Tic32 cross-linking product is indicated by an asterisk. (B) Even in the absence of exogenous calmodulin (Right), low amounts of a 48-kDa cross-linking product (∗) can be observed exclusively in the presence of CaCl2, indicating the presence of indigenous calmodulin in the inner envelope preparation.
Tic32 Is a Bona Fide NADPH-Dependent Dehydrogenase.
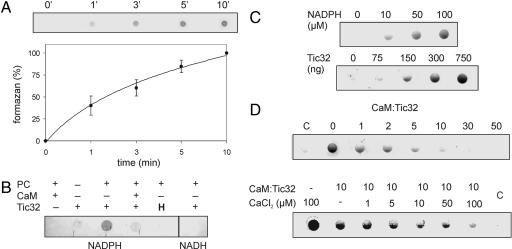
On the basis of its amino acid sequence, Tic32 was placed into the family of SDRs (5), but no dehydrogenase activity had thus far been described for the native or heterologously expressed protein. SDRs are known to accept nonspecific substrates like nitroblue tetrazolium (NBT) or FeCN for their reaction, but activity of heterologously expressed or purified protein sometimes requires conditions mimicking a lipid environment (14). Because the native Tic32 is tightly associated with the inner envelope membrane (5), we tried to recover dehydrogenase activity by combining purified heterologously expressed Tic32 with phosphatidylcholine (PC) lipids (Fig. 4A and B). No formazan formation occurred in either the absence of Tic32 or PC lipids (Fig. 4B). In combination with the PC lipids, a NADPH-dependent reduction of NBT to formazan could be observed, visible by the lilac precipitation. Under these conditions, the protein exhibited NADPH-dependent reduction of NBT in a time-dependent manner (Fig. 4A). Furthermore, enzyme activity could be correlated to the amounts of Tic32 as well as NADPH in the assay (Fig. 4C). Heat-inactivation of the purified protein abolished dehydrogenase activity (Fig. 4B) as well as the replacement of NADPH by NADH. Together these data provide experimental evidence that Tic32 indeed is a bona fide member of the subfamily of NADPH-dependent SDRs.
Fig. 4.
Tic32 is a bona fide NADPH-dependent dehydrogenase. (A) Dehydrogenase activity of heterologously expressed Tic32 was measured by using NBT as substrate. A time course of the reduction of NBT to formazan by heterologously expressed Tic32 was determined. (Upper) The outcome of a typical experiment. (Lower) The curve was calculated by measuring the color intensity of the formed formazan from four independent experiments. (B) Formazan formation was observed only in the presence of both PC and NADPH. The latter could not be substituted by NADH. Heat-treatment (H) of the proteins as well as preincubation with 100 μM calmodulin (CaM) abolished the dehydrogenase activity. (C) Dehydrogenase activity of Tic32 increased in correlation to the amount of NADPH as well as Tic32 protein. (D) Inhibition of dehydrogenase activity by increasing molar ratio of calmodulin to Tic32. Calcium dependency of calmodulin inhibition of NADPH-dependent dehydrogenase activity of Tic32 was analyzed by addition of increasing amounts of CaCl2 in the presence of a 10-fold excess of calmodulin over Tic32 in the assay. C, control without protein.
We then tested whether addition of calmodulin had any influence on the dehydrogenase activity of Tic32 by preincubating the protein with 100 μM calmodulin. Under these conditions, no reduction of NBT could be observed (Fig. 4B), indicating that interaction of calmodulin with Tic32 interferes with the dehydrogenase activity. A more detailed analysis showed that the inhibition of Tic32 by calmodulin depends on the molar ratio of Tic32 to calmodulin in the assay (Fig. 4D Upper). Already at a 1:1 ratio of Tic32 to calmodulin, a significant reduction in the formation of formazan occurs, and with 2.5 times the amount of calmodulin, the dehydrogenase activity of Tic32 is nearly abolished. We furthermore could show that the affect of calmodulin on Tic32 is calcium-dependent. By using a 10-fold excess of calmodulin, no activity is observed with 100 μM CaCl2 in the assay (Fig. 4C Lower). Decreasing the amount of CaCl2 resulted in an increased formation of formazan. The inhibitory effect of calmodulin could not be abolished in this assay most likely because of the vast excess of calmodulin paired with an inability to get rid of all of the calmodulin-bound calcium.
NADPH and Calmodulin Binding to Tic32 Is Mutually Exclusive.
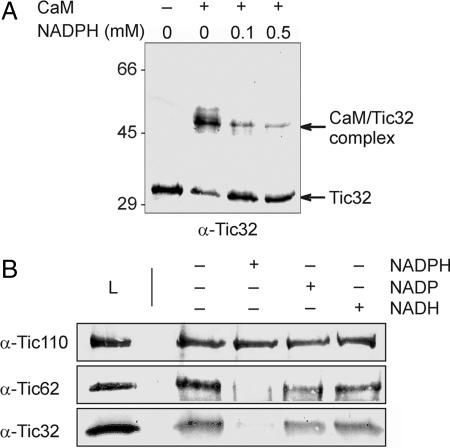
To investigate how calmodulin might affect the dehydrogenase activity of Tic32, we analyzed a potential interference of calmodulin binding with that of NADPH. The calmodulin-binding site of Tic32 does not overlap directly with any of the sites required for NADPH-binding or enzyme activity (Figs. 1B and 2A). Nevertheless, it cannot be excluded that binding of calmodulin affects the interaction of Tic32 with NADPH and vice versa. To test this possibility, we performed cross-linking experiments with inner envelope vesicles and calmodulin in the absence and presence of NADPH. The addition of NADPH significantly lowered the amount of cross-linking between Tic32 and calmodulin (Fig. 5A), an effect that became stronger when increasing amounts of NADPH were used. Thus, it appears likely that Tic32 can bind only to either of the two molecules at a given time.
Fig. 5.
Differential interaction of Tic32 with NADPH, calmodulin, and Tic110. (A) Influence of NADPH on Tic32-calmodulin interaction. Inner envelope vesicles were solubilized in 1% OG and incubated with increasing amounts of NADPH before the addition of 5 μM calmodulin. Cross-linking with EDC was performed as before. Increasing amounts of NADPH abolished the cross-link between Tic32 and calmodulin (CaM/Tic32 complex). (B) The interaction of Tic110 with Tic62 and Tic32 was analyzed by coimmunoprecipitation in the absence or presence of NADPH, NADP, or NADH. Coimmunoprecipitation was nearly abolished in the presence of NADPH but not affected by either NADP or NADH.
Influence of NADPH on the Interaction of Tic32 with Tic110.
Tic32 was originally identified as a Tic component by its interaction with Tic110 (5). We thus analyzed whether binding of NADPH to Tic32 had any influence on its interaction with Tic110 (Fig. 5B) by coimmunoprecipitation experiments with inner envelope vesicles by using an antiserum against Tic110. Reactions were carried out in the presence of NADPH, NADH, or NADP+. The presence of NADPH nearly abolished the coimmunoprecipitation of Tic32 with Tic110 (Fig. 5B Bottom), whereas NADP and NADH had little to no effect on the interaction between these proteins. Because Tic32 is only one of two Tic components containing an NADPH binding site, we also analyzed the coimmunoprecipitation of Tic62 with Tic110 under these conditions (Fig. 5B Middle). The protein displayed a similar behavior, indicating that the interaction of both proteins with Tic110 is affected by binding of the NADPH cofactor.
Discussion
Considering that calcium is one of the most prominent secondary messengers in eukaryotes, it is not surprising that the endosymbiotic organelle chloroplast was integrated into the calcium-signaling network of the cell. We have shown previously that the import of nuclear-encoded chloroplast proteins that translocate by way of an N-terminal, cleavable presequence is regulated by calcium (6). Here we show that Tic32, a component of the Tic translocon, represents the predominant calmodulin-binding protein of the inner envelope membrane and that the binding occurs in a calcium-dependent manner. These results not only support the calcium regulation of the protein translocation process, but also they corroborate the function of calmodulin in mediating this regulation. Our results furthermore indicate that the regulation occurs from inside the organelle because Tic32 is associated with the Tic translocon on the stromal side of the inner envelope (5). Besides, binding of external calmodulin to Tic32 only occurred after solubilization of right-side-out inner envelope vesicles. At the same time, we could obtain evidence that some indigenous calmodulin is present in the inner envelope membrane preparation. Together these data provide additional support for the presence of calmodulin-regulation within chloroplasts.
The fact that calmodulin regulates protein import by binding to Tic32 is especially intriguing because the protein has been shown to be a vital component of the Tic translocon (14). In contrast, Tic32 itself does not require the Tic translocon for insertion into the inner envelope membrane (15) and thus is itself not inhibited in its translocation by calmodulin or calcium (8). Although the exact function of Tic32 in the translocation process so far remains enigmatic, the protein has been proposed as part of a redox regulation circuit of the translocation processes. Other components of the Tic translocon, namely Tic55 and Tic62, have been suggested to sense the redox status of the chloroplast, the latter by binding both FNR and NADP (4, 6). According to its amino acid sequence, Tic32 belongs to a group of SDRs that are found ubiquitously in eukaryotic as well as prokaryotic organisms and that possess a great number of different enzymatic activities. We were able to establish that Tic32 indeed has NADPH-dependent dehydrogenase activity, and NADH could not substitute for NADPH in this assay. Specificity for either NADPH or NADH is a characteristic of SDRs that can be deduced from specific features in their sequence (7), and sequence analysis of Tic32 had placed the protein within the group of NADPH-dependent SDRs. Dehydrogenase activity of Tic32 depended on a lipid environment, a feature that had been shown before for other SDRs (16). Our results show that Tic32 indeed represents a bona fide NADPH-dependent SDR. Because of its NADPH-dependency, the dehydrogenase activity of Tic32 should be tightly associated with the redox status of the chloroplasts. In contrast, it appears much less likely that Tic32 is connected to the ROS-signaling pathway via NADPH oxidase activity. All results indicate that the enzyme represents a classical SDR, and no homology to the described class of NADPH oxidases could be found.
Interestingly, our results indicate that NADPH binding of Tic32 governs its interaction with the Tic translocon. We could show that this interaction is significantly reduced in the presence of NADPH but neither NADP nor NADH. This feature is shared with Tic62, which is a further redox-regulated Tic component. It has been speculated that the chloroplast contains different subsets of Tic complexes having different subunit compositions. Thus, by affecting the interaction between Tic110, Tic62, and Tic32, the NADPH content of the chloroplast could mediate the redox-regulation of chloroplast protein import by influencing the assembly/disassembly of such a subcomplex. Even more, because binding of calmodulin and NADPH to Tic32 appear to be mutually exclusive, binding of calmodulin to Tic32 would promote its interaction with Tic110. Thus, Tic32 could serve as a switch to differentially integrate redox signals from the inside of the chloroplast with calcium signals from outside the organelle. Tic32 could convey these signals by association/disassociation from Tic110, thereby altering either the activity and/or the substrate specificity of the translocon.
Materials and Methods
Materials.
Bovine brain calmodulin and biotinylated bovine brain calmodulin were obtained from Alexis (Grünberg, Germany). Calmodulin-agarose, PC, and streptavidin-peroxidase conjugate were from Sigma-Aldrich Chemie (Munich, Germany). The cross-linking reagents EDC and sulfo-N-hydroxysulfosuccinimide (sulfo-NHS) were from Pierce (Bonn, Germany).
Affinity Chromatography on Calmodulin-Agarose.
Unless otherwise stated, all procedures were carried out at 4°C. Inner envelope vesicles were purified from intact chloroplasts isolated from leaves of 10- to 12-day-old pea plants (Pisum sativum L., cultivar Arvika) as described in ref. 17. Inner envelope vesicles corresponding to 1 mg of protein were solubilized with 1% decylmaltoside in calmodulin-binding buffer (20 mM Tris·HCl, pH 7.5/150 mM NaCl/0.1 mM CaCl2) for 15 min on ice. For control experiments, the calcium was substituted with 5 mM EGTA. Nonsolubilized membrane components were removed by centrifugation at 21,000 × g for 10 min. The supernatant was diluted to 0.1% decylmaltoside with calmodulin-binding buffer, and 100 μl of calmodulin-agarose slurry was added. The suspensions were incubated for 16 h. The agarose beads were collected by a brief low-spin centrifugation and washed with 10 column volumes of 0.5 M NaCl in calmodulin-binding buffer. Bound proteins were eluted by 20 μM calmodulin in the respective binding buffer. Alternatively, the column was washed with calmodulin-binding buffer without CaCl2, and bound proteins were eluted with 5 mM EGTA. Fractions were analyzed by SDS/PAGE followed by silver staining or Western blot analysis using an antibody raised against Tic32 from pea.
Construction and Expression of Various Tic32 Constructs.
The full-length Tic32 clone was constructed as described in ref. 5. Several constructs comprising different parts of Tic32 were obtained by PCR and cloned into pET21d (Novagen, Schwalbach, Germany) with a C-terminal His6 tag. After cloning, all constructs were verified by DNA sequencing. For heterologous expression, the clones were introduced into E. coli BL21(DE), and expression was induced by addition of 1 mM isopropyl β-D-thiogalactoside (IPTG) for 3 h at 37°C. Purification of expressed proteins was performed on Ni2+-nitrilotriacetic acid agarose (Qiagen, Hilden, Germany) following the manufacturer's instructions.
Calmodulin-Binding Assay.
Purified proteins were separated by SDS/PAGE and transferred onto PVDF membrane (Immobilon; Millipore, Schwalbach, Germany). For renaturation of immobilized proteins, the membrane was incubated for 16 h at 4°C in assay buffer [50 mM Tris·HCl, pH 7.5/150 mM NaCl/1% (wt/vol) BSA/1% (vol/vol) Brij35/1 mM CaCl2]. For control experiments, the calcium was substituted by 5 mM EGTA. The membrane subsequently was incubated with 0.1 μg/ml biotinylated calmodulin in assay buffer for 3 h at room temperature. After washing for 10 min three times with assay buffer, the membrane was treated with 1 μg/ml streptavidin-peroxidase conjugate in assay buffer for 1 h. Bound calmodulin was visualized by using an ECL detection system (Amersham Biosciences, Little Chalfont, U.K.) according to the manufacture's instructions.
Cross-Linking of Pea Inner Envelope Proteins with EDC.
Isolated inner envelope membrane vesicles were incubated in the presence or absence of 5 μM calmodulin in 50 mM Hepes/KOH (pH 7.5) and 0.1 mM CaCl2 supplemented with 1% OG for 1 h at 4°C. Control experiments were performed with 5 mM EGTA and 5 mM EDTA instead of CaCl2. Cross-linking with EDC and sulfo-N-hydroxysulfosuccinimide was performed as described (18). The cross-linking reactions were separated by SDS/PAGE, followed by Western blot analysis using α-Tic32, α-Tic55, and α-Tic40 antisera.
To determine the influence of NADPH on Tic32-calmodulin interaction, inner envelope vesicles were incubated with different concentrations of NADPH for 20 min at 4°C before the addition of 5 μM calmodulin. After a further incubation for 40 min, cross-linking was performed with EDC as described above.
Dehydrogenase Activity Assay.
For dehydrogenase activity assays, Tic32 was heterologously expressed in E. coli at 12°C overnight. The protein was purified by using Ni-chelating chromatography followed by size-exclusion chromatography on Superdex S-200 in 10 mM Tris·HCl (pH 8.0) and 200 mM NaCl. Dehydrogenase assays with NBT chloride as substrate were adapted from NADPH-NBT zymograms (19). In short, 1 μg of purified Tic32 (at 0.1 mg/ml) was incubated with PC lipids (1 mg/ml) at 30°C for 30 min before its use in the enzyme assay. If applicable, calmodulin was added to the Tic32-lipid mixture and incubated for 45 min at room temperature. If not otherwise stated, 0.5 μg of Tic32 was used for each assay. Dehydrogenase assays were carried out in 110-μl reactions containing 10 mM Tris·HCl (pH 8.0), 33 μg of NBT, and 100 μM NADPH, NADP, or NADH if not otherwise stated. The reaction was stopped after 10 min at room temperature by spotting the solution onto a nitrocellulose membrane by using a 96-well vacuum manifold (Bethesda Research Laboratories, Bethesda, MD) and thoroughly washing the membrane with H2O. Formazan formation was analyzed by scanning the blot membranes and quantification of the signal using the software included in the AIDA package (Version 3.52.046).
Coimmunoprecipitation of Tic110, Tic62, and Tic32.
Inner envelope vesicles were solubilized in 1% OG for 1 h at 4°C in a buffer containing 50 mM Hepes/KOH (pH 7.5), 0.05% egg albumin, 0.1 mM CaCl2, and either 500 μM NADPH, 500 μM NADH, or 500 μM NADP. Coimmunoprecipitation with antiserum against Tic110 was performed as described in ref. 5. Samples were analyzed by SDS/PAGE and Western blot analysis using antisera against Tic110, Tic62, and Tic32.
Supplementary Material
Acknowledgments
The technical assistance of Claudia Sippel is gratefully acknowledged. The pea vipp1 clone was a kind gift from Dr. K. Keegstra (Department of Energy Research Laboratory, Michigan State University, East Lansing, MI). This work was supported by Deutsche Forschungsgemeinschaft Grants SFB-TR1 (to U.C.V.) and SFB594 (to J.S.).
Abbreviations
- SDR
short-chain dehydrogenase
- OG
n-octylglycoside
- EDC
1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride
- NBT
nitroblue tetrazolium
- PC
phosphatidylcholine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Abdallah F, Salamini F, Leister D. Trends Plant Sci. 2000;5:141–142. doi: 10.1016/s1360-1385(00)01574-0. [DOI] [PubMed] [Google Scholar]
- 2.Vothknecht UC, Soll J. Gene. 2005;354:99–109. doi: 10.1016/j.gene.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 3.Schleiff E, Jelic M, Soll J. Proc Natl Acad Sci USA. 2003;100:4604–4609. doi: 10.1073/pnas.0730860100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caliebe A, Grimm R, Kaiser G, Lubeck J, Soll J, Heins L. EMBO J. 1997;16:7342–7350. doi: 10.1093/emboj/16.24.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hormann F, Kuchler M, Sveshnikov D, Oppermann U, Li Y, Soll J. J Biol Chem. 2004;279:34756–34762. doi: 10.1074/jbc.M402817200. [DOI] [PubMed] [Google Scholar]
- 6.Kuchler M, Decker S, Hormann F, Soll J, Heins L. EMBO J. 2002;21:6136–6145. doi: 10.1093/emboj/cdf621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Persson B, Kallberg Y, Oppermann U, Jornvall H. Chem Biol Interact. 2003;143/144:271–278. doi: 10.1016/s0009-2797(02)00223-5. [DOI] [PubMed] [Google Scholar]
- 8.Chigri F, Soll J, Vothknecht UC. Plant J. 2005;42:821–831. doi: 10.1111/j.1365-313X.2005.02414.x. [DOI] [PubMed] [Google Scholar]
- 9.Berridge MJ, Lipp P, Bootman MD. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 10.Yang T, Poovaiah BW. Trends Plant Sci. 2003;8:505–512. doi: 10.1016/j.tplants.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Burgess WH, Jemiolo DK, Kretsinger RH. Biochim Biophys Acta. 1980;623:257–270. doi: 10.1016/0005-2795(80)90254-8. [DOI] [PubMed] [Google Scholar]
- 12.Reddy VS, Ali GS, Reddy AS. J Biol Chem. 2002;277:9840–9852. doi: 10.1074/jbc.M111626200. [DOI] [PubMed] [Google Scholar]
- 13.Arazi T, Baum G, Snedden WA, Shelp BJ, Fromm H. Plant Physiol. 1995;108:551–561. doi: 10.1104/pp.108.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adami P, Duncan TM, McIntyre JO, Carter CE, Fu C, Melin M, Latruffe N, Fleischer S. Biochem J. 1993;292:863–872. doi: 10.1042/bj2920863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nada A, Soll J. J Cell Sci. 2004;117:3975–3982. doi: 10.1242/jcs.01265. [DOI] [PubMed] [Google Scholar]
- 16.Quiles MJ, Cuello J. Plant Physiol. 1998;117:235–244. doi: 10.1104/pp.117.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waegemann K, Soll J. Plant J. 1991;1:149–158. [Google Scholar]
- 18.Arazi T, Baum G, Snedden WA, Shelp BJ, Fromm H. Plant Physiol. 1995;108:551–561. doi: 10.1104/pp.108.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quiles MJ, Cuello J. Plant Physiol. 1998;117:235–244. doi: 10.1104/pp.117.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.