Abstract
The Saccharomyces cerevisiae DNA helicase Rrm3p is needed for normal fork progression through >1000 discrete sites scattered throughout the genome. Here we show that replication of all yeast chromosomes was markedly delayed in rrm3 cells. Delayed replication was seen even in a region that lacks any predicted Rrm3p-dependent sites. Based on the pattern of replication intermediates in two-dimensional gels, the rate of fork movement in rrm3 cells appeared similar to wild-type except at known Rrm3p-dependent sites. These data suggest that although Rrm3p has a global role in DNA replication, its activity is needed only or primarily at specific, difficult-to-replicate sites. By the criterion of chromatin immunoprecipitation, Rrm3p was associated with both Rrm3p-dependent and -independent sites, and moved with the replication fork through both. In addition, Rrm3p interacted with Pol2p, the catalytic subunit of DNA polymerase ε, in vivo. Thus, rather than being recruited to its sites of action when replication forks stall at these sites, Rrm3p is likely a component of the replication fork apparatus.
Keywords: Rrm3p, Mrc1p, DNA replication, helicase, yeast, chromatin
DNA helicases have fundamental roles in all processes involving DNA, including replication, recombination, and repair (Caruthers and McKay 2002; Tuteja and Tuteja 2004; Eoff and Raney 2005). Rrm3p is a member of the Pif1 family of DNA helicases, a family that is highly conserved from yeasts to humans (Boule and Zakian 2006). Saccharomyces cerevisiae RRM3 was first discovered because its mutation causes increased recombination in the ribosomal DNA (rDNA) (Keil and McWilliams 1993). As assayed by two-dimensional (2D) gel electrophoresis, the absence of Rrm3p results in replication fork pausing at multiple sites in the ribosomal DNA (rDNA) (Ivessa et al. 2000), in telomeres and subtelomeric DNA (Ivessa et al. 2000, 2002), as well as at tRNA genes, inactive replication origins, centromeres, and the silent mating-type loci (Ivessa et al. 2003). By the same assay, the absence of Rrm3p results in increased replication fork pausing within plasmid borne polymerase II transcribed genes when replication and transcription move in opposite directions through the gene (Prado and Aguilera 2005). All Rrm3p-dependent sites that have been identified to date are assembled into stable, nonnucleosomal protein–DNA complexes. Disruption of these complexes makes replication through them Rrm3p independent (Ivessa et al. 2003; Torres et al. 2004a). Rrm3p acts catalytically to promote fork progression as mutations that eliminate its ability to hydrolyze ATP have the same phenotypes as an RRM3 deletion (Ivessa et al. 2000, 2002).
The role of Rrm3p in replication fork progression contributes to genomic integrity. Broken replication forks are detected in rrm3 cells within rDNA, subtelomeric DNA, and tRNA genes (Ivessa et al. 2000, 2002, 2003). These breaks trigger a checkpoint response as the effector checkpoint kinase Rad53p is constitutively phosphorylated in rrm3 cells (Ivessa et al. 2003). The DNA breaks also promote recombination, which is elevated in rrm3 cells. Cells lacking Rrm3p exhibit increased recombination in the rDNA (Keil and McWilliams 1993), which generates rDNA circles (Ivessa et al. 2000; Torres et al. 2004a) that are associated with premature aging (Sinclair and Guarente 1997). Recombination is also increased between other tandem repeats such as CUP1 genes (Keil and McWilliams 1993) and subtelomeric Y′ elements (Ivessa et al. 2002), within a 184-kb tRNA-rich region on chromosome VII (Ivessa et al. 2003; Admire et al. 2006), and between plasmid-borne RNA polymerase II-transcribed genes (Prado and Aguilera 2005).
Although rrm3 cells are viable, they require checkpoint, replication, and repair genes for normal growth (Schmidt and Kolodner 2004; Tong et al. 2004; Torres et al. 2004b). For example, rrm3 cells that lack Mec1p or Rad53p, which are, respectively, sensor and effector kinases for both the intra-S-phase and DNA damage checkpoints, are either dead or very slow growing (Ivessa et al. 2003; Torres et al. 2004b). Likewise, a rrm3 mrc1 strain is not viable nor is the defect in this doubly mutant strain rescued by deletion of RAD51, suggesting that the events that impose the lethality are upstream of recombination (Torres et al. 2004b). Although Mrc1p has both replication and intra-S-phase checkpoint functions (Alcasabas et al. 2001; Osborn and Elledge 2003), only its replication function is essential in rrm3 cells (Szyjka et al. 2005). Mrc1p travels with the replication fork (Alcasabas et al. 2001; Katou et al. 2003; Osborn and Elledge 2003), and in its absence, replication forks move at ∼50% of the wild-type rate (Szyjka et al. 2005; Tourriere et al. 2005).
Taken together, the data suggest that the catalytic activity of Rrm3p is required for efficient replication past specific, particularly stable chromatin-associated complexes. In the absence of Rrm3p, replication forks stall and break at these sites, and this damage is repaired by recombination. Rrm3p is unlikely to be necessary to restart or to repair stalled or broken replication forks as rrm3 cells are not sensitive to DNA damaging agents such as ultraviolet (UV), hydroxyurea, or X-rays (Torres et al. 2004b; S. Dunaway and V.A. Zakian, unpubl.). Rather, Rrm3p likely promotes DNA replication directly. Rrm3p could do so by being recruited to its sites of action when replication forks stall at these sites or it could be a constitutive part of the replication machinery but act only at discrete sites. Alternatively, Rrm3p could affect replication of the entire genome but be particularly important for replication through stable protein–DNA complexes.
Here we show that Rrm3p has a more global role in DNA replication than predicted from previous data. Replication of all yeast chromosomes was delayed in its absence, including a chromosomal region that contains no known Rrm3p-dependent sites. Rrm3p is probably part of the replisome as it moved with the replication fork through both Rrm3p-dependent and -independent sites and interacted with the catalytic subunit of DNA polymerase ε. These results suggest that by virtue of its role in promoting fork progression through difficult-to-replicate sites, Rrm3p is needed for the timely replication of the entire genome.
Results
The Rrm3p helicase affects replication of every yeast chromosome
Although 2D gel analysis of DNA replication in rrm3 cells indicates that replication forks stall at ∼1400 discrete sites, by the same assay, replication fork progression through a large portion of chromosome VI is not Rrm3p sensitive (Ivessa et al. 2003). Since a large fraction of the pause sites (∼900) are in the rDNA on chromosome XII (Ivessa et al. 2000), it is not clear if timely replication of all chromosomes is Rrm3p dependent.
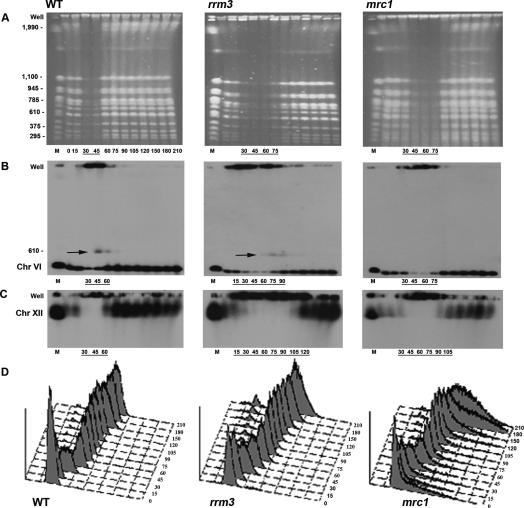
To determine if Rrm3p has global effects on DNA replication, we synchronized cells and examined the replication timing of individual chromosomes by pulsed field gel (PFG) electrophoresis. A replicating chromosome does not enter a PFG but instead is enriched in the well (Hennessy et al. 1991). We examined DNA replication in wild-type, rrm3, and mrc1 cultures. The mrc1 strain was chosen for comparison because it proceeds more slowly through S phase than wild type and because the replication function of Mrc1p is essential in rrm3 cells (see above). To obtain synchronous cultures, cells were arrested in late G1 phase by incubation with α factor. After removal from α factor (0 time point), nocodazole was added to the media in order to prevent entry into a second S phase. Cells proceeded synchronously through S phase at 24°C as monitored by flow cytometry (Fig. 1D). Samples were removed at 15-min (and later 30-min) intervals and analyzed by PFG electrophoresis. Ethidium bromide staining of the gels allowed visualization of the 16 yeast chromosomes (Fig. 1A). In wild-type cells, all chromosomes were underrepresented in the gel 30 and 45 min after α-factor release but were back to normal levels by 60 min, indicating that replication was mostly complete by this time (Fig. 1A, left panel). In the rrm3 strain, chromosomes were depleted from the gel at the 30-, 45-, 60-, and 75-min time points (Fig. 1A, middle panel), indicating a delay in replication as compared with wild-type cells. As expected, replication was also delayed in mrc1 cells (Fig. 1A, right panel).
Figure 1.
Replication of all chromosomes is delayed in rrm3 cells. All time points are indicated below the ethidium bromide-stained wild-type (WT) gel. For ease of viewing, only time points where chromosomes are replicating are indicated below all subsequent gels and Southern blots. (A) Ethidium bromide-stained PFGs of chromosomal-sized DNA molecules from wild-type, rrm3, and mrc1 strains. Chromosomal markers (M) are indicated on the left in kilobases. (B) PFGs were visualized by Southern hybridization with a chromosome VI probe (HIS2). Arrows indicate a form of chromosome VI detected in late S phase that behaves like a linear dimer. The membrane in B was stripped and reprobed sequentially for chromosome XIV (ARS1414 probe; Supplementary Fig. 1) and chromosome XII (rDNA probe; C). (D) Flow-cytometric profiles of synchronized cultures.
To investigate the replication behavior of individual chromosomes, the gels were transferred to membranes and analyzed by Southern hybridization. For each time point, we estimated the replication timing of individual chromosomes by monitoring the enrichment of signal in the well and the diminished signal of the chromosome in the gel, as compared with the signal at the 0 time point, which represents one copy of each chromosome per cell.
By the criterion of 2D gels, chromosome VI, which at 270 kb is the second-smallest yeast chromosome, contains multiple Rrm3p-dependent sites as well as long stretches that lack detectable pauses in rrm3 cells (Ivessa et al. 2003). In wild-type cells, chromosome VI replication occurred mainly in the 30- and 45-min time points (Fig. 1B, left panel). In the 45- and 60-min samples, some chromosome VI hybridizing DNA was found at a size of about twice that of chromosome VI (Fig. 1B, marked by arrow). This transient structure likely represents an almost linear dimer of chromosome VI in which the sister chromatids are held together by a replication fork near one telomere. In rrm3 cells, chromosome VI was depleted in the gel in the 30–75-min time points (Fig. 1B, middle panel). The putative chromosome VI dimer was visible in the 75- and 90-min time points. Moreover, enrichment of chromosome VI DNA in the well occurred as early as 15 min and extended up to the 90-min time point. Thus, chromosome VI replication took considerably longer in rrm3 compared with wild-type cells. Chromosome VI replication was also delayed in mrc1 cells, with most replication occurring between 30 and 75 min (Fig. 1B, right panel). A similar replication pattern was seen for a medium-sized chromosome (785 kb), chromosome XIV (Supplementary Fig. 1). Compared with wild-type cells, replication of chromosome XIV was markedly delayed in both rrm3 and mrc1 cells.
Finally, we examined replication of chromosome XII, the largest yeast chromosome. Chromosome XII contains ∼100–200 tandem copies of the 9.1-kb rDNA repeat (Skryabin et al. 1984) as well as ∼1 Mb of the non-rDNA sequence. Each rDNA repeat has six Rrm3p-dependent sites; resolution of forks converged at the replication fork barrier (RFB) is particularly delayed in rrm3 cells (Ivessa et al. 2000). In wild-type cells, chromosome XII replication was most evident in the 30- and 45-min time points but was still ongoing at 60 min (Fig. 1C, left panel). In rrm3 cells, chromosome XII replication had begun by 15 min and was not complete even at 120 min (Fig. 1C, middle panel). Chromosome XII replication was also delayed in the mrc1 strain but to a lesser extent than in rrm3 cells (complete at 120 min) (Fig. 1C, right panel).
Considering both the ethidium bromide-stained profile and the Southern analysis of chromosomes VI, XIV, and XII, we conclude that Rrm3p is needed for the timely replication of most, and probably all, yeast chromosomes. By PFG analysis, replication in rrm3 cells is initiated slightly earlier than in wild-type or mrc1 cells, and takes somewhat longer than in the mrc1 strain. The replication of chromosome XII is particularly prolonged in rrm3 compared with wild-type or even mrc1 cells.
Slow replication of chromosome XII is due largelyto delayed replication of the rDNA
We next asked if the large delay in completion of chromosome XII replication in rrm3 cells was due solely to the rDNA locus. To release the rDNA array from chromosome XII, the whole chromosome samples that were examined in Figure 1 were digested with the restriction enzyme NotI, which does not cut within the rDNA array. NotI digestion of chromosome XII generates eight fragments, including a 107-kb fragment containing the left telomere, a >1000-kb fragment containing the rDNA, and a 101-kb fragment, the penultimate NotI fragment that ends 50 kb from the right telomere and contains two tRNA genes (Fig. 2A). In wild-type cells, the 107-kb left telomere and the 101-kb right arm fragments were completely replicated by 60 min (Fig. 2A; Supplementary Fig. 2B, left panels). In rrm3 cells, the 107-kb left telomere and the 101-kb right arm fragments were present in the well from 30 to 75 min, indicating that replication of these fragments was not complete until ∼90 min (Fig. 2A; Supplementary Fig. 2B, right panels). When the NotI-digested DNA from wild-type cells was probed for rDNA, rDNA was detected in the well from 30 through 75 min, while in rrm3 cells rDNA replication was present in the well even at 120 min (Fig. 2B, cf. left and right panels). Thus, while replication of all parts of chromosome XII takes longer in rrm3 than in wild-type cells, the prolonged replication of chromosome XII in rrm3 cells (Fig. 1C, middle panel) is largely explained by slow replication of rDNA.
Figure 2.
Delayed replication of segments of chromosome XII and XIV in rrm3 cells. The DNA samples prepared for Figure 1 were digested with NotI and then analyzed by PFG and Southern hybridization. Symbols are the same as in Figure 1. The positions of NotI sites in chromosomes XII (A) and XIV (C) are indicated. Black circles represent centromeres. Time points are labeled as in Figure 1. After separation by PFGs, NotI-digested DNA was transferred to membranes and hybridized sequentially with probes that detect the 107-kb telomeric NotI fragment from chromosome XII (AYTI probe; A), the penultimate 101-kb NotI fragment from the right arm of chromosome XII (CRN1 probe; Supplementary Fig. 2B), the entire rDNA array (rDNA probe; B), and a 129-kb NotI fragment on chromosome XIV (ARS1414 probe; C).
A region with no known Rrm3p-dependent sites shows delayed replication in rrm3 cells
All chromosomes contain multiple Rrm3p-dependent sites (telomeres, centromeres, and tRNA genes). To determine the replication timing of a chromosomal region that lacks known Rrm3p-dependent sites, we monitored replication of a 129-kb NotI fragment from chromosome XIV, a fragment that contains no known Rrm3p-sensitive sites (Fig. 2C). However, this fragment has two active origins of DNA replication, and it is thus one of the earlier replicating regions on chromosome XIV (Raghuraman et al. 2001). While replication of this fragment in wild-type cells occurred in the 30- and 45-min time points, its replication in rrm3 cells was not complete until 90 min, a delay of 30 min compared with wild type (Fig. 2C, cf. left and right panels). Since this NotI fragment contains two active origins of replication, its prolonged replication is unlikely due to delayed movement of replication forks into the region.
Rrm3p associates with both Rrm3p-dependentand -independent sites
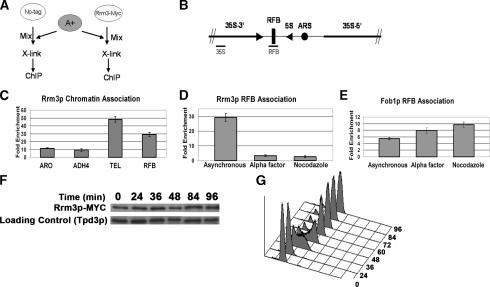
Using chromatin immunoprecipitation (ChIP), we showed previously that subtelomeric Y′ and multiple sites within the rDNA are Rrm3p associated (Ivessa et al. 2000, 2002). In contrast, ACT1 DNA was present in low amounts in the anti-Rrm3p immunoprecipitate (IP). These data led us to propose that Rrm3p associates with the subset of DNA sequences whose replication is Rrm3p dependent. However, the PFG data showing that replication of a region with no known Rrm3p-dependent site was slowed in rrm3 cells (Fig. 2C) led us to consider that Rrm3p may associate with all parts of the genome, not just with Rrm3p-sensitive sites. To test this possibility, we performed ChIP on asynchronous cells, examining the association of Rrm3p with Rrm3p-dependent and -independent sites. For these experiments, the C terminus of Rrm3p was tagged at its endogenous locus with 13 Myc epitopes (Rrm3p-MYC). The cells expressing Rrm3p-MYC and the isogenic no-tag control strain lack the distal portion of the ADH4 gene (a sequence called A+).
If Rrm3p does indeed associate with all sites in the genome, there is no DNA sequence in the same cell that can serve as an appropriate negative control. To circumvent this problem, we performed ChIP on a mixture of two strains (Fig. 3A). Prior to cross-linking, asynchronous cells from the test strains (strains expressing Rrm3p-MYC or the isogenic no-tag control) were mixed with an equal number of asynchronous cells from a strain that contains the entire ADH4 gene (A+ strain). The cell mixtures were then cross-linked with formaldehyde and processed for ChIP. The amount of A+ DNA in the IP served as a negative control for normalization.
Figure 3.
Rrm3p associates with Rrm3p-dependent and -independent sites in a cell cycle-dependent manner. (A) Schematic of cell-mixing experiment used for C and D. The A+ strain contains a portion of ADH4 that is absent from the no-tag control and Rrm3p-MYC strains. (B) Schematic of a single 9.1-kb rDNA repeat. Bars indicate positions of PCR-amplified sequences used for D (RFB) and E (RFB, 35S). (C) DNA immunoprecipitated from the Rrm3p-MYC strain was PCR-amplified for 23 cycles using primers for the RFB and for 28 cycles using primers for the single-copy TEL, ARO, or ADH sequences. Intensities of amplified fragments were quantified by densitometric analysis. The A+ sequence, which is absent from experimental strains, was used for background normalization. Fold enrichment ± standard deviations are indicated here and in D and E. (D) DNA was immunoprecipitated from asynchronous, G1 phase, and G2/M phase Rrm3p-MYC cells and amplified using 23 cycles of multiplex PCR with primers specific to the RFB. The amount of RFB sequence in the immunoprecipitate was normalized to the background intensity of the A+ sequence. (E) DNA was immunoprecipitated as described for D from a Fob1p-MYC strain except that the amount of RFB sequence in the immunoprecipitate was normalized to a sequence from within the 35S-encoding rDNA. (F) Rrm3p-MYC abundance is constant throughout the cell cycle. Synchronized Rrm3p-MYC cells were collected at the indicated time points and analyzed for Rrm3p-MYC abundance after separation by SDS-PAGE and Western blotting with MYC antibody and subsequently with Tpd3 antibody. (G) Flow-cytometric profile of synchronized cells used in F.
We monitored the presence of four test sequences as well as the A+ sequence in the anti-MYC IP. Two of the sequences, the RFB within the rDNA (Fig. 3B) and a single-copy sequence near the left telomere of chromosome VII (TEL), are sites of replication pausing in rrm3 cells (Ivessa et al. 2000, 2002). The other two sites are a segment within the ADH4 gene that is present in the Rrm3p-MYC, no-tag, and A+ strains and a segment of the ARO1 gene. Neither the ADH4 nor the ARO1 segments contain a known Rrm3p-dependent site. PCR reactions were multiplexed with each reaction containing a set of primers for the A+ sequence and one of the four test sites. Compared with the amount of A+ DNA in the IPs, the ARO1 and ADH4 fragments were enriched, respectively, 11 ± 0.6 (average ± standard deviation) and 9 ± 1.5 (Fig. 3C). The enrichment of the single-copy TEL sequence was four- to fivefold higher (48 ± 3.5-fold).The level of enrichment at the RFB (29 ± 2.7) was not directly comparable to the three single-copy sequences, as its amplification was accomplished with fewer PCR cycles due to its being multicopy. At all four sites, association of Rrm3p-MYC was dependent on both in vivo cross-linking and the presence of the MYC tag (data not shown).
If Rrm3p associates with DNA during DNA replication, we expect its association to be higher in asynchronous cells than in G1- or G2/M-arrested cells. Indeed, the enrichment of Rrm3p at the RFB was higher in asynchronous cultures (29 ± 2.6 fold) than in either α-factor-arrested (3 ± 0.6-fold) or nocodazole-arrested cells (2.7 ± 0.6) (Fig. 3D). In contrast, the RFB-binding Fob1p protein showed robust association with the RFB in asynchronous, G1, and G2/M cells (Fig. 3E). Taken together, these data suggest that Rrm3p associates with many genomic sites, including those that are not known to be Rrm3p dependent, and this association likely occurs during S phase.
To determine if Rrm3 protein levels fluctuate throughout the cell cycle, α-factor-arrested Rrm3p-MYC-expressing cells were released into the cell cycle at 24°C and time points were processed for flow cytometry (Fig. 3G) and protein analysis (Fig. 3F). We find that Rrm3p-MYC abundance was similar from late G1 phase through the end of the cell cycle. Likewise, RRM3 transcription is not cell cycle regulated (Spellman et al. 1998).
Rrm3p loads onto origins at the beginning of S phase and migrates with Mcm4p and Pol2p through adjacent DNA, even in regions that contain no known Rrm3p-dependent sites
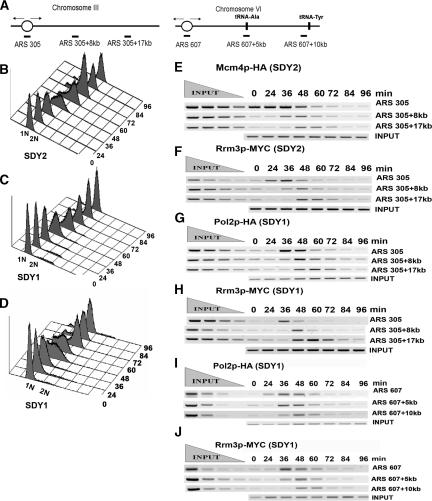
The observation that Rrm3p is associated with Rrm3p-dependent and -independent sites in a cell cycle-dependent manner (Fig. 3) suggested that Rrm3p may be a component of the replication fork machinery. To address this question, we used the ChIP assay in synchronized cells. For these experiments, we constructed two strains, SDY2 and SDY1. Both strains expressed Rrm3p-MYC and either Mcm4p-HA (SDY2) or Pol2p-HA (SDY1) tagged at their C termini with three HA epitopes (Aparicio et al. 1997). Mcm4p, a component of the prereplication complex (pre-RC) (Labib et al. 2001), associates with origins in G1 phase and then moves with the replication fork upon origin firing (Aparicio et al. 1997). Pol2p, the catalytic subunit of DNA polymerase ε, which is thought to be the leading-strand polymerase, loads onto replication origins at the time of origin firing and then moves with the replication fork (Aparicio et al. 1997; Kawasaki and Sugino 2001). These doubly tagged strains allowed us to compare in the same cells the spatial and temporal chromatin association of Rrm3p with that of known components of the replication complex.
Cells released from an α-factor-induced G1-phase arrest (0 time point) at 18°C proceeded synchronously through the cell cycle as monitored by flow cytometry (Fig. 4B–D). Samples were taken throughout the cell cycle and assayed by ChIP. In strain SDY2, we examined protein association at ARS305 on chromosome III and at two sites 8 and 17 kb to the right of ARS305 (Fig. 4A, left). This region was chosen because the movement of Mcm4p and Pol2p through these sites has been studied previously (Aparicio et al. 1997). This region has no predicted Rrm3p-dependent sites and no detectable replication pauses in rrm3 cells (data not shown).
Figure 4.
Rrm3p loads onto origins at the beginning of S phase and migrates with the replication fork through both Rrm3p-dependent and -independent sites. (A) Diagram of the ARS305 region on chromosome III (left) and the ARS607 region on chromosome VI (right). The oval indicates the origin of DNA replication. Vertical bars indicate the positions of two tRNA genes to the right of ARS607. Horizontal bars indicate the positions of the segments amplified by PCR. (B-D) Flow-cytometric profiles for strains used for E and F (SDY2 cells, express Mcm4p-HA and Rrm3p-MYC), G and H (SDY1 cells, express Pol2p-HA and Rrm3p-MYC), and I and J and Supplementary Figure 3 (SDY1 cells, express Pol2p-HA and Rrm3p-MYC). (E,F) SDY2 cells were arrested in late G1 phase and released synchronously into the cell cycle at 18°C. Samples were collected for flow cytometry, ChIP, and Western blot analysis at 12-min intervals. Formaldehyde-fixed cells were immunoprecipitated with anti-HA (Mcm4p, E) or anti-MYC (Rrm3p, F) antibodies. DNA was purified, PCR-amplified, and resolved on a 2.8% ethidium bromide agarose gel. Each primer set was used to amplify both input and immunoprecipitated DNA but only one representative input reaction per site is shown. (G,H). Synchronized SDY1 cells were processed for ChIP and analyzed for association of Pol2p-HA (G) and Rrm3p-MYC (H) with the ARS305 region as described for E and F. (I,J) Synchronized SDY1 cells were processed for ChIP and analyzed for association of Pol2p-HA (I) and Rrm3p-MYC (J) with the ARS607 region as described for E and F.
As reported previously (Aparicio et al. 1997), Mcm4p-HA was associated with ARS305 in G1-arrested cells (Fig. 4E, row one, 0-min time point) and moved along the chromosome as S phase progressed. Mcm4p-HA was associated with ARS305 + 8 kb at 36 and 48 min after release from α factor (Fig. 4E, second row), and with ARS305 + 17 kb at 48 and 60 min (Fig. 4E, third row). In the same cells, Rrm3p-MYC was not origin associated in G1-arrested cells (Fig. 4F, row one, 0-min time point), indicating that Rrm3p is not part of the pre-RC. Rrm3p-MYC was ARS305 associated at 36 min, which by flow cytometry (Fig. 4B) corresponds to the beginning of S phase. The timing and pattern of Rrm3p-MYC association with sites ARS305 + 8 kb and ARS305 + 17 kb was indistinguishable from that of Mcm4p-HA.
Next, we used strain SDY1 to compare the association of Pol2p-HA (Fig. 4G) and Rrm3p-MYC (Fig. 4H) through the ARS305 region. Pol2p-HA and Rrm3p-MYC showed similar patterns of association: Neither was ARS305 bound in G1-arrested cells, but both loaded onto ARS305 at the beginning of S phase (36 min) (Fig. 4C). As S phase progressed, both proteins dissociated from origin DNA. Both proteins were maximally associated with ARS305 + 8 kb at 48 min and with ARS305 + 17 kb at 60 min. As a negative control, we used a strain expressing Est2p-MYC, the catalytic subunit of telomerase, which is not expected to be associated with the replication fork. We saw no association of Est2p-MYC with the ARS305 region at any time in the cell cycle (data not shown). In addition, Pol2p-HA and Rrm3p-MYC associated similarly with the ARS1 region on chromosome IV, which, like the ARS305 region, lacks any known Rrm3p-sensitive sites (Supplementary Fig. 3). Neither protein was ARS1 associated in G1 phase (0 min), both were ARS1 associated at the start of S phase (36 min) (Fig. 4D) and both moved with similar kinetics through sites that are 3 and 12 kb to the left of ARS1.
We also examined Pol2p-HA and Rrm3p-MYC association with ARS607 and sites that are 5 and 10 kb to the right of ARS607 (Fig. 4A, right). The ARS607 + 5-kb segment contains a tRNA gene, tA(AGC)F, that is a site of dramatic replication pausing in rrm3 cells (see also Fig. 7B, below; Ivessa et al. 2003). There is a second tRNA gene, tY(GUA)F2, ∼70 base pairs (bp) to the right of the ARS607 + 10-kb site that is surely Rrm3p dependent as transcription and replication proceed in opposite directions through this gene. As at the other sites, Pol2p-HA (Fig. 4I) and Rrm3p-MYC (Fig. 4J) bound to ARS607 at the beginning of S phase (Fig. 4D), and the two proteins comigrated through ARS607 + 5 kb and ARS607 + 10 kb.
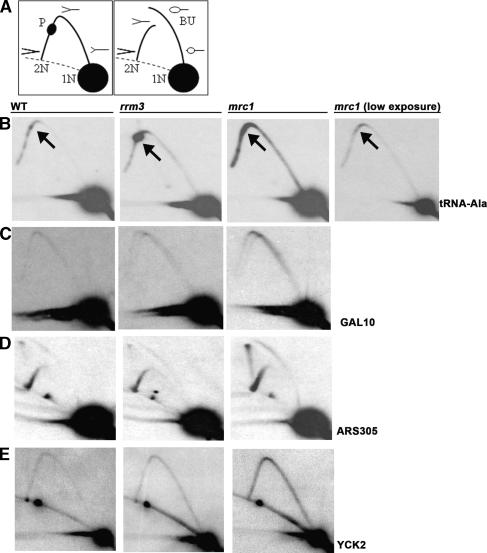
Figure 7.
Abundance of replication intermediates in wild-type (WT), rrm3, and mrc1 strains. (A) Schematics of replication intermediates as visualized in 2D gels. (1N) Nonreplicating fragment; (2N) almost fully replicated fragment right before sister chromatids separate; (P) replication pause; (BU) bubble-shaped replication intermediates. Dotted lines indicate arc formed by linear DNA molecules. (B–E) DNA from glucose-grown asynchronous cells from the indicated strains was restriction enzyme-digested, separated on 2D gels, and analyzed by Southern blotting with the indicated probes (restriction enzymes and hybridization probes are noted in parentheses). (B) tRNAA (BglII, HIS2); arrows indicate the position of the tRNA gene on the arc of replication intermediates; a lower exposure of the sample from the mrc1 strain is shown to provide better visualization of fork movement through the tRNA gene. (C) GAL10 (PvuII, GAL10). (D) ARS305 (EcoRV, ARS305). (E) YCK2 (BglII, YCK2).
Thus, Rrm3p-MYC was associated with three different origins at the beginning of S phase but not in G1 phase. Rrm3p-MYC comigrated with Pol2p-HA through three different regions, including two regions that do not contain Rrm3p-dependent sites. We conclude that Rrm3p moves with the replication fork machinery through both Rrm3p-dependent and -independent sites.
Rrm3p associates with Pol2p in vivo
To provide further support for the interpretation that Rrm3p is part of the replication machinery, we asked if Rrm3p-MYC is associated in vivo with Pol2p-HA (Fig. 5A). Strain SDY1, which expresses Rrm3p-MYC and Pol2p-HA, or control strains expressing only one of the two tagged proteins were arrested with α factor, released into the cell cycle at 24°C, and collected at 40 min when cells were in mid-S phase as determined by flow cytometry (data not shown). Lysates were prepared from each strain and precipitated with anti-MYC antibody (Fig. 5A, left panel) or with anti-HA antibody (Fig. 5A, right panel). The proteins in the IP were separated by SDS-PAGE and analyzed by Western blotting with both anti-HA and anti-MYC antibodies.
Figure 5.
Rrm3p associates with Pol2p during S phase. Protein extracts were prepared from mid-S-phase cultures of strains SDY1 (A) and SDY2 (B), treated with DNase I (+) or not (−), and immunoprecipitated (IP) with either anti-C-MYC Agarose-conjugated beads or anti-HA Agarose-conjugated beads. Input and IP samples were separated on SDS-PAGE gels, and analyzed by Western blotting. Blots were probed with anti-HA (top) or anti-MYC (bottom).
In strain SDY1, an interaction of Pol2p-HA and Rrm3p-MYC was readily detected in the S-phase extracts (Fig. 5A). This interaction was seen when extracts were precipitated with either anti-MYC (Fig. 5A, lanes 3,4) or anti-HA (Fig. 5A, lanes 7,8), and was maintained even when the lysate was treated with DNase I prior to immunoprecipitation (Fig. 5A, lanes 4,8). In the control strain expressing Rrm3p-MYC or Pol2p-HA alone, there were no cross-reacting proteins in either the anti-MYC or anti-HA IPs (Supplementary Fig. 4A,B)
The same experiment was performed using strain SDY2, which expresses both Rrm3p-MYC and Mcm4p-HA. Although an interaction between Rrm3p-MYC and Mcm4-HA was seen in the absence of DNase I treatment (Fig. 5B, lanes 4,8), this interaction was lost when the lysates were pretreated with DNase I (Fig. 5B, lanes 5,9). The coimmunoprecipitation (co-IP) in a DNA-dependent manner of Rrm3p-MYC with Mcm4p-HA probably reflects the fact that although both proteins are at the fork in mid-S phase, they are not in close proximity.
In the absence of Rrm3p, replication forks slowat Rrm3p-dependent sites
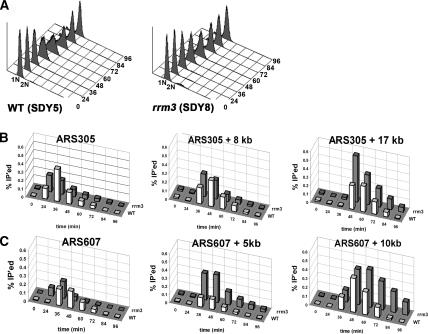
To determine if Pol2p progression through DNA is affected by the absence of Rrm3p, we constructed two strains, SDY5 and SDY8, both of which express Pol2p tagged at its C terminus with 13 MYC epitopes (Pol2p-MYC). The two strains were isogenic except that RRM3 was deleted from strain SDY8. Both strains were arrested with α factor and released synchronously into the cell cycle at 18°C, and samples were processed for flow cytometry (Fig. 6A) and ChIP. In the same synchrony, we examined replication through the ARS305 region, which contains no known Rrm3p-dependent sites (Fig. 4A, left) and the ARS607 region that contains two Rrm3p-dependent sites, one at 5 kb and the other at 10 kb to the right of ARS607 (Fig. 4A, right). If replication forks move more slowly through a given site in rrm3 compared with wild-type cells, there should be more of that sequence in the anti-MYC IP from the mutant cells.
Figure 6.
Pol2p migrates more slowly in the absence of Rrm3p through Rrm3p-dependent sites. Strains SDY5 (expresses Pol2p-MYC) and SDY8 (rrm3 expresses Pol2p-MYC) were synchronized and processed for ChIP as described in Figure 4. (A) Flow-cytometric profiles for synchronized SDY5 and SDY8. (B) The association of Pol2p-MYC with the ARS305 region (diagrammed in Fig. 4A) with (white bars) and without (gray bars) Rrm3p was examined. (C) DNA from the same samples used in B was analyzed for the association of Pol2p-HA with the ARS607 region, which contains two Rrm3p-dependent sites (diagrammed in Fig. 4A, right), with (white bars) and without (gray bars) Rrm3p. In B and C, the intensities of amplified fragments were quantified by densitometric analysis. The percentage of input DNA precipitated (% IP’ed) is graphed for one of the two independent experiments performed with each strain.
By flow-cytometric analysis, both strains were in early S phase at the 36-min time point (Fig. 6A). At both ARS305 and ARS607, Pol2p-MYC was detected at the origin even at 24 min in rrm3 cells (Fig. 6B,C), consistent with evidence from PFG experiments for earlier initiation in rrm3 cells. However, this earlier association of Pol2p-MYC with both origins was seen in only one of two experiments. The levels of Pol2p-MYC association with ARS305 + 8 kb were similar in wild-type and rrm3 cells (Fig. 6B, middle panel) in two of two experiments. The relatively modest increase in Pol2p-MYC association with ARS305 + 17 kb in rrm3 compared with wild-type cells was also seen in both experiments (Fig. 6B, right panel).
More dramatic differences in Pol2p-MYC progression between the wild-type and rrm3 strains were seen in the ARS607 region (Fig. 6C), and these differences were observed in two of two experiments. There was more Pol2p-MYC associated at ARS607 + 5 kb in rrm3 cells compared with wild type (Fig. 6C, middle panel). The association of Pol2p-MYC with ARS607 + 10 kb was even higher and persisted for longer in rrm3 than in wild-type cells (Fig. 6C, right panel). Indeed, in rrm3 cells, association of Pol2p-MYC was readily detected at this site at 84 and even 96 min, when flow-cytometric analysis shows that replication of most of the genome was complete (Fig. 6A). These data suggest that in the absence of Rrm3p, the replication fork moves more slowly at sites that are Rrm3p dependent.
In the absence of Rrm3p, replication fork progression is similar in wild-type and rrm3 cellsat Rrm3p-independent sites
The rate of fork progression can also be estimated from the abundance of replication intermediates in 2D gels. Slower fork movement causes the fraction of DNA in replication structures to increase in vivo. We used 2D gels to examine replication intermediates at multiple loci in wild-type, rrm3, and mrc1 cells (Fig. 7; data not shown). Of the four loci examined in Figure 7, only one contains a known Rrm3p-dependent site (Fig. 7B, tRNA-Ala). At each of the four loci, the intensity of the arc of forked replication intermediates was similar in wild-type and rrm3 cells. As shown previously (Szyjka et al. 2005), there were more replication intermediates in mrc1 cells at each site. Whereas increased replication pausing at tRNA-Ala was readily detected in rrm3 cells, there was no detectable pausing at this site in mrc1 cells (Fig. 7B). Thus, although replication of all chromosomes is slowed in both rrm3 and mrc1 cells (Figs. 1, 2), the mechanism leading to this effect is different in the two mutants.
Discussion
Here we show that lack of Rrm3p delays replication of all yeast chromosomes (Fig. 1). The extent of this delay was particularly dramatic at the rDNA locus on chromosome XII (Fig. 2B). However, the impact on other chromosomes such as chromosomes VI and XIV was also striking, as complete replication of these chromosomes took roughly twice as long in rrm3 compared with wild-type cells (Fig. 1B; Supplementary Fig. 1). The extent of this replication delay was even greater than for mrc1 cells (Fig. 1) where replication forks are estimated to move at 50% of wild-type rates. We cannot rule out that some of the delay in chromosomes entering the PFGs in the rrm3 strain was due to the presence of recombination intermediates rather than to replication intermediates. However, even though recombination is increased in rrm3 cells, the recombination rate is still sufficiently low that recombination should affect the behavior of only a small number of chromosomes. For example, the rate of recombination in rrm3 cells in a tRNA-rich 184-kb interval on chromosome VII is only 1.5 × 10−5 events per cell division (Ivessa et al. 2003).
Not only was replication of all chromosomes Rrm3p dependent, replication of a region without known Rrm3p-dependent sites, a 129-kb NotI fragment on chromosome XIV, was dramatically slowed in rrm3 cells (Fig. 2C). These results suggest that there are additional Rrm3p-dependent sites that have not yet been identified. By the criterion of 2D gels, we did not see strong replication pausing at the ∼50 RNA polymerase II-transcribed genes within a 105-kb region of chromosome VI nor during galactose-induced transcription of the GAL1, GAL7, or GAL10 genes (Ivessa et al. 2003). However, using 2D gels, others detect replication pauses within genes transcribed from the GAL1 promoter when these genes are carried on plasmids, and this pausing is increased in rrm3 cells (Prado and Aguilera 2005). Regions like the 129-kb NotI fragment may contain one or more genes whose replication is Rrm3p sensitive, even if this sensitivity cannot be detected with 2D gels when the genes are in their normal chromosomal context. In addition, Rrm3p may be needed for efficient resolution of converged replication forks throughout the genome, as it is in the rDNA (Ivessa et al. 2000). Consistent with this possibility, X-shaped molecules are more abundant within subtelomeric Y′ elements, the silent mating type loci, and on plasmid-borne RNA polymerase II-transcribed genes in rrm3 cells (Ivessa et al. 2000, 2002, 2003; Prado and Aguilera 2005). Since the 129-kb NotI fragment contains two early-firing origins, its delayed replication in rrm3 cells may be due in part to decreased efficiency in resolution of converged forks. A role for Rrm3p in the resolution of converged forks may also explain the modest increase in Pol2-MYC at ARS305 + 17 kb in the absence of Rrm3p (Fig. 6B). This site is midway between ARS305 and ARS306, a region where replication forks are expected to converge.
One model for the mechanism of Rrm3p action is that it is recruited to the fork when the fork stalls at a difficult-to-replicate site. However, in asynchronous cells, Rrm3p was associated with both Rrm3p-dependent and Rrm3p-independent sites (Fig. 3C). Moreover, in synchronized cells, Rrm3p loaded onto three different origins, ARS305, ARS1, and ARS607 at the same time as Pol2p. Rrm3p movement through the DNA adjacent to the three ARSs paralleled that of both Pol2p and Mcm4p, even though only one of these regions, ARS607, contained Rrm3p-sensitive sites (Fig. 4; Supplementary Fig. 3). In addition, using co-IP, we found that Rrm3p was associated with Pol2p in vivo (Fig. 5). Thus, our data argue strongly against a model in which Rrm3p is recruited to specific sites upon stalling of the replication fork. Rather, our data suggest that Rrm3p is a constitutive part of the replication apparatus. Alternatively, Rrm3p could be part of a chromatin remodeling complex that moves immediately ahead of the replication fork and makes chromatin more accessible to the replication machinery.
Our results disagree with another study that examined replication through the ARS305 region into which two ectopic RFB sites were inserted (Calzada et al. 2005). Because this group detected Rrm3p at the RFB but not at ARS305 nor at a site distal to the ectopic RFB, they concluded that Rrm3p is not part of the replisome but rather is recruited to stalled forks. In contrast, in the unmodified ARS305 region, as well as at two additional genomic loci, we saw Rrm3p moving with the fork (Fig. 4; Supplementary Fig. 3). We think a possible explanation for the discrepancy between the data from the two labs is that there is more Rrm3p associated with the RFB as compared with the number of Rrm3p molecules that travel with the replication machinery. We speculate that more Rrm3p molecules may be recruited to particularly stable protein–DNA complexes such as the complexes at the RFB and telomeres (Fig. 3). The number of Rrm3p molecules required to facilitate replication past specific protein–DNA complexes may depend on the stability of the complex in front of the fork.
Is replication fork stalling at stable chromatin structures and difficulty in resolving converged forks sufficient to explain the large replication delay seen in rrm3 cells? A decrease in the number of active origins can also prolong S phase. However, origin usage is similar in wild-type and rrm3 cells in the rDNA (Ivessa et al. 2000), at ARS605 (Ivessa et al. 2003), and at ARS305 (Fig. 7D), while normally inactive origins are actually more active in rrm3 cells (Ivessa et al. 2002, 2003). A decrease in the speed of replication forks can also prolong S phase. Slower fork movement in mrc1 cells results in more replication intermediates, as assayed by 2D gels, a result confirmed here (Fig. 7). However, increased fork pausing at Rrm3p-dependent sites such as tRNA genes was not seen in mrc1 cells (Fig. 7B; data not shown). In contrast to the results in mrc1 cells, the fraction of DNA in replication intermediates in rrm3 and wild-type cells was similar (Fig. 7), except at Rrm3p-dependent sites such as at a tRNA gene (Fig. 7B).
We conclude that neither reduced origin usage nor a genome-wide slowing of replication forks explains the longer S phase in rrm3 cells. Rather, the global delay in DNA replication observed in this strain (Figs. 1, 2) is due largely or entirely to site-specific delays in replication, including compromised resolution of converged forks. Others have speculated that the lethality of mrc1 rrm3 cells reflects a requirement for Rrm3p in the restart of forks that are damaged as a result of replication in the absence of Mrc1p (Szyjka et al. 2005). However, we find that while replication takes longer in both mrc1 and rrm3 cells (Fig. 1), the mechanism causing slow replication was different in the two strains (Fig. 7). We think that the most likely explanation for the lethality of mrc1 rrm3 doubly mutant cells is the additive effects of two different types of defects in DNA replication.
Although there are several examples of prokaryotic helicases that help replication forks move past protein–DNA complexes (Bedinger et al. 1983; Yancey-Wrona and Matson 1992; Barry and Alberts 1994), Rrm3p is the first eukaryotic enzyme shown to function in this way. Since these difficult-to-replicate protein–DNA complexes are encountered in every S phase, they may pose an even greater threat to genome stability than exogenous DNA damage. Genetic assays using mutants in replication and repair genes have found high frequencies of genomic instability at Ty1 elements, tRNA genes, and converging replication forks, sites that include Rrm3p-dependent loci (Scholes et al. 2001; Cha and Kleckner 2002; Lemoine et al. 2005; Admire et al. 2006). Some of these sites overlap with Rrm3-dependent sites, suggesting that forks stall and collapse at preferred locations within the genome that are difficult to replicate. The rrm3 system provides a unique opportunity to study the mechanism of replication fork stalling and breakage at sites that are natural impediments to fork progression. Analysis of Rrm3p may provide insights into the fate of replication fork arrest and breakage at difficult-to-replicate loci in human chromosomes, which become fragile sites in response to replication stress and whose breakage can initiate the types of chromosomal aberrations found in human tumors.
Materials and methods
Strains
Yeast strains are described in Supplementary Table 1. Precise deletions of the RRM3 and MRC1 ORFs were created using the pRS303-HIS3 plasmid (Sikorski and Hieter 1989) as a template to generate HIS3 PCR products with overhangs that targeted the product to either RRM3 or MRC1 (Ivessa et al. 2000). Similarly, the BAR1 ORF was precisely deleted using pRS304-TRP1 (Sikorski and Hieter 1989). The bar1Δ∷TRP1 wild-type, rrm3Δ, and mrc1Δ strains were constructed by B. Lenzmeier. The bar1Δ allele was introduced into rrm3Δ and mrc1Δ strains by mating followed by sporulation and identification of doubly mutant spore clones. Rrm3p, Fob1p, and Pol2p were tagged at their C termini with 13 MYC epitopes using the plasmid pFA6-TRP-13MYC as a template (Longtine et al. 1998). The Rrm3p-MYC and Fob1p-MYC had wild-type function as assayed by 2D gel analysis of rDNA replication intermediates (data not shown). The growth rate of the Pol2p-MYC strain was indistinguishable from wild type, suggesting that the Pol2p-MYC also had wild-type function (data not shown). To generate VPS106-UT, VPS106 was transformed with the 2.4-kb SalI–EcoR1 fragment of pADH4UCAIV. Integration of this fragment results in deletion of the terminal ∼15 kb of chromosome VII-L and its replacement with URA3 adjacent to a new telomere (Gottschling et al. 1990). The strains for the fork migration studies were rrm3Δ∷HIS3 or Rrm3p-MYC13 derivatives of strains generously provided by O. Aparicio and S. Bell that express Pol2p-3HA (strain OAy618) or Mcm4p-3HA (strain OAy535) (Aparicio et al. 1997). In all cases, tagged proteins were expressed from their endogenous promoters, and expression was verified by Western analysis.
Cell synchronies
For PFG, ChIP, and co-IP experiments, single colonies were inoculated in 5 mL of YEPD, grown for 6 h at 30°C, diluted into YEPD, and grown overnight at 30°C to an OD660 of 0.15. α Factor (Princeton University) was added to cultures at a concentration of 0.015 ng/mL, and cultures were incubated for 3 h at 24°C until microscopic examination indicated that ∼95% of cells were unbudded. Cells were washed in YEPD; released into fresh YEPD containing 70 μg/mL Pronase (Sigma) at 24°C for co-IP, PFG, and asynchronous ChIP experiments, and at 18°C for synchronous ChIP; and samples were collected at the indicated time points. For PFG experiments, 15 μg/mL nocodazole (Sigma) was added to the releasing media to arrest cells in G2/M and prevent their entry into a second cell cycle. The quality of each synchrony was monitored by flow cytometry. Briefly, 500 μL of cells from each time point were resuspended in water, sonicated, and fixed in 70% ethanol at room temperature for 3 h. Cells were resuspended in 50 mM sodium citrate and incubated with 0.125 mg of RNase A overnight at 50°C. One milligram of Proteinase K (Roche) was added and cells were incubated for 2 h at 50°C. Cells were sonicated and incubated in the dark with Sytox Green (2 μM) (Invitrogen) for 45 min at room temperature. Samples were analyzed in a FACScan single-laser fixed-alignment benchtop analyzer with an emission at 488 nm. A 530/30 bandpass filter was used to collect emissions in front of the detector.
Gel methods
To process samples for PFGs, cultures were killed in 0.1% sodium azide, pelleted, and washed in ddH2O and then in SPE (0.5 M Sorbitol, 100 mM NaPO4 at pH 7.5, 10 mM EDTA). Cells were resuspended in 1.5 mL of 1 M sorbitol, pelleted, and resuspended in SPEM (SPE containing 30 mM β-mercapthenol) and Zymolyase (100 μg/mL; ICN Biomedicals). After cells were incubated for 10 min at 30°C, an equal volume of InCert agarose (BMA) was added to each cell suspension, and the mixture was pipetted into disposable CHEF plug molds (Bio-Rad) and allowed to solidify on ice. Plugs were incubated in SPEM containing 0.05 mg/mL Zymolyase for 3 h at 37°C, rinsed in 0.5 M EDTA (pH 9.0), resuspended in lysis solution (10 mM Tris-HCl at pH 8.0, 50 mM EDTA at pH 9.0, 1% w/v Sarkosyl) containing proteinase K (Roche) at a final concentration of 1 mg/mL, and incubated overnight at 55°C. Plugs were washed twice with 2.5 mL 0.5× TBE (1 mM EDTA at pH 8.0, 0.044 mM boric acid, 0.045 mM Tris Base), resuspended in 2.5 mL 0.5× TBE Buffer, and stored at 4°C. Chromosomes were separated using 1% gels (Bio-Rad Pulsed Field Certified Agarose) in 0.5× TBE buffer using the Bio-Rad CHEF-DR III System. Running conditions for PFGs were 60–120-sec switch time, 6 V/cm voltage gradient, and 120° angle for 26 h at 14°C. NE Biolaboratories yeast chromosome PFG Marker was run as a reference. Gels were stained in 0.5 μg/mL ethidium bromide for 30 min, destained in deionized water for 20 min, and photographed using the AlphaImager system (Alpha Innotech). Chromosomes were UV-cross-linked and transferred to nylon Hybond membranes (Amersham) for Southern analysis. All probes, with the exception of the BglII-B rDNA probe, were PCR products amplified from yeast genomic DNA (Supplementary Table 2). PCR products were gel-purified using Zymoclean DNA column (Zymo Research), and 10 ng of product was used as a template for a second round of PCR. The second PCR products were gel-purified and labeled using the RediPrime II Random Prime Labeling System (Amersham) and 10 μL of 6000 μCi α-P32 dCTP (IDT). Membranes were subsequently reprobed after stripping with boiling 0.25% SDS solution. For NotI-digested samples, DNA agarose plugs were digested with 10,000 U of NotI (NE Biolaboratories) for 12 h at 37°C and analyzed as described above. Methods of DNA preparation and conditions for running 2D gels to detect replication intermediates are as described (Ivessa et al. 2000).
ChIP
ChIP analyses with synchronized cells were performed essentially as described (Taggart et al. 2002) except samples were not multiplexed. Monoclonal anti-HA (12CA5; Roche or Santa Cruz Biotechnology) or anti-Myc (BD Biosciences) antibodies were used. The number of PCR cycles varied from 25 to 30, depending on the sequence being amplified and on the protein that was analyzed. PCR products were resolved in 2.8% ethidium bromide agarose gels. Amplified fragment intensities were visualized using the Alpha Imager system. Ethidium bromide gels of PCR products were quantified by densitometric analysis using Scion Image 4.0.3.2 software (http://www.scioncorp.com). The percent immunoprecipitated is the relative recovery of immunoprecipitated DNA relative to the amount of input DNA based on the quantification of the respective PCR products and the dilution factor of immunoprecipitated and input DNA. Each ChIP experiment was done at least twice, with similar results.
For ChIPs on asynchronous cultures, cells were grown to an OD660 of 0.7. The Rrm3p-MYC (JZT200) or the untagged control strain (VPS106UT) was mixed with an equal number of VPS106 cells. Strains VPS106UT, JZT200, and JZT515 lack a portion of the ADH4 gene, called the A+ sequence, that is present in VPS106. The amount of A+ DNA in the IP served as a negative control for a sequence that is not Rrm3p-MYC associated. Cross-linking and ChIP were carried out as described except that magnetic protein G Dynabeads (Dynal Biotech) were used. DNA immunoprecipitated from Rrm3p-MYC was amplified using 23 cycles of PCR with primers that amplify the RFB and 28 cycles with primers that amplify the ARO1, ADH4, TEL, and A+ sequences. PCR products were resolved on 2.8% ethidium bromide agarose gels and bands were quantified by densitometric analysis (NIH Image1.60). Fold enrichment of Rrm3p-bound DNA was determined as follows: (MYC-tagged B/no-tag B) × (no-tag A+/MYC-tagged A+), where B denotes tested sequence. For G1 (α-factor-arrested) and G2/M (nocodazole-arrested) cells, cultures were grown to an OD660 of 0.3 and arrested with a final concentration of 2 μg/mL α factor for 2 h at 30°C or with a final concentration of 15 μg/mL nocodazole for 3 h at 30°C prior to mixing and in vivo cross-linking. To determine Fob1p-MYC association with the RFB, DNA was amplified using 23 cycles of PCR with primers that amplify the RFB and a 407-bp sequence internal to the 35S rDNA (35S). Fold enrichment of Fob1p-MYC with the RFB was determined as follows: (MYC-tagged RFB/no-tag RFB) × (no-tag 35S/MYC-tagged 35S).
Co-IP of Rrm3p with replication proteins
Cell cultures were synchronized with α factor at 24°C as described, collected at the 40-min S-phase time point, killed with 0.1% sodium azide on ice, pelleted, resuspended in cell lysis buffer (50 mM HEPES at pH 7.5, 10% v/v glycerol, 140 mM NaCl, 1 mM EDTA, 0.5% IGEPAL CA-630, 1 mM EDTA, 1 mM PMSF, Roche Complete EDTA-free Protease inhibitors), and frozen as pellets in liquid nitrogen. Cell pellets were processed using a Freezer Mill (SPEX CertiPrep) to >95% lysis efficiency. An aliquot was taken and resuspended in protein sample buffer (Fig. 5, Input). Cell lysates were spun down at 4°C to clear lysates, which were divided in half for control and DNase I-treated samples. Twenty millimolar MgCl2 and 1 mM PMSF were added to all lysates, and 150 U of DNase I (Sigma) were added to DNase I samples. Lysates were incubated on ice for 15 min. Aliquots of cell extracts were taken after DNase I incubation and resuspended in protein sample buffer as protein controls and for phenol-chloroform extraction of DNA to assay efficient digestion by DNase I. Fifty microliters of anti-C-MYC Agarose-conjugated beads (Sigma) for anti-MYC immunoprecipitation or 50 μL of rabbit anti-HA Agarose-conjugated beads (Abcam) were added to both samples and the mixtures were rotated for 1.5 h at 4°C. Beads were washed seven times in 1 mL of cell lysis buffer, spun down, resuspended in protein sample buffer, and boiled. Equal portions of each IP were separated on 7% SDS-PAGE gels and transferred to PVDF membranes (Millipore). Membranes were blocked in TBST (10 mM Tris-HCl at pH 8, 150 mM NaCl, 0.07% Tween-20) containing 5% milk, incubated in a 1:400 dilution of HA monoclonal antibody (Santa Cruz Biotechology) for anti-MYC IPs or 1:1000 dilution of MYC monoclonal antibody (BD Biosciences) for anti-HA IPs, and then incubated in a 1:3000 dilution of goat antimouse horseradish peroxidase-conjugated secondary antibody (Bio-Rad). Membranes were developed using ECL chemiluminescence system (Amersham) and exposed to Kodak Biomax XAR autoradiography film. Membranes were subsequently incubated in stripping buffer (100 mM β-mercapthenol, 2% SDS, 64 mM Tris-HCl at pH 6.8) for 30 min at 50°C, blocked in TBST containing 5% milk, and reprobed with an antibody corresponding to the one used for the IP (MYC or HA) to evaluate efficiency of the IP.
Levels of Rrm3p over the cell cycle
To determine Rrm3p levels throughout the cell cycle, cultures were synchronized with α factor and released into the cell cycle at 24°C as described above. Cells were collected at the indicated time points and crude extract was prepared using the trichloracetic acid (TCA) method described in Pellicioli et al. (1999). Protein concentration in TCA protein preparations were determined using RC DC colorimetric assay (Bio-Rad). For each time point, equal protein amounts were processed for Western analysis. Proteins were separated on 7% SDS-PAGE gels and transferred to PVDF membranes (Millipore). After probing with anti-MYC antibody, membranes were stripped and probed with 1:1000 dilution of anti-Tpd3 antibody (gift of J. Broach), and then incubated in a 1:3000 dilution of goat anti-rabbit secondary antibody (Bio-Rad). Membranes were then stained with 0.2% Ponceau Stain for 30 min, and destained in water as a second control for protein loading.
ACKNOWLEDGMENTS
We thank J. Broach for the anti-Tpd3 antibody, S. Bell and O. Aparicio for providing strains, R. Ricke and A.K. Bielinsky for advice on co-IP, A. Ivessa for assistance with 2D gels, C. Tuzon for assistance with PFGs, M. Sabourin for advice on ChIP, and S. Aubert, M. Mateyak, S. Pinter, and C. Webb for suggestions on this manuscript. This work was supported by National Institutes of Health grants R37 GM26938, T32CA09328 (to J.Z.T., A.A., and S.D.) and a predoctoral fellowship from the NJ Commission of Cancer Research (J.B.B.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1478906
REFERENCES
- Admire, A., Shanks, L., Danzl, N., Wang, M., Weier, U., Stevens, W., Hunt, E., Weinert, T. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes & Dev. 2006;20:159–173. doi: 10.1101/gad.1392506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcasabas, A., Osborn, A., Bachant, J., Hu, F., Werler, P., Bousset, K., Furuya, K., Diffley, J., Carr, A., Elledge, S. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Aparicio, O.M., Weinstein, D.M., Bell, S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Barry, J., Alberts, B. A role for two DNA helicases in the replication of T4 bacteriophage DNA. J. Biol. Chem. 1994;269:33063–33068. [PubMed] [Google Scholar]
- Bedinger, P., Hochstrasser, M., Jongeneel, C., Alberts, B. Properties of the T4 bacteriophage DNA replication apparatus: The T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983;34:115–123. doi: 10.1016/0092-8674(83)90141-1. [DOI] [PubMed] [Google Scholar]
- Boule, J.B., Zakian, V.A. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 2006;4:4147–4153. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada, A., Hodgson, B., Kanemaki, M., Bueno, A., Labib, K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes & Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers, J.M., McKay, D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- Cha, R.S., Kleckner, N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- Eoff, R.L., Raney, K.D. Helicase-catalysed translocation and strand separation. Biochem. Soc. Trans. 2005;33:1474–1478. doi: 10.1042/BST0331474. [DOI] [PubMed] [Google Scholar]
- Gottschling, D.E., Aparicio, O.M., Billington, B.L., Zakian, V.A. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Hennessy, K.M., Lee, A., Chen, E., Botstein, D. A group of interacting yeast DNA replication genes. Genes & Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Ivessa, A.S., Zhou, J.-Q., Zakian, V.A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- Ivessa, A.S., Zhou, J.-Q., Schulz, V.P., Monson, E.M., Zakian, V.A. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes & Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa, A.S., Lenzmeier, B.A., Bessler, J.B., Goudsouzian, L.K., Schnakenberg, S.L., Zakian, V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- Katou, Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., Shirahige, K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Kawasaki, Y., Sugino, A. Yeast replicative DNA polymerases and their role at the replication fork. Mol. Cells. 2001;12:277–285. [PubMed] [Google Scholar]
- Keil, R.L., McWilliams, A.D. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics. 1993;135:711–718. doi: 10.1093/genetics/135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., Kearsey, S.E., Diffley, J.F. MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol. Biol. Cell. 2001;12:3658–3667. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine, F.J., Degtyareva, N.P., Lobachev, K., Petes, T.D. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Osborn, A.J., Elledge, S.J. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes & Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli, A., Lucca, C., Liberi, G., Marini, F., Lopes, M., Plevani, P., Romano, A., Di Fiore, P.P., Foiani, M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado, F., Aguilera, A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman, M.K., Winzeler, E.A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D.J., Davis, R.W., Brewer, B.J., Fangman, W.L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Ricke, R.M., Bielinsky, A.K. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol. Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Schmidt, K.H., Kolodner, R.D. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 2004;24:3213–3226. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes, D.T., Banerjee, M., Bowen, B., Curcio, M.J. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, V.P., Zakian, V.A. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair, D.A., Guarente, L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Skryabin, K.G., Eldarov, M.A., Larionov, V.L., Bayev, A.A., Klootwijk, J., de Regt, V.C., Veldman, G.M., Planta, R.J., Georgiev, O.I., Hadjiolov, A.A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984;12:2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., Futcher, B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyjka, S.J., Viggiani, C.J., Aparicio, O.M. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol. Cell. 2005;19:691–697. doi: 10.1016/j.molcel.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Taggart, A.K.P., Teng, S.-C., Zakian, V.A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., Lesage, G., Bader, G.D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G.F., Brost, R.L., Chang, M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Torres, J.Z., Bessler, J.B., Zakian, V.A. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes & Dev. 2004a;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, J.Z., Schnakenberg, S.L., Zakian, V.A. The Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: Viability of rrm3 cells requires the intra S phase checkpoint and fork restart activities. Mol. Cell. Biol. 2004b;24:3198–3212. doi: 10.1128/MCB.24.8.3198-3212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere, H., Versini, G., Cordon-Preciado, V., Alabert, C., Pasero, P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol. Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Tuteja, N., Tuteja, R. Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery. Eur. J. Biochem. 2004;271:1835–1848. doi: 10.1111/j.1432-1033.2004.04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey-Wrona, J.E., Matson, S.W. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 1992;20:6713–6721. doi: 10.1093/nar/20.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]