Abstract
SMAD4 is inactivated in the majority of pancreatic ductal adenocarcinomas (PDAC) with concurrent mutational inactivation of the INK4A/ARF tumor suppressor locus and activation of the KRAS oncogene. Here, using genetically engineered mice, we determined the impact of SMAD4 deficiency on the development of the pancreas and on the initiation and/or progression of PDAC—alone or in combination with PDAC-relevant mutations. Selective SMAD4 deletion in the pancreatic epithelium had no discernable impact on pancreatic development or physiology. However, when combined with the activated KRASG12D allele, SMAD4 deficiency enabled rapid progression of KRASG12D-initiated neoplasms. While KRASG12D alone elicited premalignant pancreatic intraepithelial neoplasia (PanIN) that progressed slowly to carcinoma, the combination of KRASG12D and SMAD4 deficiency resulted in the rapid development of tumors resembling intraductal papillary mucinous neoplasia (IPMN), a precursor to PDAC in humans. SMAD4 deficiency also accelerated PDAC development of KRASG12D INK4A/ARF heterozygous mice and altered the tumor phenotype; while tumors with intact SMAD4 frequently exhibited epithelial-to-mesenchymal transition (EMT), PDAC null for SMAD4 retained a differentiated histopathology with increased expression of epithelial markers. SMAD4 status in PDAC cell lines was associated with differential responses to transforming growth factor-β (TGF-β) in vitro with a subset of SMAD4 wild-type lines showing prominent TGF-β-induced proliferation and migration. These results provide genetic confirmation that SMAD4 is a PDAC tumor suppressor, functioning to block the progression of KRASG12D-initiated neoplasms, whereas in a subset of advanced tumors, intact SMAD4 facilitates EMT and TGF-β-dependent growth.
Keywords: Smad4, pancreatic cancer, epithelial-to-mesenchymal transition mouse models, TGF-β
PDAC (pancreatic ductal adenocarcinoma) ranks as the fourth leading cause of cancer mortality in the United States and carries a median survival of <6 mo (Li et al. 2004). Hallmarks of this disease include the infiltration of the tumor with a proliferative stroma (desmoplasia), early invasion and metastasis, and pronounced genomic instability (Solcia et al. 1995). PDAC is characterized by a highly recurrent pattern of genetic lesions consisting of activating mutations of KRAS and inactivation of INK4A (via mutation, deletion, or promoter methylation) in virtually all cases, inactivation of the p53–ARF pathway in ∼87% of cases (including tumors with deletions of the INK4A/ARF locus), and SMAD4 inactivation in ∼53% (Hansel et al. 2003). Hence, SMAD4 status can be considered as a distinguishing molecular feature of two major classes of PDAC. Significant ongoing efforts are being directed toward the elucidation of how specific signature mutations contribute to the genesis and progression of PDAC and influence its tumor biological features.
Morphological and genetic analyses have implicated three distinct ductal neoplasms as potential precursor lesions of human PDAC, designated pancreatic intraepithelial neoplasms (PanIN), intraductal papillary mucinous neoplasms (IPMN), and mucinous cystic neoplasms (MCN) (Maitra et al. 2005). The best characterized of these neoplasms, PanIN, appears to evolve in a stepwise manner through stages that display increasing cellular atypia and accumulating clonal mutations—in KRAS, p16INK4A, p14ARF, p53, and SMAD4—in the course of progressing to PDAC (Hansel et al. 2003). IPMN and MCN—distinguished by their cystic character—show a comparable mutational profile to PanIN, albeit with different frequencies of the various mutations. The interrelationship of these various premalignant neoplasms and the basis for these tumor phenotypes are not clear.
Genetically engineered mouse models have provided a system to define genotype–phenotype relationships in PDAC pathobiology. In the mouse pancreas, expression of an activated Kras (KrasG12D) knock-in allele induces PanIN lesions that can gradually progress to PDAC (average latency >1 yr) (Aguirre et al. 2003; Hingorani et al. 2003). Inactivation of the Ink4a/Arf tumor suppressor locus (encoding both p16INK4A and p19ARF), or of p53, does not induce pancreatic neoplasia; however, concurrent KrasG12D expression and inactivation of Ink4a/Arf or p53 drives the development of advanced PDAC with short latency (Aguirre et al. 2003; Hingorani et al. 2005; Bardeesy et al. 2006). The role of Smad4 has not yet been investigated in the context of these models. Mice with germline homozygous Smad4 inactivation die during early embryogenesis, while Smad4+/− mice develop gastric and duodenal polyps but have not been reported to have pancreatic pathology (Sirard et al. 1998; Yang et al. 1998; Takaku et al. 1999).
Inactivating mutations in SMAD4 are far more common in PDAC than in other cancer types (Hahn et al. 1996). This tumor suppressor gene encodes a transcription factor that is a central effector of the transforming growth factor-β (TGF-β), bone morphogenetic protein (BMP), and Activin signaling pathways (Bierie and Moses 2006). TGF-β, BMP, or Activin family ligands signal through cognate receptor serine/threonine kinases that phosphorylate receptor SMADs—SMAD1, SMAD2, SMAD3, SMAD5, and SMAD8. The activated receptor SMADs bind to SMAD4 and translocate to the nucleus, where higher-order transcriptional complexes regulate expression of broad sets of genes. These pathways can also signal independently of SMAD4 as reflected by the capacity of TGF-β to regulate JNK and Par6 in some cell types and to induce Smad2/3-TIF1γ transcriptional complexes (He et al. 2006; for review, see Bierie and Moses 2006).
TGF-β is a potent inhibitor of epithelial cell growth and survival through modulation of expression of cell cycle regulators and activation of apoptosis, although these effects are highly dependent on cellular context (Bierie and Moses 2006). The tumor suppressor role of TGF-β signaling is underscored by presence of inactivating TGF-β receptor mutations in several cancers. On the other hand, TGF-β can enhance the malignant growth of some established epithelial tumors, promoting tumor cell proliferation, migration, and the epithelial-to-mesenchymal transition (EMT)—a process by which advanced carcinomas acquire a highly invasive, undifferentiated and metastatic phenotype (Zavadil and Bottinger 2005). Therefore, TGF-β signaling can have biphasic stage-specific effects—inhibiting carcinoma initiation while promoting the high-grade advancement and dissemination of established tumors. BMP signaling also has a clear role in tumorigenesis as demonstrated by the association of juvenile polyposis syndrome with germline loss-of-function mutations in either BMPR-1A or in SMAD4 (Howe et al. 1998, 2001); the impact of these germline mutations on PDAC risk or clinical presentation is not defined.
The biological role of SMAD4 mutations in human PDAC progression is an area of active investigation, often with contrasting observations. PDAC cell lines show variable sensitivity to TGF-β-induced cytotoxicity in a manner that appears independent of SMAD4 status (Dai et al. 1999; Giehl et al. 2000; Jonson et al. 2003; Nicolas and Hill 2003; Subramanian et al. 2004). Some evidence suggests that a prominent role for SMAD4 resides in modulating the tumor microenvironment. Specifically, while SMAD4 restoration in some PDAC cell lines has minimal effects on cell growth in vitro, there is strong inhibition of PDAC xenografts perhaps through repression of angiogenesis and extracellular matrix remodeling (Schwarte-Waldhoff et al. 2000; Duda et al. 2003). On the other hand, separate studies have shown that SMAD4 restoration results in attenuated growth in soft agar of some PDAC cell lines, suggesting a cell-autonomous function (Peng et al. 2002).
Complicating interpretation of the role of SMAD4 loss in PDAC is the observation that TGF-β signaling may enhance tumorigenicity in established tumors. TGF-β ligands and receptors are frequently expressed at elevated levels in PDAC relative to normal pancreas, and this overexpression appears to correlate with decreased survival (Friess et al. 1993b; Wagner et al. 1999). TGF-β is thought to promote PDAC desmoplasia as well as contribute to proliferation of the tumor cells in an autocrine manner (Friess et al. 1993a; Wagner et al. 1999; Lohr et al. 2001); notably, blockade of TGF-β signaling by expression of soluble type II TGF-β receptor attenuates tumorigenicity of xenografts (Rowland-Goldsmith et al. 2001), and, reciprocally, exogenous addition of TGF-β enhances tumor invasion (Ellenrieder et al. 2001).
It is notable that opposing conclusions have been reached, in separate clinicopathological studies, regarding the impact of SMAD4 status on the prognosis of PDAC. One study determined that patients expressing SMAD4 had significantly worse outcomes and did not benefit from surgery (Tascilar et al. 2001), while another study indicated that SMAD4 expression predicted increased survival and improved response to surgery (Biankin et al. 2002). Overall, the complex response profiles to TGF-β suggest that TGF-β/SMAD4 signaling may have pleiotropic and context-dependent roles in PDAC. This significant genetic and biological complexity provides formidable challenges in designing therapeutic strategies directed against this pathway. In this study, we sought to understand the role of Smad4 in normal pancreas development and physiology as well as in the genesis and progression of PDAC, alone or together with other common PDAC genetic lesions.
Results
Normal development and functionof the SMAD4-deficient pancreas
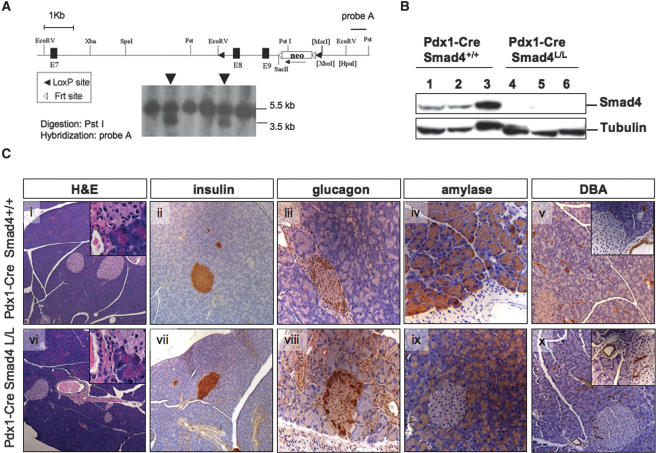
To study the role of Smad4 in pancreatic development, physiology, and malignant transformation, we generated a conditional knockout allele of Smad4 (Smad4lox) harboring loxP sites flanking exons 8 and 9 in the mouse germline (Fig. 1A). We crossed Smad4lox homozygous mice to either the Pdx1-Cre or Ptf1a-Cre transgenic mice (Gu et al. 2002; Kawaguchi et al. 2002) (hereafter, for brevity, Cre will be used for references to both strains together); these transgenes direct Cre recombinase expression to the epithelial lineages of the embryonic pancreas (Pdx1-Cre is also expressed in the duodenum and pylorus). The use of Pdx1-Cre allowed direct comparison with our existing extensively characterized Pdx1-Cre-driven models of PDAC, while the more organ-specific Ptf1a-Cre transgene obviated certain extra-pancreatic phenotypes (see below). Cre-mediated rearrangement of the Smad4lox locus in pancreas tissue from Cre Smad4lox/lox mice was documented by allele-specific PCR genotyping (data not shown) and by elimination of Smad4 protein on Western blot analysis (Fig. 1B), confirming generation of a null allele in the pancreas.
Figure 1.
Conditional deletion of Smad4 in the pancreas. (A, top) Genomic structure of the targeted Smad4 allele. Exons (black rectangles), the Pgk-Neo cassette, loxP sites, Frt sites, and the probe for embryonic stem (ES) cell screening (black bar) are depicted. (Bottom) Southern blot of targeted Smad4 locus in two ES cell clones (arrowheads). (B) Western blot analysis of Smad4 expression in pancreatic lysates from 8-wk-old Pdx1-Cre Smad4+/+ and Pdx1-Cre Smad4lox/lox mice. (C) Histological sections from 8-wk-old Pdx1-Cre Smad4+/+ (panels i–v) and Pdx1-Cre Smad4lox/lox (panels vi–x) mice stained with H&E (panels i,vi); with antibodies to insulin (panels ii,vii), glucagon (panels iii,viii), and amylase (panels iv,ix); and with DBA-lectin (panels v,x). Magnifications: Panels i,vi, 50×; panels ii,v,vii,x, 100×; panels iii,iv,viii,ix, and insets, panels v,x, 200×; insets, panels i,vi, 400×.
Mice with homozygous deletion of Smad4 in the pancreas were born at the expected frequency (data not shown), showed no evidence of any gross anatomic or physiological abnormalities, and exhibited normal pancreatic cytoarchitecture and differentiation throughout postnatal life (n = 10 mice, ages 46–70 wk) (Fig. 1C). In addition, the normal glucose tolerance tests and normal serum lipase and amylase levels at 10 wk of age were consistent with the generally healthy appearance and weight gain of these mice. Amylase, insulin, and glucagon showed normal expression by immunohistochemical analysis, and staining with the Dolichos biflorus (DBA) lectin confirmed the presence of a normal proportion of pancreatic ducts (Fig. 1C). Finally, it is notable that none of the Cre Smad4lox/lox animals developed pancreatic neoplasms (up to 70 wk of age/observation), although all Pdx1-Cre Smad4lox/lox mice were found to harbor duodenal polyps comparable to those previously reported in germline Smad4+/− mice (Takaku et al. 1999; Supplementary Fig. 1A). These data indicate that Smad4 inactivation alone does not prominently affect pancreas development or physiology or play a role in the initiation of pancreatic cancer (see Discussion).
KrasG12D-directed pancreatic tumorigenesisis enhanced in mice with Smad4 deletion
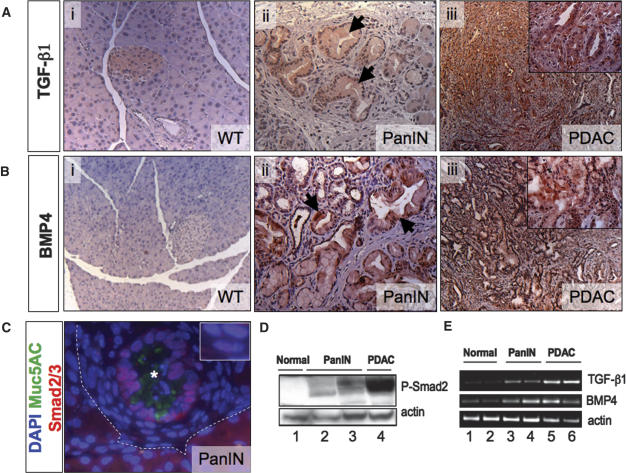
To investigate the role of Smad4 in regulating PDAC progression, we first studied the activation state of TGF-β family signaling throughout the course of tumorigenesis in the Pdx1-Cre LSL-KrasG12D model. We found that expression of TGF-β1 and BMP4 were induced in the PanINs and metaplastic ducts of Pdx1-Cre LSL-KrasG12D pancreata as demonstrated by immunohistochemical analysis (Fig. 2A,B). Moreover, we detected nuclear localization of Smad2/3 in PanIN lesions by immunofluorescence (Fig. 2C), and Smad2 phosphorylation was revealed by immunoblot analysis (Fig. 2D). RT–PCR analysis confirmed the induction and sustained expression of TGF-β, and BMP4 in PanIN and established PDAC (Fig. 2E); moreover, expression profiling analysis of PDAC samples demonstrated broad induction of TGF-β pathway components (Supplementary Fig. 2). These data indicate that TGF-β signaling is activated in PanINs and advanced cancers, consistent with observations in human specimens (Friess et al. 1993b; Wagner et al. 1999; Ito et al. 2004), suggesting that this signaling pathway may serve to support neoplastic growth in the pancreas or, alternatively, provide a checkpoint mechanism designed to constrain tumor progression.
Figure 2.
TGF-β family signaling in KrasG12D-initiated PanIN/PDAC. Immunohistochemical staining for TGF-β (A) and BMP-4 (B) in pancreata from wild-type mice (panel i), Pdx1-Cre LSL-KrasG12D mice (panel ii), and Pdx1-Cre LSL-KrasG12D Ink4a/Arflox/lox mice(panel iii) with PDAC. Note positive staining for both TGF-β and BMP4 in normal islets (panel i), in PanINs and metaplastic ducts (panel ii), and in PDAC (panel iii). (C) Immunofluorescence staining for Smad2/3 (red) and Muc5ac (green) in a PanIN arising in a Pdx1-Cre LSL-KrasG12D mouse (region above dashed line; the PanIN lumen is marked with an asterisk). The ductal epithelial cells of the PanIN show cytoplasmic and membranous Muc5ac staining and nuclear localization of Smad2/3. (Inset) The stromal cells surrounding the PanIN epithelium also show nuclear Smad2/3. Nuclei are stained with DAPI (blue). (D) Western blot showing expression of phospho-Smad2/3 in a pancreas from a wild-type mouse (lane 1), from Pdx1-Cre LSL-KrasG12D mice (lanes 2,3), and in PDAC from a Pdx1-Cre LSL-KrasG12D Ink4a/Arflox/lox mouse (lane 4). (E) RT–PCR analysis of TGF-β1, BMP4, and actin expression in pancreata from wild-type mice (lanes 1,2), Pdx1-Cre LSL-KrasG12D mice (lanes 3,4), and in PDAC from Pdx1-Cre LSL-KrasG12D Ink4a/Arflox/lox mice (lanes 5,6). Magnifications: A (panels i,iii), B (panels i,iii), 100×; A (panels ii,iii [inset]), B (panels ii,iii [inset]), 200×; C, 630×.
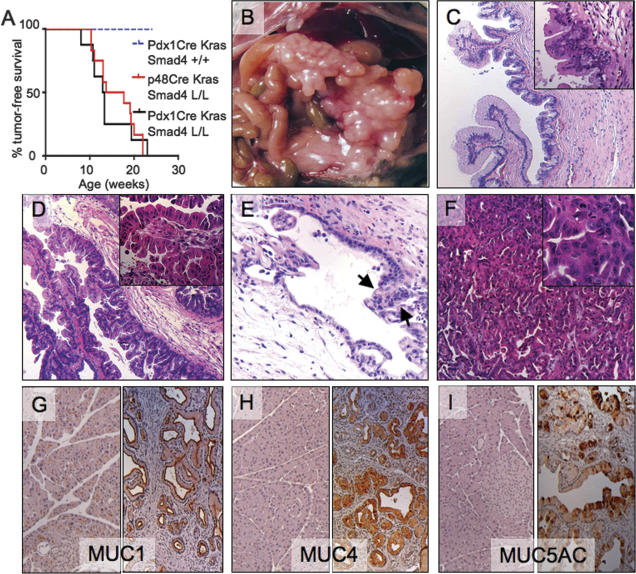
Molecular studies have identified distinct subsets of human PDAC that have common mutations in KRAS and INK4A and are distinguished by intact or loss-of-function alleles of SMAD4. These mutational profiles, coupled with the activated TGF-β signaling in KrasG12D-induced PanINs, prompted evaluation of mice harboring various combinations of Cre, LSL-KrasG12D, Smad4lox, and/or Ink4a/Arflox alleles. As reported previously, Cre-mediated activation of the LSL-KrasG12D allele in the pancreas resulted in the development of slowly progressive PanINs with uniformly good health through age 40 wk and thereafter a subset of these mice developed PDAC (Aguirre et al. 2003; Hingorani et al. 2003; Bardeesy et al. 2006). In contrast, the combination of activated KrasG12D and Smad4 deficiency resulted in a dramatic reduction in survival; all Ptf1a-Cre LSL-KrasG12D Smad4lox/lox (n = 7) and Pdx1-Cre LSL-KrasG12D Smad4lox/lox (n = 8) mice presented with a palpable abdominal mass between ages 7 and 12 wk, and reached terminal morbidity between ages 8 and 24 wk of age (p < 0.001 for both colonies compared with Pdx1-Cre LSL-KrasG12D cohort) (Fig. 3A; Table 1).
Figure 3.
Smad4 suppresses KrasG12D-driven pancreatic tumorigenesis. (A) Survival curve (Kaplan-Meier) of the Pdx1-Cre LSL-KrasG12D, Ptf1a-Cre LSL-KrasG12D Smad4lox/lox, and Pdx1-Cre LSL-KrasG12D Smad4lox/lox cohorts. (B–I) Pancreatic tumorigenesis in Ptf1a-Cre LSL-KrasG12D Smad4lox/lox mice. (B) Gross photo of cystic pancreatic tumor in a 14-wk-old mouse. C and D show H&E images of the pancreas in B demonstrating regions of IPMN adenoma (C), borderline IPMN (D), and IPMN with carcinoma (E) in situ (arrows). (F) H&E image from 17-wk-old mouse showing PDAC with moderately differentiated ductal histology. (G–I) IHC of normal pancreas (left panels) and IPMN (right panels) staining for Muc1 (G), Muc4 (H), and Muc5AC (I). Magnifications: C–I, 100×; C (inset), 200×; insets in E,F, 400×.
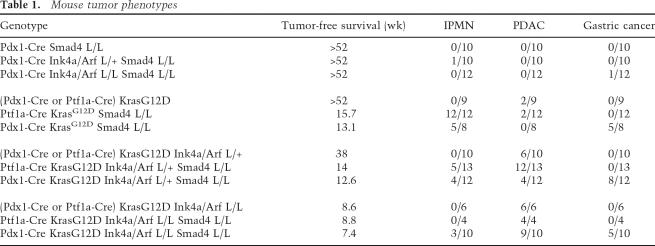
Table 1.
Mouse tumor phenotypes
Gross inspection of the Ptf1a-Cre LSL-KrasG12D Smad4lox/lox mice at necropsy revealed cystic pancreata in 12 of 12 cases (Fig. 3B). Comparable pancreatic tumors were observed in five out of eight Pdx1-Cre LSL-KrasG12D Smad4lox/lox mice with five out of eight of these mice also possessing gastric tumors, consistent with foregut expression of Pdx1 (Supplementary Fig. 1B). Histological analysis revealed large lesions reminiscent of increasing grades of human IPMN in 17 out of 20 mice (Fig. 3C–E), while PDAC with papillary and moderately differentiated ductal elements was observed in two out of 20 mice (Fig. 3F; see Table 1). Pancreatic ductal neoplasms in humans show characteristic profiles of mucin expression (Moniaux et al. 2004); assessment of the murine IPMNs revealed positive staining for Muc1, Muc4, and Muc5AC and absence of reactivity for Muc2 and Cdx2, indicating that the mouse lesions resemble the “gastric-type” IPMN in humans (Fig. 3G–I; Adsay et al. 2004). The stomach cancers in the Pdx1-Cre mice showed either squamous or adenosquamous histology (Supplementary Fig. 1B). Together, our findings provide clear genetic evidence of a role of Smad4 in blocking the progression of KrasG12D-initiated PanINs to lethal pancreatic and gastric cancer.
We have reported previously on the absence of pancreatic neoplasia in Pdx1-Cre Ink4a/Arf mutant mice (Aguirre et al. 2003). Extending those results, we observed only a modest cancer predisposition in the Pdx1-Cre Smad4lox/lox Ink4a/Arflox/lox or Pdx1-Cre Smad4lox/lox Ink4a/Arflox/+ mice. Examination of 23 mice, ages 30 and 64 wk, revealed a single case of an IPMN-like tumor and another with metastatic gastric cancer (Table 1). Thus, these observations underscore the roles of these tumor suppressors in progression rather than initiation of pancreatic cancer, as well as the requirement of coincident activated Kras signaling in order to effect malignant transformation.
Smad4 deficiency enhances developmentof KrasG12D-initiated pancreatic ductal lesions
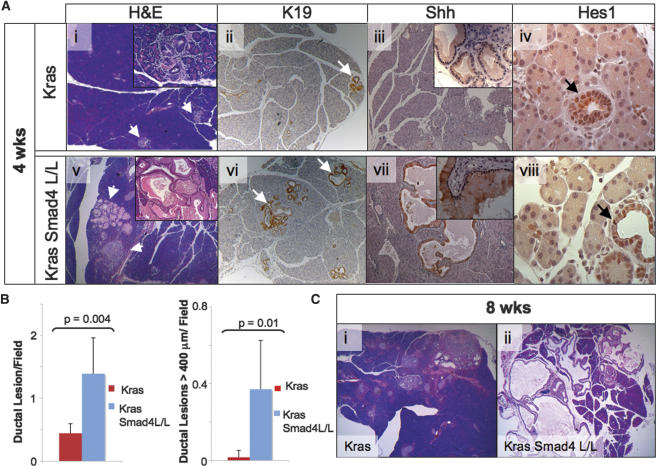
The more rapid tumor progression in mice with combined KrasG12D activation and Smad4 deficiency prompted more detailed cellular and molecular analyses to clarify the impact of Smad4 deletion on evolving pancreatic neoplasms in the KrasG12D model. In previous studies, the Pdx1-Cre LSL-KrasG12D mice developed focal PanINs by 3 wk that showed a gradual increase in size and number but no malignant progression over the next 20 wk (Aguirre et al. 2003; Hingorani et al. 2003). On the basis of these kinetics, we performed serial autopsies at 2, 4, 5, and 8 wk (n = 3–8 mice per time point). While no histological differences were evident at 2 wk (data not shown), the 4-wk time point revealed a significant impact of Smad4 deficiency. Although age-matched Cre LSL-KrasG12D and Cre LSL-KrasG12D Smad4lox/lox pancreata (n = 6 mice/genotype) both showed low-grade PanINs and acinar-ductal metaplasia, Smad4 deficiency correlated with a significant increase in both the number and size of the lesions (p = 0.004 and p = 0.01, respectively; Mann-Whitney), as demonstrated by morphometric analysis (Fig. 4A [panels i,v], B; see Materials and Methods). These more pronounced pathological changes were associated with rapid neoplastic progression in the Ptf1-Cre LSL-KrasG12D Smad4lox/lox animals, which showed extensive IPMN and advanced PanIN lesions by 8 wk, whereas their age-matched Ptf1-Cre LSL-KrasG12D counterparts had only focal low-grade PanINs (Fig. 4C).
Figure 4.
Comparison of evolving pancreatic lesions in LSL-KrasG12D mice with intact or deleted Smad4 alleles. (A) Pancreas specimens from 4-wk-old Ptf1-Cre LSL-KrasG12D (panels i–iv) and Ptf1-Cre LSL-KrasG12D Smad4lox/lox (panels v–vii) mice were stained with H&E (panels i,v) and with antibodies to cytokeratin-19 (panels ii,vi), Sonic Hedgehog (panels iii,vii) and Hes1 (panels iv,viii). The arrows point to pancreatic ductal lesions. (B) Morphometric analysis of pancreata from 4- to 5-wk-old Ptf1-Cre LSL-KrasG12D and Ptf1-Cre LSL-KrasG12D Smad4lox/lox mice (see Materials and Methods). (Left panel) Graph of number of ductal lesions per 100× field. (Right panel) Graph of number of ductal lesions per 100× field measuring >400 μm at greatest diameter. Six mice per genotype were analyzed with a minimum of eight fields counted per mouse. (C) H&E-stained pancreas from 8-wk-old Ptf1-Cre LSL-KrasG12D (panel i) and Ptf1-Cre LSL-KrasG12D Smad4lox/lox (panel ii) mice. Magnifications: A (panels i–iii,v–vii), C (panels i,ii), 50×; A (insets in panels i,v), 200×; A (insets in panels iii,vii), 400×.
We next sought to characterize potential Smad4-dependent changes in the neoplastic epithelium and in the tumor microenvironment. IHC analysis of lesions at 4 wk in both genotypes revealed comparably altered epithelial differentiation and activation of early developmental pathways implicated in PDAC development, with positive staining for cytokeratin-19; Shh1; Hes1; Pdx1; phospho-Stat3;mucins Muc1, Muc4, and Muc5AC; and lack of acinar (amylase) and islet (insulin) marker expression (Fig. 4A; Supplementary Table 1; Moniaux et al. 2004; Hezel et al. 2006; data not shown).
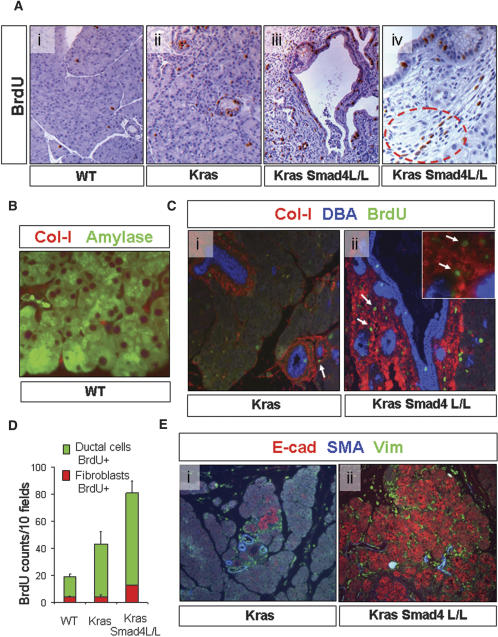
On the other hand, the PanIN and metaplastic ductal lesions in Cre LSL-KrasG12D Smad4lox/lox mice demonstrated increased proliferation relative to lesions in Cre LSL-KrasG12D Smad4+/+ mice and to controls, with prominent BrdU staining in both epithelial and stromal cells (Fig. 5A). Desmoplasia, a hallmark of PDAC in humans (Solcia et al. 1995), is a collagen-rich stromal proliferation within a tumor containing proliferative fibroblasts including so-called pancreatic stellate cells that have been implicated in promotion of tumorigenesis (Apte et al. 2004). In the normal pancreas, collagen-1 expression marks the periacinar and periductal fibroblasts (Fig. 5B). Multilabel immunofluorescence and BrdU staining was used to assess both the extent of stromal deposition and the relative rates of proliferation in the fibroblast and ductal epithelial compartments (detected by collagen-1 and DBA lectin immunofluorescence, respectively). In 4-wk old mice, the ductal lesions from both models expressed collagen 1 in their stroma, with the staining being particularly prominent in the larger lesions in the Smad4-deficient animals (Fig. 5C); quantitation of the BrdU staining confirmed that Smad4 deficiency was associated with significantly increased proliferation of both the neoplastic epithelium and the associated stromal fibroblasts (p < 0.001, unpaired t-test) (Fig. 5D).
Figure 5.
Smad4 deficiency cooperates with KrasG12D to promote epithelial and stromal expansion. (A) BrdU labeling of pancreata from wild-type (panel i), Ptf1-Cre LSL-KrasG12D (panel ii), and Ptf1-Cre LSL-KrasG12D Smad4lox/lox (panel iii) mice. (Panel iv) High-power view of proliferating epithelial and stromal cells (circled) in evolving PanIN from Ptf1-Cre LSL-KrasG12D Smad4lox/lox mice. (B) Immunofluorescence staining of wild-type mouse pancreas with antibodies against amylase (green) and collagen-1 (red) demonstrating the periacinar location of pancreatic fibroblasts. (C) Staining of ductal lesions from Ptf1-Cre LSL-KrasG12D (panel i) and Ptf1-Cre LSL-KrasG12D Smad4lox/lox (panel ii) mice with DBA lectin (blue) and with antibodies to collagen-1 (red) and BrdU (green) reveals proliferation in both the PanIN epithelium and fibroblastic components. (Panel ii) Inset is a high-power view showing fibroblast proliferation (arrows). (D) Graph of number of BrDU-positive epithelial and fibroblast cells per 200× field; three mice per genotype were analyzed. (E) Immunofluorescence staining of pancreata from 4-wk-old Ptf1-Cre LSL-KrasG12D (panel i) and Ptf1-Cre LSL-KrasG12D Smad4lox/lox (panel ii) mice with antibodies to E-cadherin (red), smooth muscle actin (blue), and vimentin (green). Magnifications: H (panels i,ii), 50×; A–C,F (panels i,ii), 100×; C, inset, 200×; D,E, 400×.
Activated stellate cells in human PDAC are characterized by induction of expression of smooth muscle actin expression (SMA), vimentin, and PDGFRβ (Apte et al. 2004). Further characterization of the fibroblast compartment revealed comparable marker profiles in the PanINs of either Smad4 genotype; strong staining was noted for vimentin and PDGFRβ, while SMA was absent in the earliest PanINs—except in the perivascular smooth muscle cells—indicating the apparent absence of stellate cell activation in these incipient lesions, whereas established PanINs showed induction of SMA regardless of Smad4 status (Fig. 5E; Supplementary Fig. 3A). Analysis of inflammatory cells revealed increases in macrophages in the Smad4 mutant lesions although these differences did not reach statistical significance (Supplementary Fig. 3B). Overall, these results indicate that Smad4 deficiency is associated with increased proliferation of both the neoplastic epithelium as well as the stromal tissue. This may suggest that Smad4 has both cell-autonomous and non-cell-autonomous functions in the pancreas, whereas alternatively, the stromal alterations may be secondary to epithelial proliferation. Finally, E-cadherin—a strongly induced protein in human PanIN—was also elevated in the murine lesions with notable increased staining intensity in the Smad4-deficient PanIN and metaplastic epithelium, suggesting Smad4-depedent alterations in the epithelial program (Fig. 5E; see below).
Smad4-deficiency promotes well-differentiated PDAC in the KrasG12D Ink4a/Arflox model
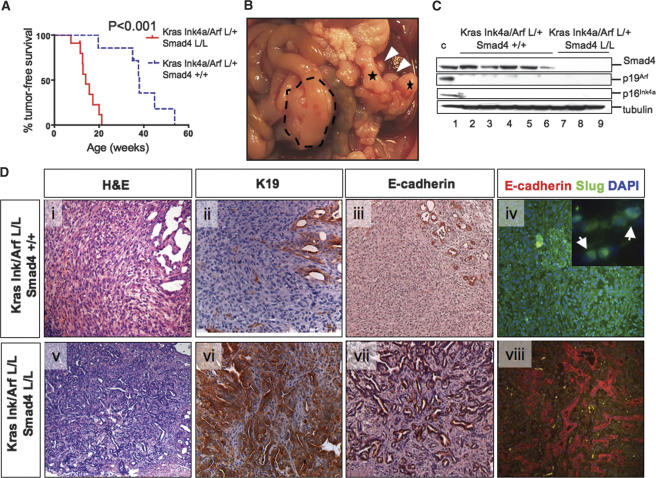
Since the mice with combined KrasG12D expression and Smad4 deletion showed rapid onset of IPMN and advanced PanIN lesions but exhibited only moderate malignant progression of pancreatic tumors, and since SMAD4 loss occurs with concurrent INK4A loss and KRAS activation in human PDAC, we assessed subsequently the combined impact of Smad4 and Ink4a/Arf mutations in Cre LSL-KrasG12D mouse PanIN/PDAC models. Similar to our previous report (Bardeesy et al. 2006), KrasG12D and Ink4a/Arf heterozygosity cooperated to promote PDAC progression in Cre LSL-KrasG12D Ink4a/Arflox/+ mice. A pronounced cooperative effect of Smad4 deletion was noted with Ptf1a-Cre LSL-KrasG12D Smad4lox/lox Ink4a/Arflox/+ mice showing significantly reduced survival relative to Cre LSL-KrasG12D Ink4a/ Arflox/+ controls (mean survival 14.0 wk vs. 38.0 wk; p < 0.001) (Fig. 6A). This reduced survival was associated with PDAC in 12/13 mice, IPMN in 1/13 mice, and concomitant IPMN and PDAC in four out of 13 animals (Fig. 6B; see Table 1). Pdx1-Cre LSL-KrasG12D Smad4lox/lox Ink4a/Arflox/+ mice also showed a shortened survival (mean 12.6 wk) associated with PDAC in four out of 12 mice, gastric cancer in eight out of 12 mice (including one with PDAC and four with IPMN) (see Table 1). Six out of 16 Smad4-deficient PDACs showed liver metastasis or invasion (Supplementary Fig. 1C).
Figure 6.
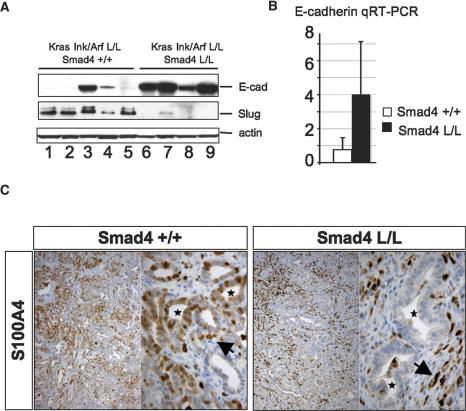
Smad4 deletion promotes the glandular PDAC in cooperation with KrasG12D activation and Ink4a/Arf deficiency. (A) Kaplan-Meier analysis showing pancreas tumor-free survival of Ptf1a-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4lox/lox mice and Ptf1a-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4+/+ mice. Twelve of 13 deaths in the Ptf1a-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4lox/lox mice were due to PDAC, and one out of 13 was due to IPMN. (B) PDAC arising in a Ptf1a-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4lox/lox mouse (indicated by dashed line); note the liver metastases (white arrowheads). This mouse also showed an IPMN (denoted by asterisks). (C) Western blot analysis of lysates from early passage PDAC cell lines from the Ptf1a-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4+/+ (lanes 2–6) and Ptf1a-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4lox/lox (lanes 7–9) models for expression of Smad4, p19Arf, p16Ink4a, and tubulin. Lane 1 shows positive controls. (D) Undifferentiated PDAC from a Ptf1a-Cre LSL-KrasG12D Ink4a/Arflox/+ mouse (panels i–iv) and a moderately differentiated PDAC from a Ptf1a-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4lox/lox mouse (panels v–viii) stained with H&E (panels i,v), or with antibodies to cytokeratin-19 (panels ii,vi) and E-cadherin (panel iii,vii), and double-labeled with antibodies to E-cadherin (red) and Slug (green) (panels iv,viii). Magnifications: All are 100× except D (panel iv, inset), which is 630×.
Western blot analysis of early passage PDAC cell lines revealed absence of Smad4 expression in all tumors from the Smad4lox/lox mice and retention of expression in all PDACs from Smad4+/+ animals (Fig. 6C). Absence of p16Ink4a and p19Arf expression was documented in each of these tumor cell lines, regardless of germline Smad4 status, and was associated with loss of the wild-type Ink4a/Arf allele (Fig. 6C; data not shown). p53 was intact in all cases as determined by p21CIP induction following exposure of the cell lines to ionizing radiation (data not shown). These genetic studies and molecular profiles show that Smad4 deficiency promotes the development of IPMN in KrasG12D mutant mice independently of Ink4a/Arf mutation, while Ink4a/Arf mutations are required for full malignant progression to PDAC in the context of KrasG12D and Smad4 inactivation.
Smad4 deficiency also altered the tumor phenotypes of mice with combined KrasG12D activation and homozygous Ink4a/Arf deletion; Pdx1-Cre LSL-KrasG12D Smad4lox/lox Ink4a/Arflox/lox mice (n = 10) had a slightly decreased survival (7.4 wk vs. 8.6 wk in Pdx1-Cre LSL-KrasG12D Ink4a/Arflox/lox mice; p = 0.01) with nine out of 10 mice displaying PDAC, five out of 10 mice harboring gastric cancers, and three out of 10 with IPMN coincident with the other tumors (Table 1). Ptf1-Cre LSL-KrasG12D Smad4lox/lox Ink4a/Arflox/lox mice (four of four) developed PDAC with a mean latency of 8.8 wk, indicating that Smad4 status did not accelerate PDAC in context of homozygous Ink4a/Arf deletion.
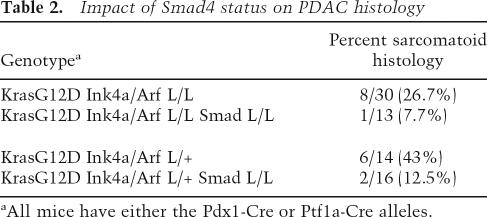
Next, as human and mouse PDACs are known to present with a range of distinct histopathological phenotypes—from well-differentiated, wherein tumor cells form discernable duct-like/glandular structures, to undifferentiated carcinomas that may arise from EMT of the glandular epithelium (Solcia et al. 1995; Hruban et al. 2006)—and Smad4/TGF-β signaling has been implicated in processes of epithelial differentiation (for review, see Zavadil and Bottinger 2005), we sought to determine whether Smad4 status influenced the morphological presentation of the Cre LSL-KrasG12D Ink4a/Arf mutant models. Upon placement on the Smad4-deficient background, there was a significant decrease in the proportion of undifferentiated carcinomas in Cre KrasG12D Ink4a/Arf lox/lox and Cre KrasG12D Ink4a/Arf lox/+ mice: from 26.7% and 43% to 7.7% and 12.5%, respectively (Table 2). The Smad4-deficient PDACs typically showed well-to-moderately-differentiated (ductal) histology and strong cytokeratin-19 staining in contrast to the weak signal detected in regions of undifferentiated histology frequently noted in the Smad4+/+ tumors (Fig. 6D, cf. panels i,ii and v,vi; note transition between undifferentiated and ductal histology in panel ii). These observations suggest that active TGF-β/Smad4 signaling may contribute the loss of epithelial differentiation in PDAC, as observed in skin and breast cancers (for review, see Bierie and Moses 2006).
Table 2.
Impact of Smad4 status on PDAC histology
aAll mice have either the Pdx1-Cre or Ptf1a-Cre alleles.
TGF-β-mediated induction of the Snail and Slug transcriptional repressors and consequent repression of E-cadherin expression contribute to EMT in several cell types (for review, see Zavadil and Bottinger 2005). Correspondingly, PDACs from Smad4+/+ mice showed frequent loss of E-cadherin and elevation of Slug expression compared with PDACs from Smad4lox/lox mice as demonstrated by immunostaining of primary tumors (Fig. 6D) and Western blot analyses of PDAC cell lines (Fig. 7A). The sustained high level of E-cadherin in the Smad4 mutant tumors was confirmed by qRT–PCR analysis (Fig. 7B). The S100A4/FSP1 protein has been shown to promote EMT in cancer cell lines while also marking a subset of fibroblasts (Okada et al. 1997). The PDAC stromal fibroblasts showed strong S100A4 staining regardless of Smad4 status; on the other hand, while the tumor cells in the Smad4+/+ tumors frequently stained positive for S100A4, staining was uncommon in the Smad4-deficient tumor epithelium (Fig. 7C). Overall, these results are consistent with a role of TGF-β–Smad4 signaling in controlling the PDAC cellular differentiation phenotype, possibly through regulation of Slug expression.
Figure 7.
Retention of epithelial differentiation in Smad4-deficient PDAC. (A) Western blot showing E-cadherin (top panel) and slug expression (middle panel) in PDAC cell lines from Cre LSL-KrasG12D Ink4a/Arflox/lox Smad4+/+ mice (lanes 1–5) and Cre LSL-KrasG12D Ink4a/Arflox/lox Smad4lox/lox (lanes 6–9) mice. (B) qRT–PCR for E-cadherin expression in primary PDAC from Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4+/+ mice and Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4lox/lox mice. (C) IHC analysis of S100A4 expression in PDAC from a Ptf1-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4+/+ mouse and a Ptf1-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4lox/lox mouse; note the staining of stromal fibroblasts in both genotypes (arrows) and staining of tumor glands in the Smad4+/+ tumors but not those lacking Smad4 (asterisks).
In addition, to the differences in tumor cell morphology, the Smad4 mutant tumors exhibited differences in stromal fibroblast content, as readily observed in histological sections. This increased fibrosis was confirmed by Western blot and IHC for collagen-1 expression (Supplementary Fig. 4A,B). There also appeared to be a trend toward an increased number of inflammatory cells as assessed by staining for macrophages and neutrophils (Supplementary Table 1; data not shown). Finally, analysis of PDAC blood vessel density failed to reveal pronounced differences between genotypes when areas of similar histopathology were compared (Supplementary Fig. 4C; data not shown).
Smad4 modulates TGF-β sensitivity in murine PDAC cell lines
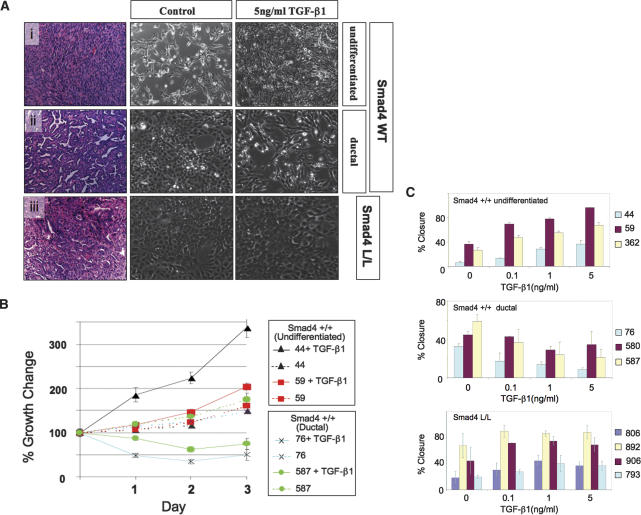
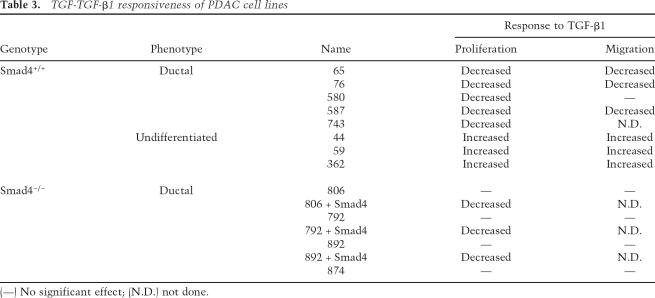
The genetically defined early passage PDAC cell lines described above, null and wild type for Smad4, provided an opportunity to understand better the cell biological impact of TGF-β–Smad4 signaling in this system. PDAC cell lines from Smad4lox/lox mice (n = 4 cell lines) were completely resistant to TGF-β-induced growth inhibition (see below), whereas those from the Smad4+/+ models showed differential sensitivity; Smad4+/+ cell lines derived from well or moderately differentiated (ductal) tumors exhibited growth inhibition and cell death (n = 4 cell lines), whereas those from undifferentiated tumors showed increased proliferation (n = 3 cell lines) (Fig. 8A; Table 3). Similar profiles were observed in scratch assays; ductal Smad4+/+ cell lines showed decreased migration upon TGF-β exposure, while enhanced migration was seen in the undifferentiated Smad4+/+ lines (Fig. 8B; Table 3). The Smad4-deficient lines also showed a modest increase in migration in response to TGF-β.
Figure 8.
Smad4 status modulates TGF-β responses in PDAC. (A) H&E-stained sections showing PDAC histology (left panels), and images of derivative cell lines either mock-treated (center) or exposed to 5 μg/mL TGF-β (right panels) from Smad4 wild-type undifferentiated tumors (panel i), Smad4 wild-type ductal tumors (panel ii), and Smad4-null tumors (panel iii). Note that TGF-β enhances growth in panel i and inhibits in panel ii while no effect is seen in panel iii. Magnifications, 100×. (B) Growth curves of Smad4+/+ PDAC cell lines in 0.5% serum in the presence or absence of 5 μg/mL TGF-β. (C) Graphs of scratch assay measuring cell migration as relative wound closure after 16 h in the presence or absence of TGF-β1.
Table 3.
TGF-TGF-β1 responsiveness of PDAC cell lines
(—) No significant effect; (N.D.) not done.
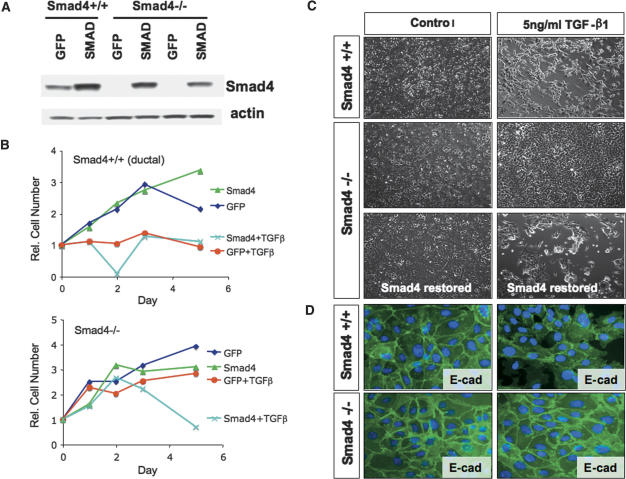
Next, we assessed whether restoration of Smad4 in Smad4-deficient PDAC cell lines affected TGF-β responsiveness. Retroviral-mediated expression of Smad4, but not GFP, in the Smad4-deficient lines (n = 3 cell lines) restored TGF-β-induced growth inhibition but had no effect on basal proliferation in the absence of exogenous TGF-β treatment (Fig. 9A,B; Table 3). In the presence of TGF-β treatment, the Smad4-reconstituted and wild-type cell lines (specifically those with a ductal phenotype) lost both their epithelial morphology and E-cadherin staining at the cell junctions before undergoing cell death, whereas no cytological changes were observed in GFP-transduced Smad4-deficient cultures (Fig. 9C,D). Overall, these results are consistent with the view that Smad4 inactivation facilitates PDAC progression by disabling TGF-β-mediated cytotoxicity while indicating that TGF-β acts to promote growth of a subset of PDAC with Smad4 intact and with an undifferentiated cellular phenotype.
Figure 9.
Smad4 expression restores TGF-β-responsiveness in Smad4-deficient PDAC cell lines. (A) Western blot analysis for Smad4 expression in PDAC cell lines from Pdx1-Cre LSL-KrasG12D Ink4a/Arflox/+ and Pdx1-Cre LSL-KrasG12D Ink4a/Arflox/+ Smad4lox/lox mice that were transduced with retroviruses expressing Smad4 or GFP. (B) Growth curves of the transduced PDAC cell lines in the presence or absence of TGF-β. (C) Morphology of PDAC cells with or without TGF-β treatment. The cells were photographed after 5 d of treatment with TGF-β or vehicle. (D) Impact of TGF-β administration on E-cadherin localization. E-cadherin staining is lost at the cellular junctions following TGF-β treatment in PDAC cell lines with intact Smad4 but not in cell lines with Smad4 inactivation.
Discussion
In this study, we have shown that Smad4 is dispensable for normal pancreas development and physiology and functions to constrain progression of KrasG12D-initiated neoplasms with a corresponding impact on the biology of tumor cells and their microenvironment. Smad4 deficiency led to rapid progression of pancreatic tumors in the context of activated KrasG12D, alone or in combination with Ink4a/Arf mutation, and was able to shift KrasG12D-initiated neoplasms toward an alternative histological pathway to advanced PDAC, one that is characterized by an IPMN presentation observed in some human cases. Along similar lines, Smad4 deficiency also influenced the differentiation state of KrasG12D Ink4a/Arf mutant PDACs, producing tumors that retained epithelial differentiation in contrast to the frequent EMT observed in pancreatic cancers with intact Smad4. This system affords an opportunity to systematically understand the role of the TGFβ–SMAD axis in many complex aspects of tumorigenesis including tumor cell proliferation, migration, survival, and differentiation as well as the tumor microenvironment including angiogenesis. As such, this genetic model provides a framework for understanding how to deploy agents targeting this pathway in human PDAC.
Impact of SMAD4 on pancreas developmentand physiology
The normal pancreatic development and function in the Pdx1-Cre Smad4lox/lox mice was not anticipated and contrasts with diverse pancreatic phenotypes associated with other experimental perturbations of TGF-β family signaling. For example, acinar-to-ductal metaplasia or autoimmune pancreatitis is observed in different transgenic models expressing a dominant-negative TGF-βRII mutant in the pancreatic acinar cells (Bottinger et al. 1997; Hahm et al. 2000). Null mutations in the Activin signaling pathway are associated with gross pancreatic defects (Kim et al. 2000; Harmon et al. 2004). Overexpression of the inhibitory Smad, Smad7, results in pancreatic malformations and β-islet cell defects (Smart et al. 2006). Finally, expression of a dominant-negative Smad4 mutant in the acinar cells results in an age-dependent increase in islet size (Simeone et al. 2006). These phenotypes, coupled with normal pancreas development and function in the Pdx1-Cre Smad4lox/lox mice, suggest roles for Smad4-independent TGF-β signaling pathways in these processes (Bierie and Moses 2006) and/or point to nonphysiological gain-of-function activities of ectopically overexpressed proteins. The dispensability of Smad4 for pancreatic organogenesis mirrors what has been observed in the liver and breast (Li et al. 2003; Wang et al. 2005); in contrast, Smad4 is required during early development to pattern derivatives of the anterior primitive streak and for formation of extraembryonic lineages (Chu et al. 2004). Overall, genetic studies have consistently revealed a more prominent role for Smad4 mediating TGF-β-regulated developmental processes relating to early specification and patterning rather than cytodifferentiation during later stages of organogenesis.
KrasG12D expression and Smad4 inactivation resultsin pancreatic ductal tumorigenesis
Our study demonstrates strong genetic interaction between KrasG12D and Smad4 in the development of IPMN and PDAC. IPMN is being increasingly recognized as an important clinical condition due to its propensity to progress to lethal PDAC (Maitra et al. 2005). Unlike PanIN, IPMN has recognizable clinical signs and can be detected noninvasively in the absence of malignant disease. Hence, improved understanding of the biology of these lesions could inform the design of treatments that prevent the onset of PDAC. While SMAD4 inactivation is associated with progression to advanced histological stages in human IPMN, our data indicate that SMAD4 can also act to restrain the initiation of these neoplasms. While Ink4a/Arf loss has been implicated in other cystic pancreatic tumors (Bardeesy et al. 2002a), it did not contribute to these IPMNs. Homozygous Smad4 deletion also cooperatively induced rapid PDAC progression, particularly in the context of KrasG12D and Ink4a/Arf heterozygosity. Together, these results indicate that Smad4 and the Ink4a/Arf locus regulate distinct pathways that contribute to PDAC tumor suppression, with Ink4a/Arf playing a more critical role in restricting malignant progression.
A related study has investigated the impact of inactivation of the Tgf-βRII on pancreatic tumorigenesis (H. Ijichi and H.L. Moses, pers. comm.). This study showed that Ptf1-Cre LSL-KrasG12D Tgf-βRIIlox/lox mice develop PDAC by 10 wk of age without exhibiting IPMN. The more rapid PDAC progression in this model compared with our Smad4-deficient model is likely to reflect a role for Smad4-independent pathways downstream from Tgf-βRII (Bierie and Moses 2006), although differences in genetic background cannot be ruled out. Likewise, the absence of IPMN in the Tgf-βRII-deficient mice may relate to the role of Smad4 in mediating signals from other classes of TGF-β family receptors; notably, BMPs and Activins are also induced in evolving PDAC, and inactivation of their receptors is associated with gastrointestinal malignancy (Howe et al. 2001; Jung et al. 2004). Hence, the IPMN phenotype may involve defects in a combination of these pathways. The detailed molecular comparison of these models may provide valuable insights into nonoverlapping PDAC tumor suppressor pathways that are differentially regulated by Smad4 and Tgf-βRII. Overall, given the more potent impact of Tgf-βRII inactivation on Pdx1-Cre LSL-KrasG12D-driven PDAC development compared with that of Smad4 inactivation, it is curious that PDACs in humans show exclusively Smad4 mutations, while Tgf-βRII mutations are restricted to a medullary pancreatic cancer, a rare variant associated with mismatch repair defects (Venkatasubbarao et al. 1998; Wilentz et al. 2000).
Smad4 regulates EMT in PDAC
Our demonstration that Smad4 deficiency promoted progression to PDAC but produced tumors that retained differentiated ductal histopathology is consistent with a bimodal role of TGF-β–Smad4 signaling in regulating the biology of this cancer. Our studies showed that, consistent with well/moderately differentiated histology, Smad4-deficient invasive tumors retained E-cadherin expression and were far less likely than Smad4 wild-type tumors to show S100A4 expression, a calcium-binding protein implicated in cell motility, EMT, and metastases (Okada et al. 1997; Helfman et al. 2005) that has been associated with poor differentiation and prognosis in PDAC (Rosty et al. 2002). In addition, in vitro studies with our early passage PDAC revealed that while Smad4 deficiency was associated with attenuated TGF-β responsiveness, Smad4-expressing lines showed two classes of prominent responses: increased proliferation and migration of undifferentiated Smad4+/+ lines and cell death in well-differentiated lines.
Although an association between SMAD4 status and tumor cell differentiation in human PDAC has not been clearly established, SMAD4 expression tracked with poor cellular differentiation in a morphometric study of human PDAC cell lines; poor differentiation was observed in four out of six SMAD4 wild-type versus zero out of five SMAD4 mutant lines (Sipos et al. 2003). Furthermore, in studies of a series of 16 human PDAC cell lines grown as xenografts, we observed that undifferentiated histology was only in tumors derived from SMAD4 wild-type cell lines, although one undifferentiated line had TGFβRII defects (data not shown). Our data suggest that Smad4 status and tumor cell differentiation may be important in determining which subset of PDAC depends on active TGF-β signaling for ongoing tumorigenic growth. An implication of our work is that Smad4 status could potentially predict which set of tumors is likely to respond to TGF-β inhibition, although further studies will be needed to validate this hypothesis.
Materials and methods
Generation of a conditional Smad4 mouse strain
A BAC clone containing the Smad4 locus was identified by screening a 129SV/ev library (Children’s Hospital of Oakland). The targeting construct was generated in the pKOII vector (Bardeesy et al. 2002b) and consisted of an 8.6-kb EcoRV–MscI fragment (long arm) containing Smad4 exons 7–9 and a 2.1-kb MscI–EcoRV fragment (short arm). A loxP site was inserted at a StuI site flanking Smad4 exons 8 and 9 (see Fig. 1A) enabling Cre-mediated deletion of a 1.4-kb fragment containing exons 8 and 9. Gene targeting in 129SV/ev embryonic stem (ES) cells and generation of chimeric animals were performed using standard techniques. The Frt-flanked PGK-neo cassette was excised by crosses to Actin-Flpe mice (Rodriguez et al. 2000), generating the Smad4Lox allele. Mice were backcrossed (N = 5) on an FVB/n background, eliminating the Actin-Flpe allele.
Mouse strains, histopathology, and establishment of primary PDAC cell lines
In addition to the Smad4Lox mice, the mouse strains in this study included the following alleles: LSL-KrasG12D (Aguirre et al. 2003), Pdx1-Cre (Gu et al. 2002), Ptf1a-Cre (Kawaguchi et al. 2002), and p16Ink4a/p19Arflox (Aguirre et al. 2003; Bardeesy et al. 2006). All experiments were performed on >96.9% FVB/n background. Tissue processing, immunohistochemical staining, and establishment of primary PDAC cell lines were performed as described (Aguirre et al. 2003). For frozen sections, tissues were immediately embedded in OCT (Sakura Finetek) and frozen on dry ice. Fixation, blocking, and antibody incubation were by standard procedures. Coverslips were mounted on fluorescently labeled tissue using Vectamount with DAPI (Vector Laboratories). Images were captured using a Leica DM1400B microsystem and Leica FW4000 version 1.2.1.
Antibodies
The antibodies used for immunoblot analyses were p16Ink4a (M-156), Smad4 (B-8) and p21 (C-19) (Santa Cruz Biotechnology), p19Arf (Ab-80, Abcam), tubulin (DM-1A) and Actin (Sigma), Slug (Abgent), E-cadherin (Invitrogen), and phospho Smad2 (Chemicon International). The antibodies for immunohistochemical analysis were amylase (Sigma); rat monoclonal cytokeratin-19 (TROMA3; kindly provided by Dr. R. Kemler, Max Planck Institute, Freiburg, Germany); E-cadherin (Zymed; Invitrogen); Glucagon (Dako); Insulin (Dako); Pdx-1 (a kind gift of Dr. C. Wright, Vanderbilt University, Nashville, TN); TGFβ1, BMP4, Muc2, Shh, Vimentin, and PDGFR-β (Santa Cruz Biotechnology); Smad2/3 and BrdU (BD; Transduction Laboratories); Muc5AC (Labvision); Muc1 (CT2 monoclonal antibody; a kind gift of Dr. S. Gendler, Mayo Clinic, Scottsdale, AZ); Muc4 (Dr. K. Carraway, University of Miami, Miami, FL); CDX2 (Biogenex); Hes1 (a kind gift of Dr. T. Sudo, Toray Scientific, Kanagawa, Japan); Collagen type I (Abcam); SMA (Novus); phospho-Stat3 (Ser727); and phospho-p44/42 (T202/Y204) and phospho-Akt (Ser473) (Cell Signaling). The following antibodies were used on frozen sections: rabbit MMP-9 (a kind gift from Dr. Z. Werb, University of California at San Francisco, San Francisco, CA); monoclonal rat anti-Meca-32 (Pharmingen); F4/80 (Serotec); PMN antibody (clone 7/4; Cedarlane Laboratories); PDGFRβ and PDGFRα monoclonal antibodies (eBioscience); and rabbit CSF1R (Upstate Biotechnology/Chemicon).
Molecular and cellular analyses
RNA and DNA isolation, protein extract preparation from PDAC cell lines and primary tumors, and Western blots were performed as described (Aguirre et al. 2003). For cell proliferation assays, cells were seeded on 96-well plates at a density of 2 × 103 to 4 × 103 per well in 100 μL of culture media with 0.5% FBS. The media were replaced every other day. To evaluate cell proliferation, cells were incubated for 1–5 d with or without TGF-β1 treatment (R&D Systems) and assayed using the WST-1 Cell Proliferation Assay reagent (Roche). For cell migration analysis, in vitro wound healing assays were used to assess cell motility in two dimensions. Cells were plated on a six-well plate and grown to confluence in their regular medium. A 200-μL pipette tip was used to create a denuded area. Cells were washed twice with PBS and kept in RPMI medium containing 0.5% FBS in the presence or absence of TGF-β1 (5 ng/mL) for 16 h. Photographs were taken at hours 0, 10, and 16, and the distance traveled was determined by subtracting the values obtained at hour 0 from 16 h.
Statistical analysis
For morphometric studies of pancreatic lesions, a series of 36 consecutive 5-μm H&-stained sections was analyzed for each pancreas, and the sections with the greatest number of visible lesions were selected for quantitation; at least eight 100× fields were. For immunofluorescence, stained tissue was examined using a Zeiss Axioskop-2+ Microscope (fluorescently labeled tissues) with Openlab software (Improvision). F4/80, PMN, PDGFRβ, and MMP-9 expression was determined by counting the staining per field. Meca-32 staining was quantitated by counting the number of vessels per field. At least four fields were measured from each slide, and an average was calculated by nonpaired t-test analysis using InStat Software (GraphPad).
Acknowledgments
We thank David Tuveson and Tyler Jacks for the LSL-KrasG12D mice, Doug Melton for the Pdx1-Cre mice, and Chris Wright for the Ptf1a-Cre mice. We thank Dr. Kwok-kin Wong for critical reading of the manuscript. This work was support by grants NIH K01 CA104647 and NIH P01 CA117969-01, and grants from the Samuel Waxman Foundation and the Harvard Stem Cell Institute to N.B.; and grants P01 CA117969-01, 5U01CA84313-08, American Cancer Society Research Professorship, Ellison Medical Foundation, and the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science to R.A.D.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1478706
References
- Adsay, N.V., Merati, K., Basturk, O., Iacobuzio-Donahue, C., Levi, E., Cheng, J.D., Sarkar, F.H., Hruban, R.H., Klimstra, D.S. Pathologically and biologically distinct types of epithelium in intraductal papillary mucinous neoplasms: Delineation of an ‘intestinal’ pathway of carcinogenesis in the pancreas. Am. J. Surg. Pathol. 2004;28:839–848. doi: 10.1097/00000478-200407000-00001. [DOI] [PubMed] [Google Scholar]
- Aguirre, A.J., Bardeesy, N., Sinha, M., Lopez, L., Tuveson, D.A., Horner, J., Redston, M.S., DePinho, R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes & Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte, M.V., Park, S., Phillips, P.A., Santucci, N., Goldstein, D., Kumar, R.K., Ramm, G.A., Buchler, M., Friess, H., McCarroll, J.A., et al. Desmoplastic reaction in pancreatic cancer: Role of pancreatic stellate cells. Pancreas. 2004;29:179–187. doi: 10.1097/00006676-200410000-00002. [DOI] [PubMed] [Google Scholar]
- Bardeesy, N., Morgan, J., Sinha, M., Signoretti, S., Srivastava, S., Loda, M., Merlino, G., DePinho, R.A. Obligate roles for p16(Ink4a) and p19(Arf)–p53 in the suppression of murine pancreatic neoplasia. Mol. Cell. Biol. 2002a;22:635–643. doi: 10.1128/MCB.22.2.635-643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy, N., Sinha, M., Hezel, A.F., Signoretti, S., Hathaway, N.A., Sharpless, N.E., Loda, M., Carrasco, D.R., DePinho, R.A. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002b;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- Bardeesy, N., Aguirre, A.J., Chu, G.C., Cheng, K.H., Lopez, L.V., Hezel, A.F., Feng, B., Brennan, C., Weissleder, R., Mahmood, U., et al. Both p16Ink4a and the p19Arf–p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl. Acad. Sci. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin, A.V., Morey, A.L., Lee, C.S., Kench, J.G., Biankin, S.A., Hook, H.C., Head, D.R., Hugh, T.B., Sutherland, R.L., Henshall, S.M. DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J. Clin. Oncol. 2002;20:4531–4542. doi: 10.1200/JCO.2002.12.063. [DOI] [PubMed] [Google Scholar]
- Bierie, B., Moses, H.L. Tumour microenvironment: TGFβ: The molecular Jekyll and Hyde of cancer. Nat. Rev. Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- Bottinger, E.P., Jakubczak, J.L., Roberts, I.S., Mumy, M., Hemmati, P., Bagnall, K., Merlino, G., Wakefield, L.M. Expression of a dominant-negative mutant TGF-β type II receptor in transgenic mice reveals essential roles for TGF-β in regulation of growth and differentiation in the exocrine pancreas. EMBO J. 1997;16:2621–2633. doi: 10.1093/emboj/16.10.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, G.C., Dunn, N.R., Anderson, D.C., Oxburgh, L., Robertson, E.J. Differential requirements for Smad4 in TGFβ-dependent patterning of the early mouse embryo. Development. 2004;131:3501–3512. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- Dai, J.L., Schutte, M., Bansal, R.K., Wilentz, R.E., Sugar, A.Y., Kern, S.E. Transforming growth factor-β responsiveness in DPC4/SMAD4-null cancer cells. Mol. Carcinog. 1999;26:37–43. doi: 10.1002/(sici)1098-2744(199909)26:1<37::aid-mc5>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Duda, D.G., Sunamura, M., Lefter, L.P., Furukawa, T., Yokoyama, T., Yatsuoka, T., Abe, T., Inoue, H., Motoi, F., Egawa, S., et al. Restoration of SMAD4 by gene therapy reverses the invasive phenotype in pancreatic adenocarcinoma cells. Oncogene. 2003;22:6857–6864. doi: 10.1038/sj.onc.1206751. [DOI] [PubMed] [Google Scholar]
- Ellenrieder, V., Hendler, S.F., Boeck, W., Seufferlein, T., Menke, A., Ruhland, C., Adler, G., Gress, T.M. Transforming growth factor β1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61:4222–4228. [PubMed] [Google Scholar]
- Friess, H., Yamanaka, Y., Buchler, M., Berger, H.G., Kobrin, M.S., Baldwin, R.L., Korc, M. Enhanced expression of the type II transforming growth factor β receptor in human pancreatic cancer cells without alteration of type III receptor expression. Cancer Res. 1993a;53:2704–2707. [PubMed] [Google Scholar]
- Friess, H., Yamanaka, Y., Buchler, M., Ebert, M., Beger, H.G., Gold, L.I., Korc, M. Enhanced expression of transforming growth factor β isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology. 1993b;105:1846–1856. doi: 10.1016/0016-5085(93)91084-u. [DOI] [PubMed] [Google Scholar]
- Giehl, K., Seidel, B., Gierschik, P., Adler, G., Menke, A. TGFβ1 represses proliferation of pancreatic carcinoma cells which correlates with Smad4-independent inhibition of ERK activation. Oncogene. 2000;19:4531–4541. doi: 10.1038/sj.onc.1203806. [DOI] [PubMed] [Google Scholar]
- Gu, G., Dubauskaite, J., Melton, D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Hahm, K.B., Im, Y.H., Lee, C., Parks, W.T., Bang, Y.J., Green, J.E., Kim, S.J. Loss of TGF-β signaling contributes to autoimmune pancreatitis. J. Clin. Invest. 2000;105:1057–1065. doi: 10.1172/JCI8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, S.A., Schutte, M., Hoque, A.T., Moskaluk, C.A., da Costa, L.T., Rozenblum, E., Weinstein, C.L., Fischer, A., Yeo, C.J., Hruban, R.H., et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- Hansel, D.E., Kern, S.E., Hruban, R.H. Molecular pathogenesis of pancreatic cancer. Annu. Rev. Genomics Hum. Genet. 2003;4:237–256. doi: 10.1146/annurev.genom.4.070802.110341. [DOI] [PubMed] [Google Scholar]
- Harmon, E.B., Apelqvist, A.A., Smart, N.G., Gu, X., Osborne, D.H., Kim, S.K. GDF11 modulates NGN3+ islet progenitor cell number and promotes β-cell differentiation in pancreas development. Development. 2004;131:6163–6174. doi: 10.1242/dev.01535. [DOI] [PubMed] [Google Scholar]
- He, W., Dorn, D.C., Erdjument-Bromage, H., Tempst, P., Moore, M.A., Massague, J. Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Helfman, D.M., Kim, E.J., Lukanidin, E., Grigorian, M. The metastasis associated protein S100A4: Role in tumour progression and metastasis. Br. J. Cancer. 2005;92:1955–1958. doi: 10.1038/sj.bjc.6602613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel, A.F., Kimmelman, A.C., Stanger, B.Z., Bardeesy, N., Depinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes & Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Hingorani, S.R., Petricoin, E.F., Maitra, A., Rajapakse, V., King, C., Jacobetz, M.A., Ross, S., Conrads, T.P., Veenstra, T.D., Hitt, B.A., et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Hingorani, S.R., Wang, L., Multani, A.S., Combs, C., Deramaudt, T.B., Hruban, R.H., Rustgi, A.K., Chang, S., Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Howe, J.R., Roth, S., Ringold, J.C., Summers, R.W., Jarvinen, H.J., Sistonen, P., Tomlinson, I.P., Houlston, R.S., Bevan, S., Mitros, F.A., et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- Howe, J.R., Bair, J.L., Sayed, M.G., Anderson, M.E., Mitros, F.A., Petersen, G.M., Velculescu, V.E., Traverso, G., Vogelstein, B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat. Genet. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- Hruban, R.H., Adsay, N.V., Albores-Saavedra, J., Anver, M.R., Biankin, A.V., Boivin, G.P., Furth, E.E., Furukawa, T., Klein, A., Klimstra, D.S., et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: Consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- Ito, D., Fujimoto, K., Doi, R., Koizumi, M., Toyoda, E., Mori, T., Kami, K., Kawaguchi, Y., Whitehead, R., Imamura, M. Chronic exposure of transforming growth factor β1 confers a more aggressive tumor phenotype through downregulation of p21(WAF1/CIP1) in conditionally immortalized pancreatic epithelial cells. Surgery. 2004;136:364–374. doi: 10.1016/j.surg.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Jonson, T., Heidenblad, M., Hakansson, P., Gorunova, L., Johansson, B., Fioretos, T., Hoglund, M. Pancreatic carcinoma cell lines with SMAD4 inactivation show distinct expression responses to TGFB1. Genes Chromosomes Cancer. 2003;36:340–352. doi: 10.1002/gcc.10179. [DOI] [PubMed] [Google Scholar]
- Jung, B., Doctolero, R.T., Tajima, A., Nguyen, A.K., Keku, T., Sandler, R.S., Carethers, J.M. Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology. 2004;126:654–659. doi: 10.1053/j.gastro.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, Y., Cooper, B., Gannon, M., Ray, M., MacDonald, R.J., Wright, C.V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kim, S.K., Hebrok, M., Li, E., Oh, S.P., Schrewe, H., Harmon, E.B., Lee, J.S., Melton, D.A. Activin receptor patterning of foregut organogenesis. Genes & Dev. 2000;14:1866–1871. [PMC free article] [PubMed] [Google Scholar]
- Li, W., Qiao, W., Chen, L., Xu, X., Yang, X., Li, D., Li, C., Brodie, S.G., Meguid, M.M., Hennighausen, L., et al. Squamous cell carcinoma and mammary abscess formation through squamous metaplasia in Smad4/Dpc4 conditional knockout mice. Development. 2003;130:6143–6153. doi: 10.1242/dev.00820. [DOI] [PubMed] [Google Scholar]
- Li, D., Xie, K., Wolff, R., Abbruzzese, J.L. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- Lohr, M., Schmidt, C., Ringel, J., Kluth, M., Muller, P., Nizze, H., Jesnowski, R. Transforming growth factor-β1 induces desmoplasia in an experimental model of human pancreatic carcinoma. Cancer Res. 2001;61:550–555. [PubMed] [Google Scholar]
- Maitra, A., Fukushima, N., Takaori, K., Hruban, R.H. Precursors to invasive pancreatic cancer. Adv. Anat. Pathol. 2005;12:81–91. doi: 10.1097/01.pap.0000155055.14238.25. [DOI] [PubMed] [Google Scholar]
- Moniaux, N., Andrianifahanana, M., Brand, R.E., Batra, S.K. Multiple roles of mucins in pancreatic cancer, a lethal and challenging malignancy. Br. J. Cancer. 2004;91:1633–1638. doi: 10.1038/sj.bjc.6602163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, F.J., Hill, C.S. Attenuation of the TGF-β–Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-β-induced growth arrest. Oncogene. 2003;22:3698–3711. doi: 10.1038/sj.onc.1206420. [DOI] [PubMed] [Google Scholar]
- Okada, H., Danoff, T.M., Kalluri, R., Neilson, E.G. Early role of Fsp1 in epithelial-mesenchymal transformation. Am. J. Physiol. 1997;273:F563–F574. doi: 10.1152/ajprenal.1997.273.4.F563. [DOI] [PubMed] [Google Scholar]
- Peng, B., Fleming, J.B., Breslin, T., Grau, A.M., Fojioka, S., Abbruzzese, J.L., Evans, D.B., Ayers, D., Wathen, K., Wu, T., et al. Suppression of tumorigenesis and induction of p15(ink4b) by Smad4/DPC4 in human pancreatic cancer cells. Clin. Cancer Res. 2002;8:3628–3638. [PubMed] [Google Scholar]
- Rodriguez, C.I., Buchholz, F., Galloway, J., Sequerra, R., Kasper, J., Ayala, R., Stewart, A.F., Dymecki, S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Rosty, C., Ueki, T., Argani, P., Jansen, M., Yeo, C.J., Cameron, J.L., Hruban, R.H., Goggins, M. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am. J. Pathol. 2002;160:45–50. doi: 10.1016/S0002-9440(10)64347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland-Goldsmith, M.A., Maruyama, H., Kusama, T., Ralli, S., Korc, M. Soluble type II transforming growth factor-β (TGF-β) receptor inhibits TGF-β signaling in COLO-357 pancreatic cancer cells in vitro and attenuates tumor formation. Clin. Cancer Res. 2001;7:2931–2940. [PubMed] [Google Scholar]
- Schwarte-Waldhoff, I., Volpert, O.V., Bouck, N.P., Sipos, B., Hahn, S.A., Klein-Scory, S., Luttges, J., Kloppel, G., Graeven, U., Eilert-Micus, C., et al. Smad4/DPC4-mediated tumor suppression through suppression of angiogenesis. Proc. Natl. Acad. Sci. 2000;97:9624–9629. doi: 10.1073/pnas.97.17.9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone, D.M., Zhang, L., Treutelaar, M.K., Zhang, L., Graziano, K., Logsdon, C.D., Burant, C.F. Islet hypertrophy following pancreatic disruption of smad4 signaling. Am. J. Physiol. Endocrinol. Metab. 2006 doi: 10.1152/ajpendo.00561.2005. Epub May 30, 2006; doi: 10.1152/ajpendo.00561.2005. [DOI] [PubMed] [Google Scholar]
- Sipos, B., Moser, S., Kalthoff, H., Torok, V., Lohr, M., Kloppel, G. A comprehensive characterization of pancreatic ductal carcinoma cell lines: Towards the establishment of an in vitro research platform. Virchows Arch. 2003;442:444–452. doi: 10.1007/s00428-003-0784-4. [DOI] [PubMed] [Google Scholar]
- Sirard, C., de la Pompa, J.L., Elia, A., Itie, A., Mirtsos, C., Cheung, A., Hahn, S., Wakeham, A., Schwartz, L., Kern, S.E., et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes & Dev. 1998;12:107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart, N.G., Apelqvist, A.A., Gu, X., Harmon, E.B., Topper, J.N., MacDonald, R.J., Kim, S.K. Conditional expression of Smad7 in pancreatic β cells disrupts TGF-β signaling and induces reversible diabetes mellitus. PLoS Biol. 2006;4:e39. doi: 10.1371/journal.pbio.0040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solcia, E., Capella, C., Kloppel, G. Tumors of the pancreas. Armed Forces Institute for Pathology; Washington, DC: 1995. [Google Scholar]
- Subramanian, G., Schwarz, R.E., Higgins, L., McEnroe, G., Chakravarty, S., Dugar, S., Reiss, M. Targeting endogenous transforming growth factor β receptor signaling in SMAD4-deficient human pancreatic carcinoma cells inhibits their invasive phenotype1. Cancer Res. 2004;64:5200–5211. doi: 10.1158/0008-5472.CAN-04-0018. [DOI] [PubMed] [Google Scholar]
- Takaku, K., Miyoshi, H., Matsunaga, A., Oshima, M., Sasaki, N., Taketo, M.M. Gastric and duodenal polyps in Smad4 (Dpc4) knockout mice. Cancer Res. 1999;59:6113–6117. [PubMed] [Google Scholar]
- Tascilar, M., Skinner, H.G., Rosty, C., Sohn, T., Wilentz, R.E., Offerhaus, G.J., Adsay, V., Abrams, R.A., Cameron, J.L., Kern, S.E., et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2001;7:4115–4121. [PubMed] [Google Scholar]
- Venkatasubbarao, K., Ahmed, M.M., Swiderski, C., Harp, C., Lee, E.Y., McGrath, P., Mohiuddin, M., Strodel, W., Freeman, J.W. Novel mutations in the polyadenine tract of the transforming growth factor β type II receptor gene are found in a subpopulation of human pancreatic adenocarcinomas. Genes Chromosomes Cancer. 1998;22:138–144. doi: 10.1002/(sici)1098-2264(199806)22:2<138::aid-gcc8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Wagner, M., Kleeff, J., Friess, H., Buchler, M.W., Korc, M. Enhanced expression of the type II transforming growth factor-β receptor is associated with decreased survival in human pancreatic cancer. Pancreas. 1999;19:370–376. doi: 10.1097/00006676-199911000-00008. [DOI] [PubMed] [Google Scholar]
- Wang, R.H., Li, C., Xu, X., Zheng, Y., Xiao, C., Zerfas, P., Cooperman, S., Eckhaus, M., Rouault, T., Mishra, L., et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Wilentz, R.E., Goggins, M., Redston, M., Marcus, V.A., Adsay, N.V., Sohn, T.A., Kadkol, S.S., Yeo, C.J., Choti, M., Zahurak, M., et al. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: A newly described and characterized entity. Am. J. Pathol. 2000;156:1641–1651. doi: 10.1016/S0002-9440(10)65035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Li, C., Xu, X., Deng, C. The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proc. Natl. Acad. Sci. 1998;95:3667–3672. doi: 10.1073/pnas.95.7.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil, J., Bottinger, E.P. TGF-β and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]