Abstract
Excessive production of proinflammatory cytokines, particularly tumor necrosis factor-α (TNFα) and interleukin-1β (IL-1β), plays a critical role in septic shock induced by bacterial endotoxin (endotoxemia). Precise control of cytokine expression depends on rapid degradation of cytokine mRNAs, mediated by an AU-rich element (ARE) in the 3′ noncoding region and by interacting ARE-binding proteins, which control the systemic inflammatory response. To understand the function of the ARE-binding protein AUF1, we developed an AUF1 knockout mouse. We show that AUF1 normally functions to protect against the lethal progression of endotoxemia. Upon endotoxin challenge, AUF1 knockout mice display symptoms of severe endotoxic shock, including vascular hemorrhage, intravascular coagulation, and high mortality, resulting from overproduction of TNFα and IL-1β. Overexpression of these two cytokines is specific, and shown to result from an inability to rapidly degrade these mRNAs in macrophages following induction. Neutralizing antibodies to TNFα and IL-1β protect AUF1 knockout mice against lethal endotoxic shock. These and other data describe a novel post-transcriptional mechanism whereby AUF1 acts as a crucial attenuator of the inflammatory response, promoting the rapid decay of selective proinflammatory cytokine mRNAs following endotoxin activation. Defects in the AUF1 post-transcriptionally controlled pathway may be involved in human inflammatory disease.
Keywords: AU-rich element, ARE, AUF1, mRNA decay, endotoxic shock, cytokines
Septic shock accounts for 9% of U.S. annual deaths and is a leading cause of hospital-related mortality (Tracey et al. 1987; Dinarello 1994; Hotchkiss and Karl 2003; Martin et al. 2003). Typically initiated by a bacteria infection, septic shock results from the uncontrolled expression of proinflammatory cytokines, primarily those produced by macrophages (Tracey et al. 1987; Dinarello 1994; Hotchkiss and Karl 2003; Martin et al. 2003), which leads to an overwhelming systemic inflammatory response culminating in multiple organ failure. Lipopolysaccharide (LPS) endotoxin, a component of the bacterial cell wall, stimulates macrophages to produce proinflammatory cytokines such as tumor necrosis factor-α (TNFα) and interleukin-1β (IL-1β), which are critical mediators of septic shock (Glauser et al. 1991). The excessive production of these proinflammatory cytokines leads to systemic capillary leakage and vascular hemorrhage, tissue destruction, and ultimately lethal organ failure and death (Tracey et al. 1987; Glauser et al. 1991; Dinarello 1994; Hotchkiss and Karl 2003; Martin et al. 2003). Thus, the expression of TNF-α and IL-1β cytokines needs to be tightly regulated during an inflammatory response.
Regulation of cytokines occurs at multiple levels, including the stability of their encoding mRNAs (Chen and Shyu 1995; Guhaniyogi and Brewer 2001; Wilusz et al. 2001). The mRNAs encoding most inflammatory cytokines are short-lived, with instability conferred by an AU-rich element (ARE) in the 3′ noncoding region (Chen and Shyu 1995; Guhaniyogi and Brewer 2001). The ARE consists of multiple copies of an AUUUA pentamer, either adjacent or interrupted by other sequences, or even noncanonical AU-rich motifs (for review, see Chen and Shyu 1995; Guhaniyogi and Brewer 2001). The ARE promotes rapid cytoplasmic degradation of mRNAs (Guhaniyogi and Brewer 2001) and in some cases translation arrest (Grosset et al. 2004). It is thought that the different arrangements and types of ARE sequences may confer these different activities and may be responsible for different types of control of ARE-mRNA decay (Guhaniyogi and Brewer 2001). ARE-mRNAs can also be rapidly stabilized upon exposure to certain signals including immune stimulation, ultraviolet (UV) and ionizing radiation (Chen and Shyu 1995), and heat shock (Laroia et al. 1999). Aberrant stabilization of short-lived ARE-mRNAs may also be a factor in certain human malignancies (Lebwohl et al. 1994; Dixon et al. 2001), experimentally induced tumors (Nair et al. 1994), and inflammatory disorders (Jacob et al. 1996; Kontoyiannis et al. 1999). For example, prolonged stabilization of the TNFα mRNA, caused by targeted deletion of the ARE in experimental mice, leads to TNFα overproduction, resulting in chronic inflammatory arthritis and Crohn’s-like inflammatory bowel disease (Kontoyiannis et al. 1999).
The ARE controls the stability of ARE-mRNAs through interaction with ARE-binding proteins (Guhaniyogi and Brewer 2001). A number of such proteins have been identified, and at least five families appear to control ARE-mRNA decay (Guhaniyogi and Brewer 2001). Hu ARE-binding proteins, such as HuR, stabilize ARE-mRNAs and can inhibit mRNA translation in certain cases when experimentally overexpressed in a variety of physiological settings (Fan and Steitz 1998; Brennan and Steitz 2001; Katsanou et al. 2005). HuR and HuB members of the Hu family bind to a variety of ARE sequences (Ma et al. 1996), and gene silencing of HuR destabilizes ARE-mRNAs (Levy et al. 1998; Rodriguez-Pascual et al. 2000; Wang et al. 2000). In contrast, AUF1 (or hnRNP-D) (Brewer 1991; Zhang et al. 1993), tristetraprolin (TTP) (Carballo et al. 1998), BRF1 (Stoecklin et al. 2002), and KSRP (Chen et al. 2001) promote the rapid decay of various mRNAs, indicating that cytokine ARE-mRNAs are controlled by proteins with opposing stabilizing and destabilizing activities, in tissue culture studies. It is not understood how the different ARE-binding proteins function at a molecular level to control ARE-mRNA stability, either independently or collectively. None of these proteins possess inherent enzymatic activities, indicating that they presumably function by interaction with other proteins as well as ARE sequences.
TTP is the best studied ARE-destabilizing protein, which has been shown to promote destabilization of GM-CSF and TNFα mRNAs in tissue culture studies (Lai et al. 1999; Carballo et al. 2000) and in knockout mice (Taylor et al. 1996; Carballo et al. 2000). KSRP is also involved in mediating ARE-mRNA decay as determined by expression in tissue culture (Chen et al. 2001) and gene silencing, which stabilized IL-2, c-Fos, and TNFα mRNAs (Gherzi et al. 2004). AUF1 is a family of related ARE-binding destabilizing factors, consisting of four isoforms (37, 40, 42, and 45 kDa) generated by alternative splicing of one encoding mRNA (Brewer 1991; Zhang et al. 1993). Increased ectopic expression of AUF1 has been correlated with more rapid ARE-mRNA degradation in various types of cell lines in culture (Buzby et al. 1999; Loflin et al. 1999; Sarkar et al. 2003; He and Schneider 2006), with the p37 AUF1 isoform exhibiting the highest destabilizing activity and the strongest interaction with the ARE (Loflin et al. 1999; Wang et al. 2000; Sarkar et al. 2003). The four AUF1 isoforms display different affinities for different ARE sequences in vitro (Kajita et al. 1995; Wagner et al. 1998). Although the AUF1 isoforms are thought to interact with each other, there is little understanding of the stoichiometry or function of complex formation (Wilson et al. 1999). The evidence that AUF1 promotes ARE-mRNA degradation is largely based on binding of specific isoforms to AREs in vitro (Pende et al. 1996; Sirenko et al. 1997) and studies correlating ectopic overexpression of AUF1 isoforms in cell lines with rapid degradation of ARE-mRNA (Loflin et al. 1999; Sarkar et al. 2003; He and Schneider 2006). Moreover, several studies have provided conflicting results suggesting that certain AUF1 isoforms might also be stabilizers of ARE-mRNAs (Loflin et al. 1999), at least in certain contexts. This is consistent with studies identifying specific AUF1 isoforms as components of the exosome, which carries out mRNA decay (Chen et al. 2001), and participants in the β-globin mRNA stabilization complex (Kiledjian et al. 1997). The roles of AUF1 in the regulation of inflammatory cytokine expression and control of cytokine mRNA stability have not been examined in animals or in a pathophysiological setting.
To examine the role of AUF1 in controlling or promoting inflammatory mRNA degradation in vivo, we generated homozygous AUF1-null mutant mice and studied their response to LPS-induced septic-shock-like responses (endotoxemia). We show that AUF1−/− mice are acutely susceptible to endotoxin. At typically sublethal doses of endotoxin, AUF1 knockout mice demonstrated an fivefold lower survival rate. AUF1 knockout mice overexpress the proinflammatory cytokines TNFα and IL-1β following LPS treatment, which is shown to result from abnormal stabilization of TNFα and IL-1β mRNAs in macrophages. Importantly, we show that AUF1 appears to have a selective function, targeting TNFα and IL-1β mRNAs for rapid decay, but not IL-6 mRNA, which differs in the arrangement of its ARE and fails to strongly interact with AUF1 in vivo. Our results provide the first in vivo evidence implicating AUF1 in regulation of the inflammatory response, and they provide a molecular understanding of AUF1 action in selective targeted degradation of inflammatory cytokine mRNAs.
Results
Generation of AUF1-null mutant mice
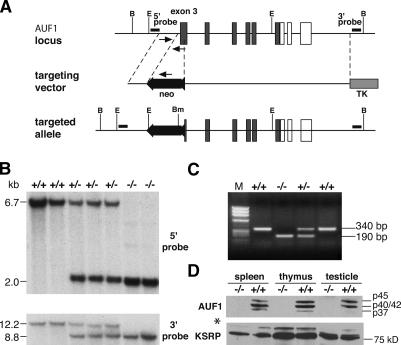
To assess the function of AUF1 in vivo, we generated AUF1 knockout mice through homologous recombination in mouse embryonic stem cells. The construct targeted the third exon of AUF1, which contains two RNA-binding motifs, and disrupted the remainder of the reading frame by homologous recombination (Fig. 1A). The wild-type and targeted AUF1 alleles were identified by Southern blot DNA hybridization analysis using probes 5′ and 3′ to the homology arm employed in the targeting construct (Fig. 1B), and by PCR amplification of genomic DNA (Fig. 1C). Furthermore, a Southern blot analysis using a probe specific for the neomycin resistance (neor) cassette confirmed the presence of only one copy of the neor cassette in the genome after homologous recombination, ruling out the possibility of a random integration (data not shown). Immunoblot analysis of protein extracts of mouse organs with high levels of AUF1 (Lu and Schneider 2004) demonstrated abrogation of AUF1 expression, while the level of another RNA-binding protein (KSRP) remained unaltered (Fig. 1D), indicating the successful disruption of the AUF1 locus. Matings of AUF1+/− mice produced homozygous AUF1 mutant animals in a Mendelian ratio (∼25% of progeny) (Supplementary Fig. S1a) with no embryonic lethality detected. AUF1−/− newborns were indistinguishable from wild-type littermates except they were smaller and had reduced body weight (Supplementary Fig. S1b). The growth retardation phenotype was equally distributed in both sexes. AUF1−/− mice survived to adulthood and were fertile. Other than smaller size and reduced body weight, macroscopic and histological examination did not reveal any morphological abnormalities in major organs, of young adult mice. Further analysis of AUF1−/− embryos found no evidence for major developmental defects (Supplementary Fig. S2) or significant alteration in fetal hematopoietic organs including thymus, spleen, and liver (data not shown). Therefore, disruption of AUF1 did not cause any severe defect in mouse development.
Figure 1.
Generation of AUF1-null mice. (A) The wild-type AUF1 allele, the targeting vector, and targeted allele are shown. Arrows indicate primers used in PCR genotyping. (B) BglI; (Bm) BamHI; (E) EcoRI; (neo) neomycin-resistant cassette; (TK) thymidine kinase cassette. (Filled box) coding region; (open box) noncoding region. (B) Southern blot analysis of genomic DNA from second-generation (F2) mice. (Top panel) EcoRI-digested DNA hybridized with the 5′ probe. (Bottom panel) BamHI- and BglI-double-digested DNA hybridized with the 3′ probe. (C) The genomic DNA fragments were amplified from wild-type (340-base-pair [bp]) and targeted (190-bp) alleles by PCR. (M) Marker. (D) Immunoblot confirmation of the absence of AUF1 proteins in AUF1−/− mice. Four AUF1 isoforms (p37, p42/p40, p45) were detected in protein extracts from wild-type mouse organs by an AUF1 polyclonal antibody. Note the p40/42 proteins comigrate. All AUF1 protein isoforms were absent in extracts from AUF1−/− mice. (Bottom panel) The same blot was probed with a polyclonal antibody against KSRP (∼75 kDa) as the control. The asterisk indicates a nonspecific cross-reactivity of KSRP antibody to an unknown protein with a molecular weight of ∼100 kDa.
Disruption of AUF1 causes a fivefold increase in mortality during LPS-induced endotoxemia
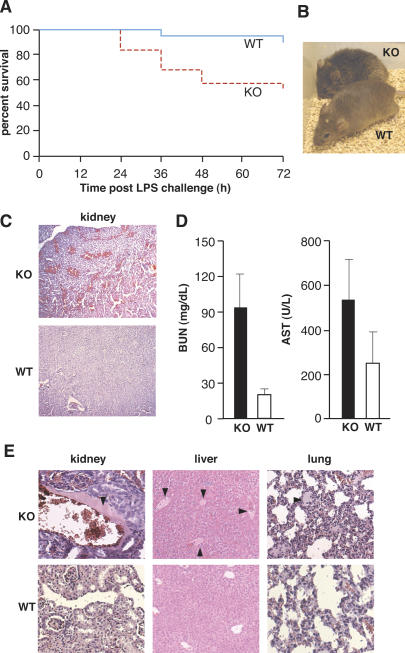
To determine whether AUF1 regulates the inflammatory response, we examined the effect of AUF1 deficiency on the survival of mice subjected to endotoxin (LPS)-induced endotoxemia. AUF1 knockout mice were injected intraperitoneally with a sublethal dose (20 mg/kg) of bacterial LPS endotoxin. At this dose, LPS potently stimulates proinflammatory cytokine expression and induces a systemic inflammatory response, but typically without provoking significant mortality. While the majority of wild-type mice (90%) survived the endotoxin challenge and showed only a slight reduction in activity, AUF1 knockout mice displayed severe manifestations of endotoxemia, including diarrhea, tachypnea, lethargy, and piloerection (Fig. 2B). By 72 h post-LPS challenge, AUF1−/− mice displayed about a fivefold increase in mortality compared with wild-type mice (47.4% vs. 10%) (Fig. 2A). Survival analysis showed that the survival rate of AUF1 knockout mice is statistically significantly lower than that of the wild-type mice after LPS challenge (Fig. 2A, p < 0.01). The surviving AUF1−/− mice required more than twice as long to recover compared with wild-type animals (3–4 d vs. 1–1.5 d, respectively) (data not shown). Thus, AUF1 knockout mice are much more susceptible to the endotoxin challenge than wild-type mice.
Figure 2.
Significantly increased susceptibility of AUF1−/− mice to endotoxin challenge. (A) Kaplan-Meier survival curves of wild-type (WT, n = 19) and AUF1 knockout (KO, n = 19) mice after sublethal LPS challenge (20 mg/kg weight, intraperitoneal injection, p < 0.01, Log-rank test). AUF1-deficient mice were more susceptible to endotoxin challenge. (B) Photograph of wild-type and knockout mice 4 h after intraperitoneal injection of a low dose of LPS (3 mg/kg body weight, n = 3). AUF1 knockout mice exhibited typical signs of endotoxemia, including lethargy, diarrhea, tachypnea, and piloerection, whereas wild-type mice showed only mild effects. (C) H&E-stained kidney sections from mice 24 h post-LPS challenge (20 mg/kg weight, intraperitoneal). Note the widespread hemorrhage in the kidney of AUF1−/− mice. (D) BUN and AST levels in mice 24 h after challenge with LPS (n = 3, p < 0.02 for the BUN level, p < 0.18 for the AST level). (E) Representative H&E-stained sections of mouse organs 24 h following LPS challenge to visualize a high degree of the intravascular coagulation in AUF1−/− mice. The arrowhead indicates blood clot formation, which was common in AUF1 knockout mice but not wild-type mice.
AUF1−/− mice undergo more severe endotoxic shock
One of the essential features of endotoxic shock is disseminated intravascular coagulation, characterized by widespread blood coagulation and vessel hemorrhage, particularly in the kidney and lung (Parrillo 1993). Disseminated intravascular coagulation is tightly associated with pathological activation of proinflammatory cytokines TNFα and IL-1β (Dinarello 1997). Histological examination found widespread hemorrhage in the kidneys of AUF1−/− mice following LPS challenge (Fig. 2C), indicating a massive capillary leakage. In contrast, there was only mild hemorrhage in the kidney of wild-type control mice. To assess the intensity of hemorrhage, the mean number of hemorrhage lesions per field at 10× magnification was determined. Compared with wild-type mice, AUF1−/− mice had a significantly greater number of hemorrhagic lesions in the kidney (mean lesion number/field ± SD: 9.8 ± 2.9 for AUF1−/− and 4.1 ± 1.6 for wild-type mice, p < 0.001), which were more extensive in size. Moreover, sampling of peripheral blood of AUF1−/− mice demonstrated a fivefold increase in blood urea nitrogen (BUN) levels and a 2.5-fold increase in the serum levels of released liver enzyme aspartate aminotransferase (AST) (Fig. 2D), markers of tissue destruction that correlate with a severe loss of renal and liver function, respectively. Further, higher resolution showed that intravascular coagulation was common in kidney, lung, and liver of AUF1−/− mice, but not in wild-type mice following endotoxin challenge, with an average of four coagulation lesions per 10× magnification field in AUF1−/− mice and less than one lesion per field in control animals (Fig. 2E). Loss of AUF1 expression therefore leads to widespread intravascular coagulation associated with organ failure, a major cause of fatal endotoxic shock after endotoxin challenge.
Loss of AUF1 promotes excessive production of TNFα and IL-1β
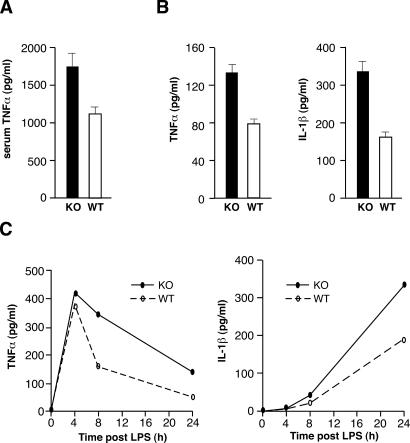
We determined whether the increased susceptibility to endotoxemia of AUF1−/− mice is due to deregulation of proinflammatory cytokine expression, particularly TNFα and IL-1β, two critical mediators of endotoxic shock. For this purpose, we measured the serum TNFα levels in wild-type and AUF1−/− mice after LPS challenge. The level of TNFα was elevated by 30% in peripheral blood of AUF1−/− mice after endotoxin injection (Fig. 3A). The serum TNFα level is likely to be an underestimate of the level of TNFα produced and locally accumulated because of rapid retention in tissues, catabolism, and incomplete release into circulating blood (Finkelman et al. 1993). In addition, the high level of blood clotting in endotoxin-challenged AUF1−/− mice makes sampling difficult. In unchallenged wild-type and AUF1 knockout mice, there was a barely detectable level of TNFα (data not shown). These data indicate that there is an increase in the overall production of TNFα in AUF1−/− compared with wild-type mice following LPS challenge.
Figure 3.
Overproduction of proinflammatory cytokines TNFα and IL-1β due to loss of AUF1. (A) Mouse serum TNFα level 75 min after LPS injection (20 mg/kg weight, p < 0.02, n = 4). (B) Accumulated cytokine production by primary macrophages. TNFα and IL-1β secretion were elevated (p < 0.05) in AUF1−/− cells after LPS endotoxin stimulation. (C) Kinetics of TNFα and IL-1β expression in LPS-stimulated primary macrophages. Sustained expression of proinflammatory cytokines led to elevated levels of cytokine production in AUF1-deficient macrophages compared with those in wild-type cells, even at 24 h post-LPS challenge. Secreted cytokines were measured by ELISA.
To directly examine cytokine production from a homogeneous cell population, primary macrophages, which express AUF1 (Fig. 5E, below), were isolated from wild-type and AUF1−/− mouse peritoneal cells and placed in primary culture. Levels of ARE-binding and control proteins were examined by immunoblot analysis of whole-cell lysate of primary macrophages derived from wild-type and AUF1 knockout mice. No significant differences were found, apart from the absence of AUF1 in knockout mice (Fig. 5E, below). Primary macrophages were stimulated with LPS and the levels of cytokines secreted into medium were determined by ELISA. Comparison of the accumulated levels of cytokine secretion after LPS stimulation showed that production of TNFα and IL-1β were increased by 40% and 70%, respectively, in AUF1-deficient macrophages (Fig. 3B). Therefore, loss of AUF1 increased TNFα and IL-1β levels in response to endotoxin challenge.
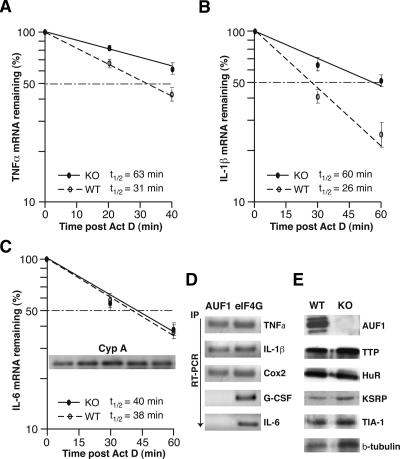
Figure 5.
Abnormal stabilization of TNFα and IL-1β mRNAs caused by loss of AUF1. (A–C) Primary peritoneal macrophages derived from AUF1 knockout or wild-type mice were stimulated with LPS (1 μg/mL) for 4 h followed by addition of actinomycin D (2 μg/mL) to block transcription. Cells were collected at the indicated time points, and cytoplasmic RNA was extracted and analyzed by real-time quantitative RT–PCR (Roche Light Cycler) and normalized to the housekeeping Cyp A mRNA. The plots average three independent experiments for each mRNA to determine mRNA half-lives for TNFα (A), IL-1β (B), and IL-6 (C), as well as the standard deviation of results. The inset in C shows the level of Cyp A mRNAs determined by qRT–PCR at different time points (0, 15, 30, 45, and 60 min post-Actinomycin D). (D) Intracellular AUF1 proteins selectively bind to IL-1β, TNFα, and Cox2 mRNAs but not IL-6 mRNA or a non-ARE G-CSF mRNA. Cultured macrophages were lysed after LPS stimulation of cytokine expression and RNA bound by AUF1 was immunoprecipitated by AUF1-specific antibody. Immunoprecipitation with translation initiation factor eIF4G was used as a control. The amount of target mRNA bound by AUF1 or eIF4G was determined by real-time quantitative RT–PCR. (E) Primary macrophages derived from peritoneal cells were subjected to immunoblot analysis using antibodies against AUF1 and the other ARE-binding proteins as indicated. Disruption of AUF1 did not significantly alter the expression level of ARE-binding proteins examined here.
The kinetics of TNFα and IL-1β cytokine expression was also examined to monitor the dynamic change of cytokine levels at time points following LPS challenge. Compared with wild-type cells, TNFα production in LPS-stimulated AUF1−/− macrophages was increased by 2.5-fold, and IL-1β by twofold overall (Fig. 3C). Importantly, TNFα and IL-1β expression in AUF1−/− macrophages remained elevated at 24 h, therefore for a longer duration of time (Fig. 3C) compared with the wild-type macrophage controls. While the expression level of TNFα declined by 24 h in wild-type macrophages, it was still almost threefold higher in AUF1−/− macrophages. IL-1β expression continued to increase over 24 h, to a level twofold greater in AUF1−/− macrophages than in wild-type cells. Expression of IL-1β reached a plateau by 30 h in wild-type macrophages but continued to increase slowly in AUF1 knockout mice (data not shown). Thus, there is an increased and more sustained expression of TNFα and IL-1β in AUF1−/− macrophages compared with wild-type macrophages, which is associated with the severe pathophysiological reaction of AUF1−/− animals to endotoxin. These data suggest that AUF1 functions to attenuate the proinflammatory cytokine response after endotoxin challenge.
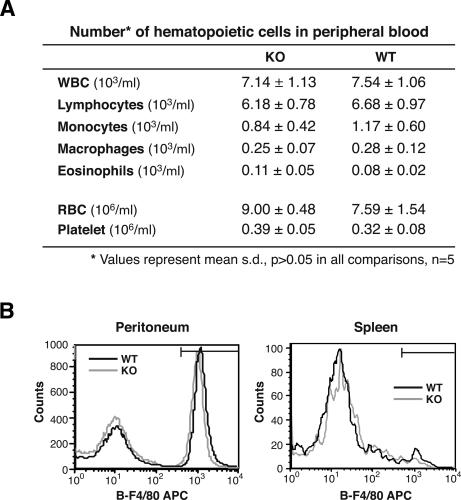
Excessive production of proinflammatory cytokines is not due to an increase in the monocyte/macrophage population in AUF1−/− mice
Hematopoietic cells, especially monocytes and macrophages, are major sources of proinflammatory cytokines TNFα and IL-1β. To determine whether lack of AUF1 affected the development and circulating levels of hematopoietic cells, we performed a complete blood count and a differential count of blood cells for wild-type and AUF1−/− mice, and analyzed their macrophage population in the peritoneum and spleen by flow cytometric analysis. Red blood cell parameters, leukocyte, and platelet counts in peripheral blood were comparable and within normal ranges in AUF1−/− and wild-type littermates. Our analysis also confirmed the presence of all major hematopoietic lineages in AUF1−/− mice without significant alteration in macrophage numbers (Fig. 4A). Flow cytometry analysis of the macrophage population (F4/80 staining for murine macrophages) in peritoneum and spleen did not reveal significant difference between AUF1−/− and wild-type mice (Fig. 4B). Thus, the excessive production of proinflammatory cytokines in LPS challenged AUF1−/− mice is not due to an increase in the number of source cells.
Figure 4.
Analysis of peripheral blood and macrophage population in AUF1−/− mice. (A) Red blood cell parameters and numbers of leukocyte subpopulations were obtained by complete blood cell count and differential cell count of peripheral blood. No significant difference was detected between AUF1 knockout and wild-type mice. (B) Numbers of macrophages in peritoneum and spleen were determined by flow cytometry analysis of F4/80 surface expression (a cell surface marker specific for murine macrophages). Representative histograms are shown (n = 5 in each group). No significant difference was observed.
Abnormal stabilization of TNFα and IL-1β mRNAs results from disruption of AUF1
Given the reported function of AUF1 in promoting rapid degradation of ARE-mRNAs (Guhaniyogi and Brewer 2001), we asked whether increased TNFα and IL-1β expression is due to an alteration in mRNA stabilities. To monitor the decay of ARE-mRNAs, actinomycin D was applied to block transcription after primary macrophages were stimulated with LPS. The level of remaining mRNA transcripts was determined by real-time quantitative RT–PCR at times post-inhibition of transcription. We found that TNFα and IL-1β mRNAs in wild-type macrophages were unstable and rapidly degraded with half-lives of 31 and 26 min, respectively (Fig. 5A–B). However, in AUF1−/− macrophages, the half-lives of TNFα and IL-1β mRNAs were 2.5-fold (60 min for IL-1β mRNA) and twofold greater (63 min for TNFα mRNA) compared with wild-type macrophages. The levels of TNFα and IL-1β mRNAs in unstimulated wild-type and AUF1 knockout cells were barely detectable (data not shown). There was no change in the stability of ARE-containing IL-6 mRNA in AUF1−/− macrophages (Fig. 5C), or in the stability of cyclophilin A (Cyp A) mRNAs (Fig. 5C, inset; data not shown), which was used as a control for normalization. These results indicate that AUF1 specifically promotes decay of select mRNAs including TNFα and IL-1β. To confirm the binding of AUF1 to TNFα and IL-1β mRNAs, immunoprecipitation-quantitative RT–PCR (IP qRT–PCR) was performed to determine the intracellular AUF1 protein–RNA interaction. AUF1 proteins were immunoprecipitated from cytoplasmic lysates of macrophages, from which mRNAs were isolated. The amount of each target mRNA bound by AUF1 was determined by real-time qRT–PCR using specific primers. TNFα, IL-1β, and Cyclooxygenase 2 (Cox2) mRNAs were detected in both AUF1 and eIF4G immunoprecipitate (Fig. 5D), the latter of which was consistent with the interaction of eIF4G with actively translated mRNAs. In contrast, IL-6 mRNA and granulocyte-colony stimulating factor (G-CSF) mRNA, a non-ARE mRNA, were found associated only with eIF4G, but not with AUF1 (Fig. 5D). These results indicate that AUF1 proteins selectively bind to TNFα, IL-1β, and Cox2 mRNAs, all of which contain overlapping AUUUA pentamers. The 3′ untranslated regions of TNFα and IL-1β mRNAs differ from that of IL-6 in containing multiple consecutive copies of the AUUUA pentamer, whereas IL-6 contains only two pentamers, and they are interrupted by intervening sequences. It is likely that in a natural physiological setting in the absence of AUF1 ectopic overexpression, AUF1 binding requires the consecutive placement of multiple AUUUA sequences in vivo.
To determine whether disruption of AUF1 affects the expression of other ARE-binding proteins that have been implicated in regulating ARE-mRNA stability, we examined their expression levels in isolated peritoneal macrophages by the immunoblot analysis, as described above. The expression levels of major ARE-binding proteins (TTP, HuR, KSRP, TIA-1) and control protein (β-tubulin) remained unaltered in AUF1−/− macrophages (Fig. 5E). Thus, the increased stability of TNFα and IL-1β mRNAs appears to be a direct consequence of loss of AUF1. Collectively, these data demonstrate that AUF1 governs the inflammatory response and prevents the excessive production of proinflammatory cytokines TNFα and IL-1β by promoting decay of their mRNAs.
Blocking TNFα and IL-1β activity protects AUF1 knockout mice from severe endotoxic shock
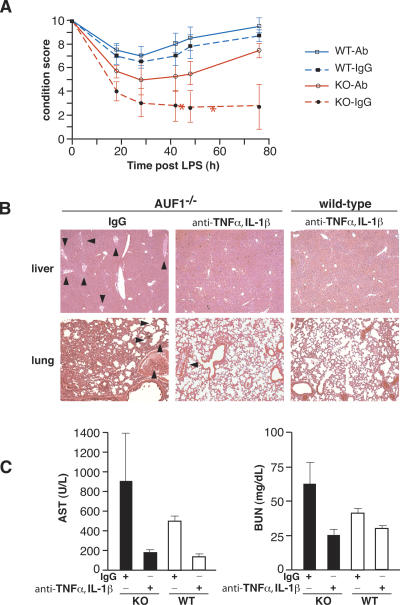
To confirm that the LPS-induced endotoxic shock phenotype in AUF1−/− mice is due to overproduction of TNFα and IL-1β, animals were injected with neutralizing antibodies against TNFα and IL-1β prior to a sublethal LPS challenge. The general condition of each mouse was monitored, and a condition score was derived based on numeric manifestations of endotoxemia, including lethargy, diarrhea, tachypnea, piloerection, and death, as described in the Materials and Methods section. Whereas IgG-injected AUF1−/− mice showed typical signs of endotoxic shock observed earlier, animals treated with antibodies against TNFα and IL-1β were almost fully protected and had their conditions significantly improved (Fig. 6A, p < 0.05). Thus, neutralizing antibody injection completely protected AUF1−/− mice against lethality, whereas half of the IgG-treated AUF1−/− group failed to survive the LPS challenge.
Figure 6.
Protection of AUF1−/− mice from severe endotoxic shock by TNFα and IL-1β antibodies. (A) The condition score of IgG- or antibody-injected AUF1 knockout (KO-IgG; KO-Ab) and wild-type (WT-IgG; WT-Ab) mice after LPS challenge. Mice were given either IgG or antibody against TNFα and IL-1β prior to LPS challenge as described in Materials and Methods (n = 4 in each group). The general condition of each mouse was monitored and scored based on the manifestations of endotoxemia including lethargy, diarrhea, tachypnea, and piloerection (score scale 0–10: 10 for normal activity of wild-type mice and 0 for death). The condition score of IgG-treated AUF1−/− group is significantly lower than those of the other three groups at the 42-, 48-, and 76-h time points (p < 0.01). Mean value of condition score is plotted. Asterisks indicate death of two out of four mice in IgG-injected AUF1−/− group. (B) Representative H&E-stained sections of organs 24 h following LPS challenge to visualize intravascular coagulation in mice injected with antibodies to TNFα, IL-1β, or IgG (n = 4 in each group). Arrowheads indicate blood clot formation. Note the dramatically reduced blood clot formation in AUF1−/− mice treated with TNFα and IL-1β antibody compared with those treated with IgG. (C) BUN and AST levels in mice 24 h following challenge with LPS (n = 3, p < 0.08 for AST levels in AUF1 knockout mice, p < 0.06 for BUN levels in AUF1 knockout mice).
Histological examination found that neutralizing antibody treatment dramatically prevented the development of widespread intravascular coagulation in AUF1−/− mice. Inhibition of clotting is shown by the reduction of blood clotting in the lung and liver, which in contrast was highly evident in IgG-injected AUF1−/− mice (Fig. 6B). The mean number of intravascular coagulation lesions per 10× magnification field in livers of control IgG-injected AUF1−/− mice was significantly higher than that of other three groups (IgG-injected AUF1−/− mice: 8.5 ± 2.4; antibody-injected AUF1−/− mice: 3.8 ± 1.6; IgG-injected wild-type mice: 1.9 ± 1.6; antibody-injected wild-type mice: 1.4 ± 1.3, mean ± SD, p < 0.005) (Supplementary Fig. S3). Treatment with neutralizing antibody against TNFα and IL-1β also reduced the AST and BUN levels by 5.6-fold and 2.5-fold, respectively, both of which were reduced to levels similar to those of antibody-injected wild-type animals (Fig. 6C). Blocking TNFα and IL-1β activity therefore alleviated otherwise severely impaired liver and renal function in AUF1−/− mice. Collectively, these data demonstrate that AUF1−/− mice excessively produce specific proinflammatory cytokines due to loss of AUF1, which is a result of a failure to selectively and rapidly degrade TNFα and IL-1β mRNAs, and is responsible for development of endotoxic shock in AUF1−/− mice after LPS challenge.
Discussion
In this study we report that AUF1 protects animals from fatal progression of endotoxemia to endotoxic shock by attenuating the expression of proinflammatory cytokines TNFα and IL-1β through facilitated degradation of their mRNAs. This was shown by the abnormal stabilization of TNFα and IL-1β mRNAs in AUF1−/− macrophages after LPS stimulation compared with wild-type macrophages. These data indicate that AUF1 specifically promotes TNFα and IL-1β mRNA decay. We further confirmed that excessive proinflammatory cytokine production is due to loss of AUF1, and is responsible for development of endotoxic shock in AUF1−/− mice. The role of AUF1 in regulating inflammatory cytokine expression in vivo was shown by targeted disruption of the mouse AUF1 gene, which resulted in increased mortality during experimentally induced endotoxemia. The increased lethality of AUF1 knockout animals was shown to result from excessive production of TNFα and IL-1β, two critical mediators of endotoxic shock. Overexpression of these two proinflammatory cytokines following LPS challenge was found to be due to the inability to rapidly degrade TNFα and IL-1β mRNAs following their induction. Thus, we have identified AUF1 as a critical attenuator of proinflammatory cytokine expression.
Notably, loss of AUF1 did not cause a severe defect in mouse development. Targeted disruption of TTP, another ARE-binding protein that also regulates TNFα mRNA stability, causes a syndrome of cachexia, arthritis, and autoimmunity (Taylor et al. 1996). Although AUF1 and TTP are both ARE-binding proteins involved in regulating ARE-mRNA stability and both are found in immune cells (Carballo et al. 1998; Loflin et al. 1999; Sarkar et al. 2003; Lu and Schneider 2004), they demonstrate different phenotypes in knockout mice. TTP knockout mice develop several abnormalities in hematopoietic systems, particularly a smaller thymus without cortical/medullary organization (Carballo et al. 1998). In contrast, thymus development in AUF1-deficient mice is normal (data not shown). It has also been reported that in peripheral blood of TTP−/− mice, the white blood cell count is elevated by more than twofold over that of wild-type mice, and there is a marked increase in the number of circulating neutrophils and macrophages (Taylor et al. 1996). In contrast, in AUF1−/− mice, similar numbers of hematopoietic cells are found in peripheral blood compared with wild-type mice, and no change was observed in macrophages in peritoneum and spleen (Fig. 4). These observations suggest that AUF1 may have different regulatory effects and different functions than TTP in the ARE-mRNA decay pathway. The phenotype of TTP−/− mice is likely the consequence of chronic excessive TNFα circulating in the blood (Taylor et al. 1996). Thus, TTP may constitutively target TNFα mRNA for degradation, and loss of TTP leads to a chronic and constant accumulation of TNFα mRNA and its overproduction. Our data from AUF1 knockout mice suggests that AUF1 may be a regulatory factor that normally does not execute its function until the activation of proinflammatory cytokine expression. In particular, in the absence of LPS-challenge, AUF1 knockout mice did not display elevated levels of TNFα or IL-1β, and AUF1 promoted the rapid decay of TNFα and IL-1β mRNAs only after their induction in response to endotoxin challenge. AUF1-directed mRNA degradation may therefore serve as a protective means to attenuate the inflammatory response post-stimulation, and to protect against deleterious effects of excessive inflammation.
The increased susceptibility of AUF1−/− mice to endotoxin challenge is reminiscent of the phenotype of mice lacking the TIA-1 gene (Piecyk et al. 2000), which encodes an ARE-binding protein that specifically binds the TNFα–ARE and represses TNFα translation. Increased production of TNFα in TIA-1−/− mice is mainly due to derepression of TNFα mRNA translation, as no alteration in TNFα mRNA stability is detected. In contrast, we found that abnormal stabilization of TNFα and IL-1β mRNAs contributes to their overexpression in AUF1−/− mice. The difference in the mechanisms by which AUF1 and TIA-1 control TNFα production implies that a precise control of cytokine expression demands regulation at multiple levels.
At this time, we cannot formally rule out a hypothetical role for AUF1 in regulating both the stability and translation of TNFα and IL-1β mRNAs. However, previous work on TTP and TIA-1 (Kontoyiannis et al. 1999; Piecyk et al. 2000) indicates that the translation repression and stability of ARE-mRNAs are independently controlled by different ARE-binding proteins. These findings suggest that AUF1 may mainly control the stability of target mRNAs. Given the overlapping tissue distribution of decay-promoting AUF1 and stability-promoting HuR (Lu and Schneider 2004), as well as the binding of AUF1 and HuR to common target mRNAs (Lal et al. 2004), loss of AUF1 might function in part by allowing HuR to play a more active role in stabilizing target mRNAs and, consequently, causing their overexpression.
Despite previous reports that mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 signaling regulates IL-6 mRNA stability and its biosynthesis via the AU-rich 3′ noncoding region (Winzen et al. 1999; Neininger et al. 2002), we did not detect a significant alteration in IL-6 mRNA stability in AUF1-deficient cells (Fig. 5C). Our results suggest that AUF1 more selectively regulates TNFα and IL-1β mRNA stability, and that IL-6 mRNA may not be controlled by AUF1 under normal physiological conditions (without AUF1 overexpression). mRNAs targeted for rapid decay by ARE elements, such as TNFα and IL-1β mRNAs, often contain three to five uninterrupted copies of an AUUUA pentamer in the 3′ noncoding region (Guhaniyogi and Brewer 2001). IL-6 mRNA, however, contains no consecutive AUUUA pentamers. Instead, its 3′ noncoding region contains two pentamers interspersed in an AU-rich region with varying U stretches of 2–5 nucleotides in length. We speculate that multiple consecutive AUUUA pentamers may be important for AUF1 to interact with target ARE-mRNAs and regulate their stability. Consistent with this hypothesis, Cox2 mRNA, a class II ARE-mRNA harboring multiple overlapping AUUUA pentamers, was found to associate with AUF1 in vivo (Fig. 5D) and its half-life was increased at least 50% in AUF1-deficient macrophages derived from AUF1 knockout mice (N. Sadri and R. Schneider, unpubl.). It is also possible that IL-6 mRNA stability is regulated by different AUF1 isoforms with opposing functions so that inactivation of all four AUF1 isoforms may obscure the effect on the target mRNA. It has been suggested that the different AUF1 isoforms possess different, or perhaps even opposing functions in regulating ARE-mRNA decay (Laroia et al. 1999; Loflin et al. 1999; Sarkar et al. 2003; Raineri et al. 2004). With the availability AUF1-deficient cells, these questions can now be experimentally addressed.
In summary, our results provide the first in vivo evidence for a novel post-transcriptional mechanism critical for controlling proinflammatory cytokine expression and protecting against fatal endotoxic shock after endotoxin exposure. Our findings indicate that after proinflammatory cytokine induction, AUF1 promotes decay of TNFα, IL-1β, and possibly other inflammatory cytokine mRNAs as a means for controlling and tempering the inflammatory response. Defects in this post-transcriptional regulation may be involved in human inflammatory diseases.
Materials and methods
Generation of AUF1−/− mice
A 1.4-kb genomic fragment upstream of mouse AUF1 exon 3, and a 7.9-kb genomic fragment downstream from AUF1 exon 3, were used as short and long recombination arms, respectively, in the construction of the targeting vector. A loxP-flanked neor expression cassette was inserted between the two regions, resulting in a vector designed to delete the two RNA-binding motifs in exon 3. W4 embryonic stem (ES) cells were electroporated with the NotI-linearized targeting construct, selected in G418 (150 μg/mL) and gancyclovir (2 mM), and used for Southern blot DNA analysis. Five positive clones were identified using Southern blot and PCR analysis. Blastocyst injection was performed using two independent targeted ES cell clones. Germline transmission was obtained on further crossing of male chimeras with C57BL/6J females.
Animal studies and histology
Experiments were carried out in accordance with NIH guidelines for animal treatment, housing, and euthanasia. Mice were challenged by intraperitoneal LPS injection (low dose: 3 mg/kg weight; sublethal dose: 20 mg/kg) and monitored for general condition and survival. At indicated times, mice were euthanized and blood serum was collected. Blood BUN and AST levels were determined using Infinity BUN Reagent and AST Reagent (Sigma Diagnostics), respectively. For histological analysis, tissues were fixed in 10% buffered formaldehyde overnight at 4°C and paraffin-embedded. Sections (5 μm) were stained with hematoxylin and eosin (H&E). To determine the general condition of mice after LPS challenge, mice were monitored blindly (without knowing their genotypes) and individual scores were given based on the symptoms of endotoxemia (motor activity/coordination (0–2, normal being 2), strength (0–2, normal being 2), food intake (0–1, normal being 1), ocular exudate (0–2, no exudates denoting 2), diarrhea (0–1, no diarrhea being 1), piloerection/coat appearance (0–1, normal being 1), and breath rate (0–1, 0 denoting slowed rate)). The condition score of a particular mouse is the summed score of individual symptom scores (10 being normal activity and 1 denoting close to death).
Quantification of cytokine expression
Serum cytokine levels and cytokines secreted from primary macrophages were determined by ELISA. Equal numbers of peritoneal macrophages derived from three mice were pooled and seeded in three replicative wells (5 × 105 cells/well). Peritoneal cells were incubated at 37°C in an atmosphere of 5% CO2 for 3 h to allow peritoneal macrophages to adhere. Nonadherent cells were removed by washing with PBS twice, and then macrophages were stimulated with 1 μg/mL LPS (Sigma) for the times indicated. Levels of TNFα and IL-1β were measured by ELISA (eBioscience) using an equal amount of culture supernatant at indicated time points (Fig. 3C), or at 8 h after LPS treatment for TNFα and 24 h after LPS treatment for IL-1β (Fig. 3B).
Determination of mRNA half-life
For decay studies, peritoneal macrophages were stimulated with LPS (1 μg/mL) for 4 h. After 4 h of LPS stimulation, actinomycin D (2 μg/mL) was added to block transcription, and total RNA was isolated after 0, 20, and 40 min post-actinomycin D treatment for TNF-α measurements, or after 0, 30, and 60 min post-actinomycin D treatment for IL-1β and IL-6 measurements. mRNA levels were determined by quantitative real-time RT–PCR (qRT–PCR, Roche Light Cycler), and were normalized to Cyp A mRNA as a control. PCR reactions were performed with gene-specific primers (primer sequences available upon request). The plots (Fig. 5A–C) average three independent experiments for each mRNA to determine mRNA half-lives and standard deviation of results.
TNF-α and IL-1β antibody administration studies
AUF1−/− and AUF1+/+ male mice each received intraperitoneal injection of 200 μg of hamster monoclonal antibody to mouse TNFα (clone TN3-19.12, BD PharMingen) and 100 μg of hamster monoclonal antibody to mouse IL-1β (clone B122, BioLegend). Mice of each genotype in the control group received 100 μg of hamster isotype IgG control (clone A19-3, BD PharMingen). Six hours post-antibody injection, mice in both groups received an intraperitoneal injection of a sublethal dose of LPS (20 mg/kg). Mice were monitored for general condition and survival, as described in the above animal studies and histology section. Measurement of BUN and AST levels, and histological analyses were performed as described above.
Immunoblot analysis and IP qRT–PCR
Immunoblot analysis was performed following standard procedures. Polyclonal antibodies against AUF1, KSRP (from D. Black), and TIA-1 (from P. Anderson), and specific antibodies for HuR (Santa Cruz Biotechnology), TTP (Santa Cruz Biotechnology), and β-tubulin (Sigma) were used. ECL Western Blotting Detection Kit (Amersham Pharmacia) was used for detection. IP qRT–PCR was performed using anti-AUF1 and anti-eIF4G polyclonal antibody according to a protocol previously described (Sarkar et al. 2003).
Analysis of immune cells
Peripheral blood analysis was performed by Anilytics, Inc. Spleen and peritoneal cells were made into single-cell suspensions in RPMI 1640 medium by passing through a 70-μm nylon cell strainer. The cell suspensions were depleted of erythrocytes by osmotic lysis, and cells were incubated on ice for 15 min with anti-CD16/CD32 (BD PharMingen) to block the Fc receptor. Subsequently, these cells were incubated with anti-F4/80-biotin (BM8, eBioscience) for 30 min. The cells were then washed with PBS and incubated with streptoavidin-APC (BD PharMingen) on ice for 15 min. Cells were washed, fixed, and analyzed on a FACSCalibur (Becton Dickson) using a live lymphocyte gate. Data were analyzed using CellQuest software (Becton Dickson).
Statistical analysis
Data are presented as mean ± SD for statistical comparison of two samples, the Student t-test was used for evaluation. Survival curves were generated using the Kaplan-Meier method, and significance was evaluated using the Log-rank test.
Acknowledgments
We thank Ralf Kist for critical advice, Tung-Tien Sun for a 129/SvJ mouse genomic library, Klaus Rajewsky for the neo plasmid, Douglas Black for anti-KSRP antibody, and Paul Anderson for anti-TIA-1 antibody. We also thank Anna Auerbach and the staff at New York University (NYU) School of Medicine Transgenic Mouse/ES Cell Chimera Facility for assistance in producing chimeric mice, and Doris Tse and the staff at Center for AIDS Research at NYU for assistance in FACS analysis. This work was supported by grant GM60428 from the NIH (R.J.S.). The authors declare that they have no competing financial interests.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1467606
References
- Brennan, C.M., Steitz, J.A. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzby, J.S., Brewer, G., Nugent, D.J. Developmental regulation of RNA transcript destabilization by A + U- rich elements is AUF1-dependent. J. Biol. Chem. 1999;274:33973–33978. doi: 10.1074/jbc.274.48.33973. [DOI] [PubMed] [Google Scholar]
- Carballo, E., Lai, W.S., Blackshear, P.J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Carballo, E., Lai, W.S., Blackshear, P.J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- Chen, C.Y., Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., Karin, M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Dinarello, C.A. The interleukin-1 family: 10 years of discovery. FASEB J. 1994;8:1314–1325. [PubMed] [Google Scholar]
- Dinarello, C.A. Proinflammatory and anti-inflammatory cytokines as mediators in the pathogenesis of septic shock. Chest. 1997;112:321S–329S. doi: 10.1378/chest.112.6_supplement.321s. (Suppl.) [DOI] [PubMed] [Google Scholar]
- Dixon, D.A., Tolley, N.D., King, P.H., Nabors, L.B., McIntyre, T.M., Zimmerman, G.A., Prescott, S.M. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X.C., Steitz, J.A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman, F.D., Madden, K.B., Morris, S.C., Holmes, J.M., Boiani, N., Katona, I.M., Maliszewski, C.R. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine–anti-cytokine antibody complexes. J. Immunol. 1993;151:1235–1244. [PubMed] [Google Scholar]
- Gherzi, R., Lee, K.Y., Briata, P., Wegmuller, D., Moroni, C., Karin, M., Chen, C.Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Glauser, M.P., Zanetti, G., Baumgartner, J.D., Cohen, J. Septic shock: Pathogenesis. Lancet. 1991;338:732–736. doi: 10.1016/0140-6736(91)91452-z. [DOI] [PubMed] [Google Scholar]
- Grosset, C., Boniface, R., Duchez, P., Solanilla, A., Cosson, B., Ripoche, J. In vivo studies of translational repression mediated by the granulocyte-macrophage colony-stimulating factor AU-rich element. J. Biol. Chem. 2004;279:13354–13362. doi: 10.1074/jbc.M308003200. [DOI] [PubMed] [Google Scholar]
- Guhaniyogi, J., Brewer, G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- He, C., Schneider, R. 14–3–3σ is a p37 AUF1-binding protein that facilitates AUF1-mediated ARE-mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss, R.S., Karl, I.E. The pathophysiology and treatment of sepsis. N. Engl. J. Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- Jacob, C.O., Lee, S.K., Strassmann, G. Mutational analysis of TNF-α gene reveals a regulatory role for the 3′-untranslated region in the genetic predisposition to lupus-like autoimmune disease. J. Immunol. 1996;156:3043–3050. [PubMed] [Google Scholar]
- Kajita, Y., Nakayama, J., Aizawa, M., Ishikawa, F. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2xRBD-Gly family. J. Biol. Chem. 1995;270:22167–22175. doi: 10.1074/jbc.270.38.22167. [DOI] [PubMed] [Google Scholar]
- Katsanou, V., Papadaki, O., Milatos, S., Blackshear, P.J., Anderson, P., Kollias, G., Kontoyiannis, D.L. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Kiledjian, M., DeMaria, C.T., Brewer, G., Novick, K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol. Cell. Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis, D., Pasparakis, M., Pizarro, T.T., Cominelli, F., Kollias, G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: Implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- Lai, W.S., Carballo, E., Strum, J.R., Kennington, E.A., Phillips, R.S., Blackshear, P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, A., Mazan-Mamczarz, K., Kawai, T., Yang, X., Martindale, J.L., Gorospe, M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia, G., Cuesta, R., Schneider, R.J. Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- Lebwohl, D.E., Muise-Helmericks, R., Sepp-Lorenzino, L., Serve, S., Timaul, M., Bol, R., Borgen, P., Rosen, N. A truncated cyclin D1 gene encodes a stable mRNA in a human breast cancer cell line. Oncogene. 1994;9:1925–1929. [PubMed] [Google Scholar]
- Levy, N.S., Chung, S., Furneaux, H., Levy, A.P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- Loflin, P., Chen, C.Y., Shyu, A.B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes & Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J.Y., Schneider, R.J. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J. Biol. Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- Ma, W.J., Cheng, S., Campbell, C., Wright, A., Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- Martin, G.S., Mannino, D.M., Eaton, S., Moss, M. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- Nair, A.P., Hahn, S., Banholzer, R., Hirsch, H.H., Moroni, C. Cyclosporin A inhibits growth of autocrine tumour cell lines by destabilizing interleukin-3 mRNA. Nature. 1994;369:239–242. doi: 10.1038/369239a0. [DOI] [PubMed] [Google Scholar]
- Neininger, A., Kontoyiannis, D., Kotlyarov, A., Winzen, R., Eckert, R., Volk, H.D., Holtmann, H., Kollias, G., Gaestel, M. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 2002;277:3065–3068. doi: 10.1074/jbc.C100685200. [DOI] [PubMed] [Google Scholar]
- Parrillo, J.E. Pathogenetic mechanisms of septic shock. N. Engl. J. Med. 1993;328:1471–1477. doi: 10.1056/NEJM199305203282008. [DOI] [PubMed] [Google Scholar]
- Pende, A., Tremmel, K.D., DeMaria, C.T., Blaxall, B.C., Minobe, W.A., Sherman, J.A., Bisognano, J.D., Bristow, M.R., Brewer, G., Port, J. Regulation of the mRNA-binding protein AUF1 by activation of the β-adrenergic receptor signal transduction pathway. J. Biol. Chem. 1996;271:8493–8501. doi: 10.1074/jbc.271.14.8493. [DOI] [PubMed] [Google Scholar]
- Piecyk, M., Wax, S., Beck, A.R., Kedersha, N., Gupta, M., Maritim, B., Chen, S., Gueydan, C., Kruys, V., Streuli, M., et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri, I., Wegmueller, D., Gross, B., Certa, U., Moroni, C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pascual, F., Hausding, M., Ihrig-Biedert, I., Furneaux, H., Levy, A.P., Forstermann, U., Kleinert, H. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J. Biol. Chem. 2000;275:26040–26049. doi: 10.1074/jbc.M910460199. [DOI] [PubMed] [Google Scholar]
- Sarkar, B., Xi, Q., He, C., Schneider, R.J. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko, O.I., Lofquist, A.K., DeMaria, C.T., Morris, J.S., Brewer, G., Haskill, J.S. Adhesion-dependent regulation of an A + U-rich element-binding activity associated with AUF1. Mol. Cell. Biol. 1997;17:3898–3906. doi: 10.1128/mcb.17.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin, G., Colombi, M., Raineri, I., Leuenberger, S., Mallaun, M., Schmidlin, M., Gross, B., Lu, M., Kitamura, T., Moroni, C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, G.A., Carballo, E., Lee, D.M., Lai, W.S., Thompson, M.J., Patel, D.D., Schenkman, D.I., Gilkeson, G.S., Broxmeyer, H.E., Haynes, B.F., et al. A pathogenetic role for TNF α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- Tracey, K.J., Fong, Y., Hesse, D.G., Manogue, K.R., Lee, A.T., Kuo, G.C., Lowry, S.F., Cerami, A. Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteraemia. Nature. 1987;330:662–664. doi: 10.1038/330662a0. [DOI] [PubMed] [Google Scholar]
- Wagner, B.J., DeMaria, C.T., Sun, Y., Wilson, G.M., Brewer, G. Structure and genomic organization of the human AUF1 gene: Alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- Wang, W., Furneaux, H., Cheng, H., Caldwell, M.C., Hutter, D., Liu, Y., Holbrook, N., Gorospe, M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, G.M., Sun, Y., Lu, H., Brewer, G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J. Biol. Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Wormington, M., Peltz, S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Winzen, R., Kracht, M., Ritter, B., Wilhelm, A., Chen, C.Y., Shyu, A.B., Muller, M., Gaestel, M., Resch, K., Holtmann, H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Wagner, B.J., Ehrenman, K., Schaefer, A.W., DeMaria, C.T., Crater, D., DeHaven, K., Long, L., Brewer, G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]