Abstract
p63 is a multi-isoform p53 family member required for epidermal development. Contrasting roles for p63 in either the initial commitment to the stratified epithelial cell fate or in stem cell-based self-renewal have been proposed. To investigate p63 function in a post-developmental context, we used siRNAs directed against p63 to down-regulate p63 expression in regenerating human epidermis. Loss of p63 resulted in severe tissue hypoplasia and inhibited both stratification and differentiation in a cell-autonomous manner. Although p63-deficient cells exhibited hypoproliferation, differentiation defects were not due to tissue hypoplasia. Simultaneous p63 and p53 knockdown rescued the cell proliferation defect of p63 knockdown alone but failed to restore differentiation, suggesting that defects in epidermal proliferation and differentiation are mediated via p53-dependent and -independent mechanisms, respectively. Furthermore, ΔNp63 isoforms are the main mediators of p63 effects, although TAp63 isoforms may contribute to late differentiation. These data indicate that p63 is required for both the proliferative and differentiation potential of developmentally mature keratinocytes.
Keywords: Epidermis, skin, differentiation, proliferation, p63, p53
The epidermis is the self-renewing stratified squamous epithelium on the skin’s outer surface. In this tissue, basal stem cell division yields a subset of daughter cells that undergo a coordinated program of cell cycle arrest, outward migration, and terminal differentiation. The epidermal differentiation program engages a variety of genes that contribute to formation of the cutaneous barrier, including involucrin, loricrin, filaggrin, keratins 1 and 10, and transglutaminase 1. These stratified epithelial differentiation genes are distinct from those expressed in simple epithelium, such as keratins 8 and 18, from which the epidermis arises during gestation (Turksen and Troy 1998). Recently, a role for p63 has been identified in the formation of stratified epithelium during development (Mills et al. 1999; Yang et al. 1999).
p63 is a member of the p53 protein family and is expressed as six distinct isoforms due to alternative transcription start sites and splicing at the C terminus (Yang et al. 1998). Three p63 isoforms contain N-terminal transcriptional activation (TA) sequences while the other three (ΔN) do not. p63 TA and ΔN isoforms are further subjected to alternative splicing at their C termini, resulting in α, β, and γ variants. While initial studies suggested ΔNp63 isoforms acted as dominant-negative molecules against both TAp63 isoforms and p53 (Yang et al. 1998), recent studies have shown that ΔNp63 also possesses transcriptional activity (Ghioni et al. 2002; King et al. 2003; Helton et al. 2006). Consistent with having both positive and negative gene regulatory roles, p63 can induce expression of some target proteins, such as JAG1 and PERP (Sasaki et al. 2002; Ihrie et al. 2005), while suppressing others, as in the case of p21 and 14–3–3σ (Westfall et al. 2003).
Targeted gene disruption studies in mice have established an important developmental role for p63 in limb morphogenesis and stratified tissue formation (Mills et al. 1999; Yang et al. 1999). At birth, p63-null mice display truncated limbs, craniofacial malformations, abnormalities in epidermal appendages, and an absence of normal stratified epithelia, including epidermis. Two contrasting models have been advanced to explain the absence of stratified epithelia (McKeon 2004). One model posits that p63 is required for simple epithelial cells to commit to a stratified epithelial lineage during development (Mills et al. 1999). The second model argues that the primary defect resides not in the acquisition of a stratified epithelial cell fate but rather in an inability of epidermal stem cells to sustain epidermal self-renewal (Yang et al. 1999). Previous studies have found that TAp63 isoforms are the first transcripts induced during embryogenesis (Koster et al. 2004); however, recent findings using in situ hybridization indicate that ΔNp63 isoforms are present prior to TAp63 expression and predominate during development (Laurikkala et al. 2006), making it unclear which p63 isoforms may be required for commitment to a stratified epithelial cell fate. Because overexpression of TAp63α in the epidermis of transgenic mice resulted in hyperplasia and inhibited differentiation (Koster et al. 2004), a balance of p63 TA and ΔN isoforms may be needed for proper development of stratified tissue.
p63-null mice die at birth (Mills et al. 1999; Yang et al. 1999), making it difficult to assess p63 function in mature epidermis. Here we have used RNA interference (RNAi) to knock down p63 expression in organotypic epidermal tissue, a regenerative setting in which mature epithelial cells undergo both stratification and differentiation to produce normal epidermal tissue. p63 loss in this context impairs epidermal stratification, prevents induction of epidermal differentiation genes, and reinduces expression of keratins 8 and 18, suggesting reversion to a simple epithelial phenotype. Moreover, we show that down-regulation of p63 causes a cell cycle arrest in keratinocytes that is p53-dependent. Simultaneous knockdown of p53 in the context of p63 loss is able to rescue cell proliferation and prevent tissue hypoplasia. However, the differentiation defect was still present, suggesting that p63 effects on differentiation are separable and distinct from its role in maintaining progenitor cell proliferation. p63 appears to regulate epidermal differentiation in a cell-autonomous manner. While the ΔNp63 isoforms are primarily responsible for p63 function in proliferation and differentiation, cDNA microarray analysis of tissue displaying isoform-specific p63 knockdown indicates that TAp63 isoforms may also provide a minor contribution to epidermal differentiation. These findings suggest that p63 isoforms are required not only for the initial formation of the epidermis during development but also for the ability of developmentally mature keratinocytes to regenerate a stratified epithelium.
Results
Down-regulation of p63 inhibits epidermal stratification and differentiation
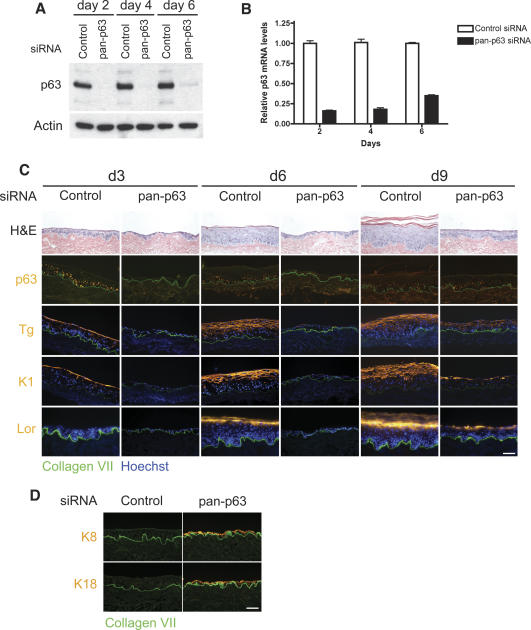
To study the functional role of p63 in a post-developmental context, we examined the ability of p63-deficient primary human keratinocytes to regenerate epidermal tissue in the presence of environmental and matrix cues sufficient to promote epidermal stratification and differentiation. Double-stranded small interfering RNA (siRNA) oligonucleotides targeting the DNA-binding domain of p63 were synthesized and used to knock down p63 expression. The DNA-binding domain is conserved in all p63 isoforms, thus resulting in a pan-isoform p63 (pan-p63) knockdown. We first determined the efficiency and duration of p63 protein and mRNA down-regulation in primary keratinocytes by immunoblot and quantitative real-time PCR, respectively. p63 protein expression was markedly decreased for 6 d following pan-p63 oligo nucleofection (Fig. 1A). mRNA levels were also substantially reduced during the same period (Fig. 1B). Both protein and mRNA levels began to recover by 6 d.
Figure 1.
p63 is required for stratification and differentiation in regenerating epidermis. (A) Western blot over a 6-d time course after introduction of siRNA oligonucleotide duplexes into primary human keratinocytes. The major band represents ΔNp63α, the predominant species in cultured keratinocytes. (Control) Control oligonucleotide; (pan-p63) oligonucleotide targeting all p63 isoforms. (B) Time course of qRT–PCR analysis of p63 mRNA levels following nucleofection with oligonucleotide duplexes targeting pan-p63 or control oligonucleotide duplexes. (C) Organotypic epidermal tissue regenerated from primary human keratinocytes after introduction of siRNA targeting all p63 isoforms (pan-p63) versus control siRNA. Tissue analyzed at days 3, 6, and 9 (d3, d6, d9, respectively) by histology and immunostaining with antibodies that recognize all p63 isoforms (p63) and the differentiation markers transglutaminase 1 (Tg), keratin 1 (K1), and loricrin (Lor), all in orange. (Green) Type VII collagen, basement membrane zone marker; (blue) Hoechst 33342 nuclear staining. Note hypoplasia and lack of differentiation marker expression in tissue lacking p63. (D) Four-day organotypic epidermal tissue generated from control or pan-p63 RNAi-treated keratinocytes was immunostained for keratins K8 and K18 (orange) and costained for type VII collagen (green); note the absence of K8/K18 in control and its appearance in p63-deficient tissue. Bar, 100 μm.
Previous studies established that p63 is required in development for proper epidermal stratification and differentiation (Mills et al. 1999; Yang et al. 1999). To determine whether loss of p63 affects the ability of mature keratinocytes to re-form a stratified epithelium, cells treated with control or pan-p63 siRNA were used to regenerate epidermal tissue in organotypic culture. In this system, cells were seeded onto intact devitalized human dermis and raised to the air/liquid interface to induce stratification and differentiation, a process that produces correctly polarized epidermis with spatially organized expression of marker proteins for differentiation (Fig. 1C). Tissue regenerated from keratinocytes treated with control siRNA began to stratify and express differentiation markers within 3 d, with formation of all epidermal layers, including the stratum corneum, evident by 6 d (Fig. 1C). Nuclear p63 protein detected in basal layer cells was down-regulated within differentiated layers, consistent with known p63 epidermal expression patterns (Yang et al. 1998). Expression of the early differentiation proteins transglutaminase 1 and keratin 1 (K1) preceded induction of the late differentiation protein loricrin, as expected in normal regenerating epidermal tissue (Fig. 1C).
In contrast, epidermis regenerated from keratinocytes with pan-p63 knockdown showed defects in both stratification and differentiation. p63-deficient epidermis was severely hypoplastic both at 3 and 6 d, and lacked proper tissue polarity (Fig. 1C). p63 expression was completely absent at 3 d but began to return by 6 d, consistent with the duration of p63 knockdown seen in cultured keratinocytes. Differentiation marker expression was undetectable in the absence of p63. However, p63 loss induced expression of keratins 8 and 18, markers of simple epithelium that are normally absent in stratified tissue (Fig. 1D). The basal keratins 5 and 14, which are expressed throughout the epithelium of regenerating tissue, was not altered in p63-deficient tissue (Supplementary Fig. 1), suggesting that p63 may not directly regulate expression of these proteins in this setting. With the return of p63 expression, epidermal stratification and differentiation were restored (Fig. 1C). These data suggest that p63 function is necessary for formation of mature stratified epithelium, and that without p63 expression, the tissue assumes features of a simple epithelial phenotype.
Differentiation defects in p63-deficient tissue are not due to hypoplasia or hypoproliferation
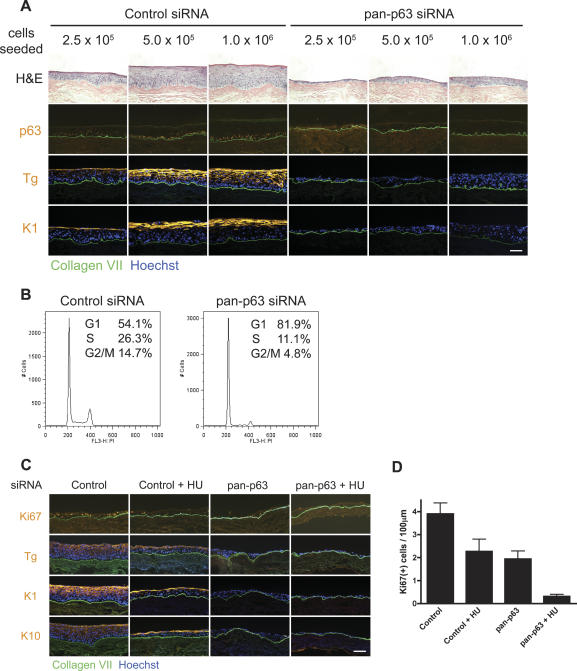
Because p63 knockdown tissue was hypoplastic, we wanted to determine whether the differentiation defects observed in p63-deficient epidermis resulted as a general consequence of tissue hypoplasia or alternatively, from the loss of a specific p63 functional program. To address this, we first seeded dermis with increasing numbers of p63 knockdown cells to approximate the cell numbers in normal tissue. Seeding 1 × 106 p63 knockdown cells resulted in epidermal thickness comparable to that obtained with 2.5 × 105 control cells. Normalization of epidermal thickness, however, failed to restore normal differentiation or tissue polarity (Fig. 2A). Furthermore, tissue with p63 knockdown still showed an up-regulation of K8 and K18 even though multiple layers of cells were present (Supplementary Fig. 2). These results confirm that the tissue effects we observed previously with p63 loss were not simply an effect of inadequate cell numbers.
Figure 2.
Effects of p63 loss on stratification and differentiation are not due to inadequate cell numbers or hypoproliferation. (A) p63 effects are independent of total cell numbers. Increasing numbers of control or p63 RNAi-treated cells were seeded onto devitalized dermis, and the tissue was harvested at 4 d. Note that even when p63 siRNA-treated cells were present in multiple layers, there was a lack in tissue polarity and organization, and differentiation marker expression was absent. Bar, 100 μm. (B) p63 knockdown results in G1 cell cycle arrest. FACS cell cycle profiles of control or pan-p63 siRNA-treated keratinocytes 60 h following transfection. Percent of cells in G1, S, or G2/M are indicated. (C) Hypoproliferation does not account for the differentiation defects observed with p63 loss. Tissue rendered hypoproliferative with 0.2 mM treatment of the DNA synthesis inhibitor HU was compared at 4 d to tissue generated from cells deficient for p63 and p53 control. Note decreased mitotic activity in HU-treated tissue but retained capacity for induction of the differentiation markers transglutaminase 1 (Tg), K1, and K10. Bar, 100 μm. (D) Quantitative mitotic index of organotypic tissue. HU indicates addition of 0.2 mM HU to the culture media.
The tissue hypoplasia observed with p63 knockdown may have resulted from either a decrease in cell viability or an inhibition of cell proliferation. A previous study has shown that p63 down-regulation leads to defects in adhesion, resulting in cell death (Carroll et al. 2006). Consistent with the previous work, we also observed a decrease in the expression of β1 and β4 integrins following knockdown of p63 protein levels (Supplementary Fig. 3A). This decrease in integrin expression correlated with reduced adhesion in culture (Supplementary Fig. 3B). However, we failed to observe an increase in apoptosis in tissue generated from pan-p63 knockdown cells (Supplementary Fig. 3B), suggesting that the level of cell adhesion in tissue is sufficient to prevent anoikis.
We next examined whether loss of p63 resulted in a cell proliferation defect. Cell cycle analysis using flow cytometry revealed that p63 knockdown resulted in a large increase in the percentage of cells in G1 and a concomitant reduction of cells in S phase and G2/M (Fig. 2B), consistent with a G1 arrest. Moreover, expression of the proliferation marker protein Ki67 was reduced by ∼50% in p63 knockdown keratinocytes in organotypic cultures compared with controls (Fig. 2C,D). Because loss of p63 has previously been shown to lead to cellular senescence in tissue (Keyes et al. 2005), we stained tissue generated from control or pan-p63 siRNA-treated keratinocytes for β-galactosidase activity, a marker of cellular senescence. However, we did not observe a significant increase in the number of senescent cells (Supplemenary Fig. 3D), suggesting that these p63 knockdown cells are growth-arrested but have not become senescent.
Because p63 knockdown keratinocytes displayed a cell proliferation defect, we determined whether hypoproliferation, as distinct from tissue hypoplasia with reduced tissue cell numbers, accounted for the effects of p63 knockdown. To examine if inhibition of cell proliferation prevented differentiation, we inhibited keratinocyte proliferation in the setting of normal p63 levels using the nucleotide synthesis inhibitor hydroxyurea (HU) and analyzed stratification and differentiation in the resultant tissue. Treatment with 0.2 mM HU lowered the mitotic index of normal tissue to approximately the levels seen with p63 knockdown (Fig. 2C,D); however, rendering normal tissue hypoproliferative did not inhibit differentiation marker induction (Fig. 2C). Even in the presence of 0.5 mM HU, when the mitotic index was below levels observed in p63-deficient tissue (Supplementary Fig. 4A), differentiation markers could still be induced (Supplementary Fig. 4B). Because a previous study had reported that HU treatment alone can promote keratinocyte differentiation (Schwartz et al. 1995), we also treated tissue regenerated from pan-p63 knockdown cells with the same concentration of HU to ensure that in our system, HU treatment itself did not affect the normal differentiation program. No differentiation marker induction was observed in the p63-deficient tissue with or without HU (Fig. 2C). Thus, although p63 loss resulted in reduced cell proliferation, defects in epidermal stratification and differentiation appear to be independent of proliferation effects.
Cell cycle arrest in p63-deficient keratinocytes is p53 dependent
To examine the mechanisms responsible for the proliferation defect in p63-deficient keratinocytes, we determined whether loss of p63 expression resulted in an up-regulation of the cell cycle inhibitors p21 or p16. p63 has previously been shown to negatively regulate p21 expression (Westfall et al. 2003). Consistent with this, we observed an increase in p21 protein levels following pan-p63 knockdown (Fig. 3A,B); levels of p16, however, did not change significantly, suggesting that p21 up-regulation may be functionally important. This was indeed the case, as down-regulation of p21 expression using siRNA delayed the onset of the proliferation defect triggered by p63 knockdown as assessed by BrdU labeling of cells in culture (Fig. 3B,C) and flow cytometric analysis (Supplementary Table 1). However, by day 4, BrdU incorporation was diminished, although p21 levels remained low (Fig. 3B,C), suggesting additional pathways may be involved in mediating this growth arrest. Since p21 is also a known p53 target gene, we speculated that up-regulation of p21 in the absence of p63 may occur through a p53-dependent mechanism. Indeed, we found that double transfection with siRNAs directed against both p63 and p53 inhibited the up-regulation of p21 expression seen with p63 knockdown alone (Fig. 3B). Furthermore, we observed a complete rescue of cell proliferation at 2 and 4 d (Fig. 3C), suggesting that the cell proliferation defect of p63 requires p53. Because we observed a slight but reproducible increase in the proliferation index with p53 down-regulation alone (Fig. 3C), it is possible that endogenous levels of p53 act to inhibit keratinocyte proliferation.
Figure 3.
Hypoproliferation induced by p63 deficiency is p53 dependent. (A) p63 knockdown increases p21 protein levels. Western blot of control or pan-p63 siRNA-transfected cells at day 2 (d2) and day 4 (d4) following siRNA transfection. (B) Simultaneous knockdown of p53 or p21 expression in combination with p63 knockdown inhibits p21 induction. Western blot of keratinocyte cell lysates; the introduced siRNA is indicated along the top, and the antibodies used for each panel are shown on the left. (C) p53 knockdown rescues the cell proliferation defect in vitro. Levels of BrdU incorporation were determined at day 2 (d2) and day 4 (d4) following transfection with the indicated siRNAs. Levels were normalized to cells receiving control siRNA, and error bars indicate the standard deviation among quadruplicate samples. (D) p53 knockdown rescues cell proliferation in organotypic epidermal tissue. Mitotic index of 4-d organotypic cultures generated from cells treated with the indicated siRNA. Cells were labeled with BrdU 12 h prior to harvesting, and tissue was stained with a BrdU antibody. (E) Rescue of cell proliferation does not rescue the differentiation defect in p63-deficient epidermal tissue. Organotypic tissues generated from the indicated siRNA-transfected cells were immunostained with antibodies against BrdU and transglutaminase 1 (Tg). Note the increase in staining with BrdU in the pan-p63/p53 double-knockdown sample but the absence of differentiation marker expression. Bar, 100 μm.
Knockdown of p53 in p63-deficient cells rescues tissue hypoplasia but not differentiation
The ability to rescue cell proliferation allowed us to determine whether cell proliferation and differentiation were separable p63-regulated processes. Loss of p53 alone did not significantly affect cell proliferation or epidermal differentiation in organotypic skin cultures (Fig. 3D,E), consistent with prior observations in p53-null mice (Donehower et al. 1992; Weinberg et al. 1995). BrdU labeling revealed that down-regulation of p63 in organotypic culture resulted in a reduction in the mitotic index to about one-third that of control tissue (Fig. 3D,E). Knockdown of p53 in combination with pan-p63 knockdown was able to completely restore the mitotic index of the tissue to that of control levels. However, although p53 knockdown could rescue the cell proliferation defect as well as the tissue hypoplasia observed with p63 knockdown (Fig. 3D,E), differentiation marker expression remained absent (Fig. 3E). These results suggest that p63 effects on proliferation and differentiation occur by separable mechanisms and that only the former is p53-dependent.
p63 regulates epidermal differentiation through cell-autonomous mechanisms
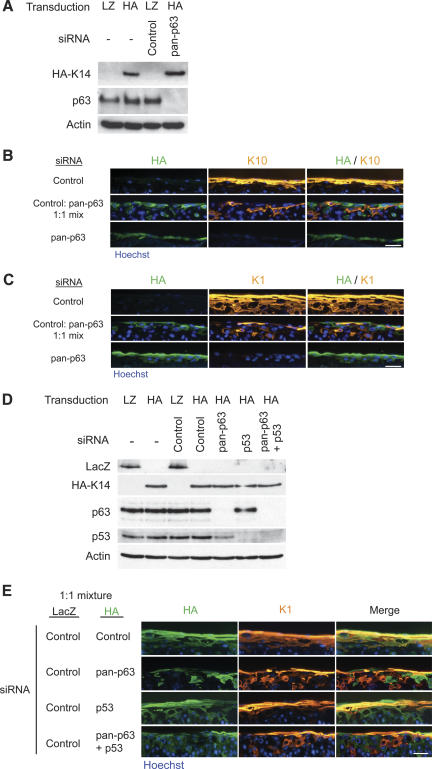
To explore how p63 functions to promote keratinocyte differentiation, we wished to determine whether the requirement for p63 in epidermal differentiation is cell autonomous as opposed to a paracrine field effect mediated by p63-dependent secreted factors. To address this question, we determined whether normal keratinocytes could rescue the differentiation defect of p63-deficient keratinocytes by establishing mosaic epidermal tissue. Retrovirally infected keratinocytes expressing LacZ were used to label cells transfected with control siRNA, while cells treated with pan-p63 siRNA were labeled with HA-epitope-tagged keratin 14 (HA-K14). K14 is normally expressed throughout regenerating epidermal tissue, and its ectopic expression has no effect on proliferation, stratification, or differentiation (Delehedde et al. 2001; Hansson et al. 2001).
Keratinocytes expressing LacZ or HA-K14 had similar p63 levels in the absence of siRNA, and p63 expression was abolished after introduction of the pan-p63 siRNA into HA-K14-expressing cells (Fig. 4A). Epidermal tissue was regenerated using a 1:1 ratio of control to pan-p63 siRNA-treated keratinocytes then harvested at 4 d and analyzed for expression of HA, K1, and K10. Interestingly, pan-p63 knockdown cells failed to occupy the basal layer and were only found among the suprabasal layers (Supplementary Fig. 5). This may reflect reduced adhesion and an inability to compete with normal cells for attachment to the basement membrane. Although pan-p63 knockdown cells were localized suprabasally, there was a complete absence of overlap between pan-p63 siRNA-treated cells labeled with HA-K14 and cells that expressed K1 or K10 (Fig. 4B,C). The inability of normal neighboring cells to rescue the differentiation defect in p63 knockdown cells indicates that p63 functions in a cell-autonomous manner to control differentiation.
Figure 4.
p63 effects on epidermal differentiation are cell autonomous. (A) Western blots of primary keratinocyte cell extracts after retroviral gene transfer and oligonucleotide nucleofection; the genes introduced are shown at the top of the panel with siRNA oligonucleotide below. Antibodies used for immunoblotting are shown at the left of each panel. LacZ (LZ)- and HA-epitope-tagged K14 were used to label cells with control and pan-p63 knockdown, respectively. (B,C) Epidermal tissue generated from LacZ-labeled control cells (top panels), HA-K14-labeled pan-p63 knockdown cells (bottom panels), or a 1:1 mixture of each (middle panels) were stained with antibodies for the differentiation markers K10 (B) and K1 (C). Tissue was harvested at 4 d. Note the complete absence of differentiation marker expression in p63 knockdown cells and the failure of immediately adjacent normal cells to rescue this. Bar, 50 μm. (D) Western blots of cell extracts following retroviral transduction and siRNA transfection. The transduced genes and siRNA oligonucleotide are indicated along the top for each sample. Antibodies detected by Western blot are indicated on the left of the panel. (E) Organotypic tissue generated from a 1:1 ratio of LacZ-expressing control cells and HA-K14-expressing cells receiving one of the following siRNAs: control, pan-p63, p53, or pan-p63 and p53. Tissue was harvested at day 4 and immunostained with antibodies recognizing HA (green) and K1 (orange). Note the presence of overlap between the HA-K14-expressing cells and K1 in the control and p53 single-knockdown mixes but the absence of overlap in tissue generated from HA-K14-expressing cells with pan-p63 knockdown alone or in combination with p53 knockdown. Bar, 50 μm.
Because p63 knockdown cells tended to enter growth arrest, it is possible that this growth arrest might render the cells unresponsive to signals from neighboring cells. We thus next examined whether simultaneous knockdown of p53 expression in the context of p63 down-regulation could rescue differentiation of p63 knockdown cells in the presence of normal keratinocytes. p53 expression was not affected by transduction with HA-K14 retrovirus, and transfection with siRNA was still efficient in the labeled cells (Fig. 4D). In mosaic tissue, HA-K14-labeled control or p53 siRNA-treated cells could be found within the basal layer, whereas no pan-p63 siRNA transfected cells were found along the basement membrane zone (Supplementary Fig. 5). This was the case even with simultaneous knockdown of p53, suggesting p53 knockdown does not rescue the adhesion defect observed in the absence of p63. Moreover, control or p53 siRNA-treated cells were able to express the differentiation protein K1, confirming that p53 knockdown did not inhibit differentiation marker expression (Fig. 4E). However, HA-K14-labeled cells receiving both pan-p63 and p53 siRNA still failed to express K1 even in the presence of normal control siRNA-treated cells, indicating that p63 effects on differentiation in this setting are p53-independent.
ΔNp63 isoforms are the main mediators of p63 function in keratinocyte proliferation and differentiation
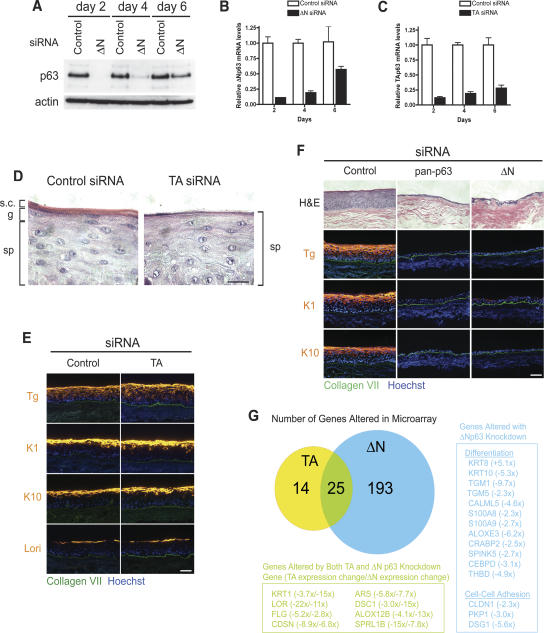
It has been proposed that, while TAp63 isoforms are responsible for initial steps toward stratification during development, their activity must be suppressed by ΔNp63 isoforms for proper differentiation to occur (Koster et al. 2004). Epidermal regeneration in organotypic culture offers an approach to determine the relative contribution of each set of p63 isoforms to epidermal stratification and differentiation in a post-developmental context. We thus designed siRNA oligonucleotides that specifically targeted each subset of p63 isoforms. TA-isospecific siRNA was designed against the transactivation domain of the protein while ΔN-isospecific siRNA targeted the unique 5′-untranslated region (UTR) of the ΔNp63 mRNA (Supplementary Fig. 6A). We first determined the efficiency and specificity of each isospecific knockdown. ΔNp63 protein was undetectable at 2 d after transfection (Fig. 5A), corresponding to a marked reduction in mRNA levels as well (Fig. 5B). Both protein and transcript levels began to rise by day 4, returning to levels approximately half that of controls by day 6 (Fig. 5A,B). TAp63 isoforms are expressed at very low levels in cultured primary keratinocytes and are not readily detectable by immunoblot; thus, we examined knockdown of transcript levels by qRT–PCR. TA p63 mRNA levels followed a similar pattern to ΔNp63 (Fig. 5C). We also confirmed that each siRNA duplex only targeted the subset of isoforms against which they were designed. As expected, the pan-p63 siRNA down-regulated expression of both the p63 TA and ΔN isoforms, while isoform-specific siRNA targeted only their respective isoforms (Supplementary Fig. 6B). Down-regulation of ΔNp63 consistently caused a slight increase in TAp63 transcript levels. Knockdown of ΔNp63, but not TAp63, isoforms reduced total p63 protein levels, confirming that ΔNp63 is the predominantly expressed isoform in cultured keratinocytes (Fig. 5A).
Figure 5.
ΔNp63 isoforms are required for epidermal stratification and differentiation. (A) Western blot after introduction of control oligonucleotide or oligonucleotide targeting ΔNp63 isoforms. (B,C) Time course of qRT–PCR analysis for cells treated with oligonucleotide targeting ΔNp63 isoforms (B) or TAp63 isoforms (C). (D) High-powered magnification (100×) of the tissue histology of 4-d organotypic cultures generated with control or TAp63 siRNA-transfected keratinocytes. The spinous layer (sp), granular layer (g), and stratum corneum (s.c.) are indicated. Note the absence of granules or stratum corneum from tissue generated from TAp63 knockdown cells. Bar, 20 μm. (E) Differentiation marker expression of 4-d organotypic epidermal tissue with control or TAp63 knockdown. Bar, 100 μm. (F) Control, pan, or ΔNp63 iso-specific RNAi-transfected keratinocytes were used to regenerate organotypic skin tissues. Tissue was harvested at 4 d and analyzed by histology and immunostaining for expression of the indicated differentiation proteins. Note the clear failure of stratification and differentiation with ΔNp63 isoform knockdown. Bar, 100 μm. (G) Venn diagram displaying the number of gene expression changes that accompanied TA or ΔNp63 knockdown in organotypic culture. Numbers in the Venn diagram represent genes that displayed a twofold or greater change with a p-value ≤0.05 by one-way ANOVA. Genes that met these criteria and are involved in differentiation or cell–cell adhesion are listed in the boxed regions. Numbers next to the gene name indicates the average fold change in expression, with (+) representing up-regulation and (−) representing down-regulation. For genes that changed in both TA and ΔN p63 knockdown tissue, the first number corresponds to TA knockdown samples, and the second number corresponds to ΔN knockdown samples.
To determine the functional roles of p63 isoforms in epidermal stratification and differentiation, we regenerated epidermal tissue with keratinocytes treated with each isospecific siRNA. Knockdown of TAp63 isoforms produced no detectable effects on proliferation (Supplementary Fig. 7) and only subtle abnormalities in differentiation, manifested consistently by incomplete development of the granular layer and stratum corneum (Fig. 5D,E). However, knockdown of ΔNp63 isoforms resulted in the same dramatic defects as pan-p63 siRNA with tissue hypoplasia and absent differentiation marker expression (Fig. 5F). Consistent with the tissue hypoplasia, ΔNp63 knockdown reduced the level of BrdU incorporation in vitro to levels observed with pan-p63 knockdown (Supplementary Fig. 7). These findings suggest that ΔNp63 isoforms are required for maintaining both epidermal proliferation and differentiation and that TAp63 isoforms may contribute to later aspects of differentiation.
To further characterize p63 isoform-specific functions in epidermal differentiation, we examined changes in gene expression following down-regulation of either p63 TA or ΔN isoforms. mRNA expression profiling was performed on epidermal tissue regenerated from primary keratinocytes that had been treated with control, TAp63, or ΔNp63 siRNA oligonucleotides. The 3-d time point was chosen for analysis because tissue polarization and differentiation processes are initiated by this time in normal cells, and p63 protein expression is still efficiently knocked down in the siRNA-treated cells. Down-regulation of ΔNp63 isoforms resulted in the altered expression of 218 genes showing a twofold or greater change (p ≤ 0.05) relative to control siRNA tissue, while TAp63 knockdown only affected the expression of 39 genes (Supplementary Table 2). The difference in the number of gene expression changes may reflect the number of target genes regulated by each subset of isoforms or it may be a consequence of the greater abundance of p63 ΔN compared with TA isoforms. Strikingly, ΔNp63 knockdown produced a global disruption of the epidermal stratification and differentiation program, as indicated by the number of changes in genes involved in desmosome assembly, formation of the stratum corneum, and generation of the skin barrier (Fig. 5G). TAp63 loss resulted in a more modest number of gene changes; however, 25 of these genes were altered in response to both p63 TA and ΔN knockdown. In all cases, genes induced by TAp63 loss were also induced by ΔNp63 loss, while genes repressed by TAp63 loss were similarly repressed by ΔNp63 loss. Surprisingly, many of these genes are also involved in differentiation or epidermal homeostasis. In particular, many of these genes represented late differentiation markers and are associated with formation of the stratum corneum, consistent with our finding of defective granular and cornified layer formation with TAp63 knockdown (Fig. 5D). Thus, the microarray data suggest that p63 TA and ΔN isoforms may not have opposing actions in regulating differentiation in this setting but may instead cooperate to carry out the epidermal program of stratification and differentiation, with ΔNp63 clearly predominant in the regeneration setting. However, we did not find any genes involved in differentiation that were regulated uniquely by TAp63 isoforms (Supplementary Table 2), suggesting that ΔNp63 isoforms may be sufficient for the epidermal differentiation program. Importantly, only ΔNp63 down-regulation resulted in induction of the simple epithelial marker keratin 8 (Fig. 5G; Supplementary Table 2), suggesting that these isoforms may be responsible for maintaining the stratified epithelial cell fate.
Discussion
Our findings indicate that sustained expression of p63 is required to maintain the ability of normal postnatal keratinocytes to form a differentiated stratified epithelium. p63 functions to maintain both the proliferative capacity and differentiation potential of keratinocytes in regenerating epidermis. While the inhibition of cell proliferation resulting from p63 loss is p53-dependent, the differentiation defects appear to be independent of p53. In regulating differentiation, p63 functions in a cell-autonomous manner because normal cells do not restore the differentiation program in immediately adjacent p63-deficient cells. Moreover, the absence of ΔNp63 isoforms results in substantial gene expression changes during epithelial regeneration. Consistent with the dramatic effects seen with p63 knockdown, many of the genes altered encode proteins involved in differentiation. These include a variety of structural proteins, such as keratins 1 and 10, loricrin, corneodesmin, desmoglein 1, and desmocollin 1, as well as enzymes, such as transglutaminase 1 and lipoxygenase-3. The failure of these cells to initiate the epidermal differentiation program in the absence of p63 suggests a central role for p63 in sustaining epidermal tissue fate postnatally.
Our work suggests that p63 is required to maintain proliferation of epidermal cells. One model of p63 function, the “stem cell self-renewal” model, has suggested that p63 is required to maintain the proliferative potential of epidermal stem cells (Yang et al. 1999; McKeon 2004). The absence of appropriate markers that distinguish interfollicular epidermal stem cells from committed transit-amplifying cells makes it difficult to determine which population of cells is being directly affected by knockdown of p63 in regenerating human epidermis. However, the dramatic hypoplasia that results from p63 knockdown suggests that loss of proliferative ability, even among the transit-amplifying cells, may severely affect stratification. The inhibition of proliferation observed in the absence of p63 appears to be a p53-dependent process. Initial studies of p63 protein function, using reporter assays with overexpression of a single p63 isoform, suggested a dominant-negative role for p63 toward p53 transcriptional activity (Yang et al. 1998). These findings are in accordance with a later study in zebrafish, suggesting that ΔNp63 isoforms are required for epithelial cell proliferation during development by inhibiting p53 function (Lee and Kimelman 2002). Thus, although p53 levels appear slightly reduced in the presence of p63 knockdown (Fig. 3B), its functional activity may actually increase. The slight increase in the mitotic index following p53 knockdown alone (Fig. 3C,D) suggests that endogenous levels of p63 may not be sufficient to inhibit all p53 function. While it is possible that p53 and p63 act through different pathways to affect cell proliferation, the ability of both proteins to recognize and bind to similar DNA sequences (Osada et al. 2005) and to regulate the same target genes (Westfall et al. 2003; Ihrie et al. 2005) suggest that these two family members function, at least in part, through common mechanisms. It is possible that regulation among the p53 family members reflects competition for binding sites to target promoters. However, p53 function is not required to achieve cell cycle arrest in many settings. This is apparent both from previous work with p53 knockout mice (Donehower et al. 1992) as well as data presented here with p53 siRNA. Whereas p53 function is not necessary for keratinocyte growth regulation and differentiation under normal homeostatic conditions, in our system, it is a key mediator of the effects of p63 loss. Interestingly, a recent study using an inducible knockout model to conditionally ablate p63 expression in stratified tissues found that loss of p63 in adult mice was associated with increased senescence and enhanced aging (Keyes et al. 2005). While we did not observe an increase in senescence in tissue generated with p63 knockdown keratinocytes, it is possible that transient loss of p63 only results in an inhibition of cell proliferation, but prolonged loss of p63 function may lead to permanent senescence. The prominent link between p53 function and cellular senescence (Bond et al. 1996) raises the possibility that the increased senescence in the absence of p63 expression may result from enhanced p53 activity as well.
Previous work with p63 down-regulation in mammary epithelial cells has suggested that p63 regulates a general program of cellular adhesion (Carroll et al. 2006). Indeed, we have also found a decrease in expression of basal integrins following p63 knockdown. Moreover, when placed into a competitive environment with normal keratinocytes, cells exhibiting decreased levels of p63 are excluded from the basal layer and found only within the suprabasal layers, suggesting a decreased ability to adhere to the underlying dermis. Attachment to the basement membrane and high integrin expression have been correlated with epidermal progenitor cells (Jones and Watt 1993). It is possible that decreased integrin expression and defects in adhesion may contribute to the proliferation defect observed with p63 knockdown. However, since down-regulation of p53 is able to completely rescue the proliferation defect, this suggests that these signals must impinge upstream of p53. Additionally, down-regulation of integrin expression and detachment from the basement membrane have been associated with initiating the differentiation program. However, keratinocytes with p63 knockdown are still unable to engage this program even when detached, suggesting an active role for p63 in initiating terminal differentiation. Thus, in addition to maintaining cell proliferation, p63 is also necessary to maintain the capacity of developmentally mature keratinocytes to stratify and differentiate.
The second proposed model of p63 function has suggested that TAp63 isoforms are required for commitment to a stratified cell lineage during development, the “commitment” model (Koster et al. 2004). This model further suggests that ΔNp63 function is required to oppose TAp63 isoforms to achieve proper differentiation (Koster et al. 2004). While our results do not address which p63 isoform is required for the commitment decision in development, down-regulation of ΔNp63 isoforms alone is sufficient to abolish stratification and up-regulate simple keratin expression in postnatal cells, suggesting these isoforms may be important in maintaining stratified tissue identity. However, it is unclear whether p63 directly regulates expression of simple keratins. Using the P-Match software program to locate transcription factor-binding sites, we failed to find any consensus sites for p53/p63 binding within a 10-kb region upstream of the transcriptional start site for either K8 or K18. However, because p63 often binds to degenerate p53 consensus sequences, it is possible that a site does exist but was not identified by this approach. Absence of p63 in development has been shown to prevent expression of the stratified epithelial markers K5 and K14 (Mills et al. 1999). However, we did not observe a decrease in expression of either basal keratin in regenerated tissue with p63 knockdown. These results are consistent with recent findings that overexpression of the transcription factor Krüppel-like factor 5 (Klf5) in the basal layer of the epidermis led to a reduction in p63 protein levels and simultaneous up-regulation of K8 expression but no effect on K5 expression (Sur et al. 2006). Thus, it is possible that regulation of the basal keratins differs in a developmental context versus that of committed, postnatal cells, where the former process requires p63, but the latter does not.
Finally, we have performed a global assessment of gene expression changes that occur with down-regulation of either p63 TA or ΔN isoforms. While previous microarray studies have been performed with p63, these have relied on over expression of a single p63 isoform or down-regulation of p63 expression with cells in culture (Wu et al. 2003; Carroll et al. 2006). A recent study in breast epithelial cells found significant changes in the expression of adhesion molecules following p63 knockdown (Carroll et al. 2006). Although p63 is required for the development of both skin and breast tissue, we found few overlapping genes in common between the two data sets. This may reflect the use of different cell types, breast epithelium versus keratinocytes, as well as assay systems, two-dimensional culture versus three-dimensional tissue. Consistent with this, our microarray data set included many genes involved in differentiation, such as K1, loricrin, and filaggrin, which were absent from the results by Carroll et al. (2006). Furthermore, our results support a recent study suggesting a cooperative effect between p63 TA and ΔN isoforms in the formation of the epidermis (Candi et al. 2006). In developmentally mature cells undergoing tissue regeneration, p63 TA or ΔN loss had similar effects on the expression of a number of differentiation genes. Thus, our findings assessing p63 function in epidermal regeneration suggest that p63 is needed both to maintain cell proliferation in the basal progenitor cells as well as to initiate the differentiation program by inducing a gene set critical for this process. Determining the direct downstream effectors of p63 that mediate the epidermal differentiation program is of great interest for future studies.
Materials and methods
Cells, gene transfer, and organotypic culture
Normal human keratinocytes isolated from discarded surgical specimens were cultured in keratinocyte serum-free medium, KSF-M (GIBCO-BRL), supplemented with epidermal growth factor (EGF) and bovine pituitary extract (BPE). Cells were grown at 37°C in a humidified chamber with 5% CO2. All siRNA oligonucleotide duplexes were designed and synthesized by Dharmacon. The control siRNA targets a region in the 3′ intron of the p63 transcript but does not affect the expression of any subset of p63 isoforms. The regions targeted by the other p63 siRNAs are noted in Supplementary Figure 3. One nanamole of siRNA oligonucleotide was electroporated into 1 × 106 primary human keratinocytes using Amaxa nucleofection reagents according to the manufacturer’s protocol. For organotypic skin cultures, cells were first nucleofected with the indicated siRNA oligonucleotide. Twelve hours to 24 h post-nucleofection, cells were trypsinized and counted. Cells (2.5 × 105 to 10 × 105) were seeded onto devitalized dermis and raised to the air/liquid interface to induce keratinocyte stratification and differentiation (Prunieras et al. 1983). Tissue was harvested at the indicated time points. Gene transfer was performed by retroviral transduction of the pLZRS vector (Kinsella and Nolan 1996) encoding LacZ- or HA-tagged K14 cDNA. Infection was performed for 1 h at 32°C with low-speed centrifugation to enhance viral infection. Cells were washed once with PBS following centrifugation and placed in KSF media. Thirty-six hours after gene transfer, cells were nucleofected with the indicated siRNA oligonucleotide and used to generate organotypic cultures.
Protein expression and tissue analysis
For immunoblot analysis, 20 μg of control or p63 RNAi-treated cell lysates were loaded per lane and subjected to 8% SDS-PAGE. Primary and secondary antibody incubations were for 1 h each. Primary antibodies used included mouse antibodies against p63 (Santa Cruz Biotechnology), p53 (Santa Cruz Biotechnology), β-actin (Sigma), and HA (Covance), and rabbit antibodies against p21 (Abcam) and p16 (Santa Cruz Biotechnology). Secondary antibodies included sheep anti-mouse or anti-rabbit IgG-HRP (Amersham Biosciences). For immunofluorescence studies, 7-μm skin cryosections were fixed in either ice-cold methanol or acetone for 15 min at −20°C, followed by blocking in 10% horse serum in PBS for 1 h. Sections were incubated with primary antibody and then detected with Alexa Fluor secondary antibodies (Molecular Probes). Counterstaining with Hoechst dye 33342 was used to visualize cell nuclei. Antibodies used included mouse antibodies against p63 (Santa Cruz Biotechnology), BrdU (BD Biosciences), transglutaminase (Biomedical Technologies), collagen VII (Chemicon), K8 (Abcam), K18 (Abcam), and HA (Covance), and rabbit antibodies against keratin 1 (Covance), loricrin (Covance), Ki67 (Neomarkers), collagen VII (Calbiochem), and HA (Abcam). For histological analysis, skin tissue was fixed in 10% formalin (Sigma-Aldrich) and embedded in paraffin.
mRNA expression analysis
For qRT–PCR, total RNA was extracted from control or siRNA-treated cells using the TRIZOL reagent (Invitrogen). qRT–PCR analysis was performed using the Mx3000P instrument with the Brilliant SYBR Green QRT–PCR Master Mix Kit (Stratagene). Samples were run in triplicate and normalized to levels of GAPDH mRNA for each reaction. At least three separate runs were performed for each sample. One-hundred nanograms of RNA were used for analyses with 100 nM pan-p63, ΔNp63, and GAPDH primers, and 150 ng of RNA were used for analysis with 100 nM TAp63 primers. The primers used were as follows: GAPDH fwd, 5′-CCGGGAAACTGTGGCGTGATGG-3′; GAPDH rev, 5′-AGGTGGAGGAGTGGGTGTCGCTGTT-3′; pan-p63 fwd, 5′-GACAGGAAGGCGGATGAAGATAG-3′; pan-p63 rev, 5′-TGTTTCTGAAGTAAGTGCTGGTGC-3′; ΔNp63 fwd, 5′-GAGTTCTGTTATCTTCTTAG-3′; ΔNp63 rev, 5′-TGTTCT GCGCGTGGTCTG-3′; TAp63 fwd, 5′-TGGTGCGACAAACA AGATTG-3′; and TAp63 rev, 5′-ATAGGGACTGGTGGACGA GG-3′. Microarray analysis was performed on duplicate samples of organotypic cultures generated from keratinocytes treated with control, TAp63, or ΔNp63 siRNA oligonucleotide. Tissue was flash-frozen in liquid N2 then homogenized in TRIZOL reagent, and RNA was extracted using the RNeasy Lipid Tissue Kit (Qiagen). Amplification and labeling of cDNA probes and hybridization to the HG-U133A2.0 microarray chip (Affymetrix) was performed by the Stanford PAN Facility. Data analysis was performed using the GeneSpring software (Agilent). Pairwise comparisons between the TAp63 or ΔNp63 siRNA-treated samples and the control samples were performed to find genes that showed twofold or greater expression change. One-way ANOVA was performed with all three sample sets to determine genes that changed significantly with a p-value ≤0.05. The intersection of these two data sets determined genes that are included in Supplementary Table 2.
Cell cycle analysis and cell proliferation assays
Cell cycle profiles were determined by analyzing total DNA content using flow cytometry. Briefly, transfected cells were harvested at the indicated time points, washed once with PBS, and fixed and permeabilized in ice-cold 70% ethanol. Prior to FACS analysis, fixed cells were washed once with PBS then stained with PI solution (50 μg/mL PI, 3.8 mM NaCitrate, 150 μg/mL RNase A in PBS) for 30 min at room temperature. FACS was performed on a FACSCalibur (Becton Dickinson), and data were analyzed with the FlowJo software using the Watson model for cell cycle analysis. Analysis of in vitro BrdU incorporation was performed with the Cell Proliferation ELISA kit (Roche) according to the manufacturer’s protocol. Five thousand cells per well were seeded into a 96-well plate in triplicate or quadruplicate samples, and the assay was performed at the time points indicated following overnight incubation with BrdU. To determine mitotic index in organotypic cultures, BrdU was added to the media to a final concentration of 50 μM for 12 h prior to harvesting the tissue. BrdU epitopes were exposed by denaturing the DNA with 2N HCl, and labeled cells were detected by immunostaining frozen cryosections with an antibody against BrdU (BD Biosciences).
Acknowledgments
We thank T. Oro, L. Attardi, H. Chang, F. Scholl, J. Reuter, A. Adams, and Z. Siprashvili for critical reading of the manuscript and helpful discussions. We also thank J. Jensen for help with the adhesion assay and G. Sen for assistance with flow cytometry. A.B.T. is a Howard Hughes Medical Institute predoctoral fellow. This work was supported by the US Veterans Affairs Office of Research and Development and by NIH/NIAMS grant AR45192 to P.A.K.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1463206
References
- Bond, J., Haughton, M., Blaydes, J., Gire, V., Wynford-Thomas, D., Wyllie, F. Evidence that transcriptional activation by p53 plays a direct role in the induction of cellular senescence. Oncogene. 1996;13:2097–2104. [PubMed] [Google Scholar]
- Candi, E., Rufini, A., Terrinoni, A., Dinsdale, D., Ranalli, M., Paradisi, A., De Laurenzi, V., Spagnoli, L.G., Catani, M.V., Ramadan, S., et al. Differential roles of p63 isoforms in epidermal development: Selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- Carroll, D.K., Carroll, J.S., Leong, C.O., Cheng, F., Brown, M., Mills, A.A., Brugge, J.S., Ellisen, L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- Delehedde, M., Cho, S.H., Hamm, R., Brisbay, S., Ananthaswamy, H.N., Kripke, M., McDonnell, T.J. Impact of Bcl-2 and Ha-ras on keratinocytes in organotypic culture. J. Invest. Dermatol. 2001;116:366–373. doi: 10.1046/j.1523-1747.2001.01260.x. [DOI] [PubMed] [Google Scholar]
- Donehower, L.A., Harvey, M., Slagle, B.L., McArthur, M.J., Montgomery, C.A., Jr., Butel, J.S., Bradley, A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Ghioni, P., Bolognese, F., Duijf, P.H., Van Bokhoven, H., Mantovani, R., Guerrini, L. Complex transcriptional effects of p63 isoforms: Identification of novel activation and repression domains. Mol. Cell. Biol. 2002;22:8659–8668. doi: 10.1128/MCB.22.24.8659-8668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson, A., Bloor, B.K., Haig, Y., Morgan, P.R., Ekstrand, J., Grafstrom, R.C. Expression of keratins in normal, immortalized and malignant oral epithelia in organotypic culture. Oral Oncol. 2001;37:419–430. doi: 10.1016/s1368-8375(00)00089-0. [DOI] [PubMed] [Google Scholar]
- Helton, E.S., Zhu, J., Chen, X. The unique NH2-terminally deleted (ΔN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the ΔN variant of p63. J. Biol. Chem. 2006;281:2533–2542. doi: 10.1074/jbc.M507964200. [DOI] [PubMed] [Google Scholar]
- Ihrie, R.A., Marques, M.R., Nguyen, B.T., Horner, J.S., Papazoglu, C., Bronson, R.T., Mills, A.A., Attardi, L.D. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Jones, P.H., Watt, F.M. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- Keyes, W.M., Wu, Y., Vogel, H., Guo, X., Lowe, S.W., Mills, A.A. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes & Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.E., Ponnamperuma, R.M., Yamashita, T., Tokino, T., Lee, L.A., Young, M.F., Weinberg, W.C. ΔNp63α functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- Kinsella, T.M., Nolan, G.P. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- Koster, M.I., Kim, S., Mills, A.A., DeMayo, F.J., Roop, D.R. p63 is the molecular switch for initiation of an epithelial stratification program. Genes & Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala, J., Mikkola, M.L., James, M., Tummers, M., Mills, A.A., Thesleff, I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- Lee, H., Kimelman, D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- McKeon, F. p63 and the epithelial stem cell: More than status quo? Genes & Dev. 2004;18:465–469. doi: 10.1101/gad.1190504. [DOI] [PubMed] [Google Scholar]
- Mills, A.A., Zheng, B., Wang, X.J., Vogel, H., Roop, D.R., Bradley, A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Osada, M., Park, H.L., Nagakawa, Y., Yamashita, K., Fomenkov, A., Kim, M.S., Wu, G., Nomoto, S., Trink, B., Sidransky, D. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol. Cell. Biol. 2005;25:6077–6089. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunieras, M., Regnier, M., Woodley, D. Methods for cultivation of keratinocytes with an air–liquid interface. J. Invest. Dermatol. 1983;81 (Suppl.):28s–33s. doi: 10.1111/1523-1747.ep12540324. [DOI] [PubMed] [Google Scholar]
- Sasaki, Y., Ishida, S., Morimoto, I., Yamashita, T., Kojima, T., Kihara, C., Tanaka, T., Imai, K., Nakamura, Y., Tokino, T. The p53 family member genes are involved in the Notch signal pathway. J. Biol. Chem. 2002;277:719–724. doi: 10.1074/jbc.M108080200. [DOI] [PubMed] [Google Scholar]
- Schwartz, P.M., Barnett, S.K., Milstone, L.M. Keratinocytes differentiate in response to inhibitors of deoxyribonucleotide synthesis. J. Dermatol. Sci. 1995;9:129–135. doi: 10.1016/0923-1811(94)00370-t. [DOI] [PubMed] [Google Scholar]
- Sur, I., Rozell, B., Jaks, V., Bergstrom, A., Toftgard, R. Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J. Cell Sci. 2006;119:3593–3601. doi: 10.1242/jcs.03070. [DOI] [PubMed] [Google Scholar]
- Turksen, K., Troy, T.C. Epidermal cell lineage. Biochem. Cell Biol. 1998;76:889–898. doi: 10.1139/bcb-76-6-889. [DOI] [PubMed] [Google Scholar]
- Weinberg, W.C., Azzoli, C.G., Chapman, K., Levine, A.J., Yuspa, S.H. p53-mediated transcriptional activity increases in differentiating epidermal keratinocytes in association with decreased p53 protein. Oncogene. 1995;10:2271–2279. [PubMed] [Google Scholar]
- Westfall, M.D., Mays, D.J., Sniezek, J.C., Pietenpol, J.A. The ΔNp63α phosphoprotein binds the p21 and 14–3–3σ promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell. Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Nomoto, S., Hoque, M.O., Dracheva, T., Osada, M., Lee, C.C., Dong, S.M., Guo, Z., Benoit, N., Cohen, Y., et al. ΔNp63α and TAp63α regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003;63:2351–2357. [PubMed] [Google Scholar]
- Yang, A., Kaghad, M., Wang, Y., Gillett, E., Fleming, M.D., Dotsch, V., Andrews, N.C., Caput, D., McKeon, F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang, A., Schweitzer, R., Sun, D., Kaghad, M., Walker, N., Bronson, R.T., Tabin, C., Sharpe, A., Caput, D., Crum, C., et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]