Abstract
For paramyxoviruses, two viral glycoproteins are key to the entry process: an attachment protein (HN, H, or G) and the fusion protein (F). The F protein folds to a metastable state that can be triggered to undergo large conformational rearrangements to a fusogenic intermediate and a more stable postfusion state. The triggering mechanism that controls paramyxovirus fusion has not been elucidated. To correlate the molecular structure of a soluble form of the prefusion F (PIV5 F-GCNt) with the biological function of F, soluble F protein was triggered to refold. In the absence of HN, heat was found to function as a surrogate F trigger, and F associated with liposomes and aggregated on sucrose density gradients. Electron microscopy data showed that triggered F formed rosettes. Taken together these data suggest that release and membrane insertion of the hydrophobic fusion peptide require both cleavage of F and heat. Heating of cleaved F causes conversion to a postfusion form as judged by its “golf tee” morphology in the electron microscope. Heating of uncleaved F also causes conversion of F to a morphologically similar form. The reactivity of the F protein with conformation-specific mAbs and peptide binding suggest that soluble F-GCNt and membrane-bound F proteins refold through a comparable pathway.
Keywords: conformational change, fusion peptide, metastable, trigger
To enter a host cell, paramyxoviruses, like other enveloped viruses such as influenza virus and HIV, require fusion of the viral membrane with a cellular membrane. For paramyxoviruses, two viral glycoproteins are critical for this process: an attachment protein (HN, H, or G) and a more conserved fusion protein (F) (1). The attachment proteins interact with different cellular receptors. For example, parainfluenza virus 5 (PIV5, formerly known as SV5) HN binds sialic acid, measles H interacts with CD46 or CDw150/SLAM (2, 3), Hendra and Nipah virus G binds to Ephrin B2 (4, 5), and respiratory syncytial virus (RSV) G binds heparin sulfate (6). Although the cellular receptors differ, in most paramyxoviruses the homotypic attachment protein is required to trigger F-mediated membrane fusion at the right place and right time (7, 8). Paramyxovirus fusion does not require the acidic pH found in the lumen of endosomes for fusion to occur (1).
F proteins are thought to drive membrane fusion by coupling irreversible protein refolding to membrane juxtaposition by initially folding into a metastable form that subsequently undergoes discrete/stepwise conformational changes to a lower energy state (7, 8). F proteins assemble into homotrimers that are proteolytically cleaved, priming the protein for membrane fusion, and similar steps occur for other class I viral fusion proteins, such as influenza virus HA, HIV gp160, retrovirus Env, Ebola GP, and severe acute respiratory syndrome coronavirus S (9–11). The uncleaved precursor (F0) is processed into a larger C-terminal fragment (F1) and smaller N-terminal fragment (F2). F1 contains a hydrophobic fusion peptide at its N terminus and two hydrophobic, heptad repeat regions (HRA and HRB). HRA is immediately adjacent to the fusion peptide, and HRB is proximal to the transmembrane (TM) domain, with ≈250 residues separating the two heptad repeats (1).
After activation by the attachment protein, by analogy to data obtained for HA (12, 13), F inserts its fusion peptide into target membranes and forms transient intermediates that can be inhibited by HRA- and HRB-derived peptides (14). Subsequent refolding and assembly of HRA and HRB into a six-helix bundle (6HB) occurs, placing the fusion peptides and the TM domains in proximity in the same membrane (11, 15). The formation of the 6HB and the associated free energy change are tightly linked to the merger of the viral and cellular membranes (14, 16, 17). The isolated F 6HB structure, generated from HRA and HRB peptides (15), is stable up to 100°C and is thought to represent the lowest-energy conformation of the protein after membrane fusion has occurred.
Recently, we reported the atomic structure of the paramyxovirus F protein in both its uncleaved prefusion conformation and its uncleaved postfusion conformation (18, 19). The prefusion structure contains a globular head attached to a trimeric coiled-coil stalk formed by the C-terminal HRB region. The globular head contains three domains (DI-DIII). The fusion peptides at the N terminus of HRA are sequestered between adjacent subunits, with cleavage sites exposed at the protein surface (19). The F structures reveal profound conformational differences between the prefusion and postfusion states, involving transformations in secondary and tertiary structure. The postfusion form of F contains the 6HB and DI and DII repositioned as rigid modules, whereas major refolding occurs in DIII. Unanticipated findings relating to the structure of the postfusion form of F were that F protein cleavage is not required to attain a postfusion conformation and that the F TM domain and/or the cytoplasmic tail are important for the folding to, or stability of, the prefusion metastable state (18). Although in the postfusion form no electron density was observed for 47 residues that included the cleavage site, the fusion peptide and residues that form part of HRA, these residues were present in the protein and structural constraints indicate they would be draped flexibly on the exterior of the structure and would contain little secondary structure (18).
The triggering mechanism that controls paramyxovirus fusion has not been elucidated. Unlike most paramyxoviruses, PIV5 F can mediate the fusion of transfected cells in the absence of HN; however, this fusion is enhanced by the coexpression of HN (14) and mutations within F can impart HN-dependence to the fusion process (20, 21). To generate a soluble form of the PIV5 F protein in its prefusion conformation, the TM domain of PIV5 F was substituted with a trimerization domain (F-GCNt) (19). To correlate the biological function of F with its molecular structure, the soluble version of prefusion F was triggered to refold to its postfusion form.
Results
The atomic structure of prefusion F was determined for the PIV5 F protein, and the atomic structure of uncleaved postfusion F was determined for the human parainfluenza virus 3 (hPIV3) F protein. Because of sequence homology and conserved alignment of cysteine residues we hypothesize that the structures are representative for all paramyxovirus F proteins, at least in the large part. This is borne out in that the structure of a proteolyzed fragment of the F protein of Newcastle disease virus (22) is very similar to the hPIV3 uncleaved postfusion structure (18).
Liposome Association Is Triggered by Heat.
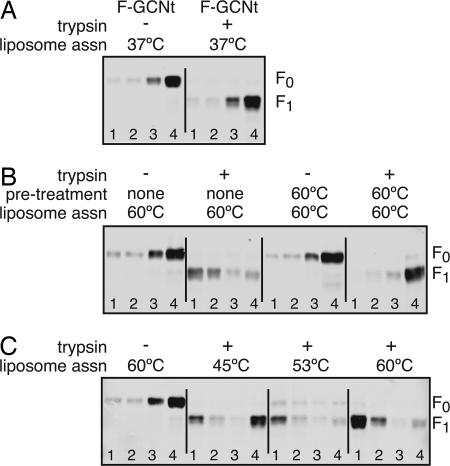
To examine an F protein transition from prefusion to postfusion, we used a soluble form of prefusion PIV5 F (F-GCNt) protein. To determine whether cleavage of purified F-GCNt is sufficient to trigger the exposure and release of the fusion peptide, F-GCNt was cleaved with trypsin and added to liposomes for 30 min at 37°C (≈100% cleavage efficiency) (see Fig. 1A). The samples were underlaid on sucrose gradients and subjected to ultracentrifugation, and fractions were collected and analyzed by immunoblotting. The opacity of the top fraction indicated that the liposomes floated to this fraction. Neither uncleaved nor cleaved F-GCNt was found in the top fraction (Fig. 1A), indicating that trypsin cleavage alone is insufficient to trigger liposome association.
Fig. 1.
Liposome association requires cleavage and elevated temperature. (A) Liposomes were added to purified uncleaved F-GCNt or trypsin-cleaved F-GCNt at 37°C for 30 min. The samples were loaded under sucrose gradients and subjected to ultracentrifugation. Fractions were collected from the top of the gradients, and total protein was precipitated and analyzed by SDS/PAGE and immunoblot. (B) Samples were treated as above, except that they were heated to 60°C after the addition of liposomes. In addition, some samples were heated to 60°C for 30 min before adding the liposomes. (C) Samples were treated as above, except that the samples were heated to 45°C, 53°C, or 60°C after the addition of liposomes.
PIV5 virus–cell fusion and PIV5 F-mediated cell–cell fusion are enhanced at elevated temperatures (21, 23). To determine whether heat would serve as a surrogate trigger of fusion peptide exposure and release, uncleaved or cleaved F-GCNt was added to liposomes and the mixture was heated to 60°C and subjected to sucrose gradient analysis. Cleaved F-GCNt was found in the top fraction of the sucrose gradient whereas uncleaved F-GCNt stayed at the bottom in the loading zone (Fig. 1B). Heat pretreatment of cleaved F-GCNt before liposome addition prevented association of F-GCNt with liposomes. The data suggest that heat triggers cleaved F-GCNt to release the fusion peptide and membrane insertion occurs when liposomes are present during heating.
To determine the temperature requirements for liposome association, the liposome association assay was performed at various temperatures. Whereas the majority of F-GCNt associated with liposomes after 53°C or 60°C treatment, less than half of the protein was liposome associated after 45°C treatment (Fig. 1C).
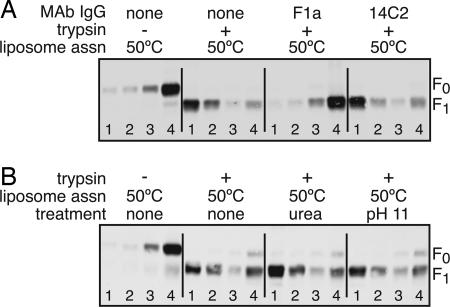
To demonstrate specificity of the liposome association, an antibody specific for F was added to F-GCNt before the addition of liposomes and 50°C treatment. Liposome association was blocked by the neutralizing mAb F1a but not by a control mAb (14C2) (Fig. 2A). The mechanism of F1a neutralization is unknown but its inhibition of F-GCNt liposome association could be specific to the F protein refolding or it could be due to the F-mAb complex hindering in a steric manner access of the fusion peptide to the liposomes.
Fig. 2.
Liposome association is inhibited by mAbs. (A) Samples were treated as in Fig. 1, except that they were incubated with mAb IgG for 15 min at 25°C before adding the liposomes. F1a is a neutralizing mAb specific for PIV5 F. 14C2 is a control mAb specific for influenza virus M2 protein. Samples were incubated with liposomes at 50°C. (B) Samples were treated as above, except that they were exposed to 6 M urea or pH 11 for 10 min after heating.
To investigate the stability of the interaction of F with liposomes, the assay was performed as above, but the samples were treated with 6 M urea or pH 11 for 10 min before sucrose gradient analysis. Neither treatment reduced liposome association (Fig. 2B), suggesting that the fusion peptide inserts stably into the membrane and is not peripherally associated with membranes.
F-GCNt Aggregation Correlates with Liposome Association.
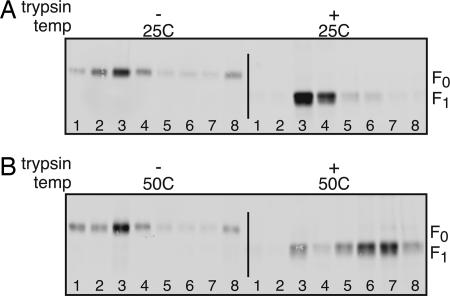
The inactivation of liposome association caused by preheating F-GCNt in the absence of liposomes (Fig. 1B) could be explained by fusion peptide release causing aggregation. To determine whether F-GCNt aggregates after cleavage and heating in the absence of lipid, the sedimentation profile of the F protein was analyzed by rate zonal ultracentrifugation through a sucrose gradient (Fig. 3). Uncleaved F-GCNt sedimented slowly and was found in upper fractions, regardless of the incubation temperature. Cleaved F-GCNt held at 25°C showed a slightly altered sedimentation pattern, but the majority of the protein was found in the upper fractions. In contrast when F-GCNt was cleaved and heated, the protein sedimented toward the bottom of the gradient, suggesting that the protein was aggregated.
Fig. 3.
Aggregation of F-GCNt requires cleavage and elevated temperature. Purified uncleaved or trypsin-cleaved F-GCNt was incubated for 30 min at 25°C (A) or 50°C (B). Samples were overlaid on sucrose gradients and subjected to rate zonal ultracentrifugation. Fractions were collected from the top of the gradients and analyzed by SDS/PAGE and immunoblot.
Electron Microscopy of F-GCNt and Rosette Formation.
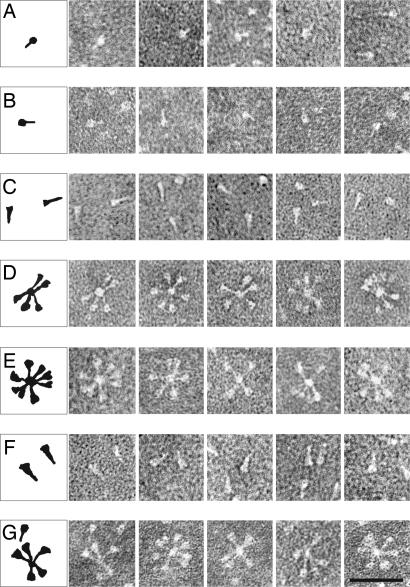
Electron microscopy was used to analyze the morphology of F-GCNt after various treatments (Fig. 4). The majority of uncleaved F-GCNt trimers were observed to resemble a “ball-and-stem” (Fig. 4A). These F molecules are ≈12 nm in length and their shape resembles that of the atomic structure of F-GCNt (19). After cleavage with trypsin, the shape of the trimers was not altered noticeably (Fig. 4B).
Fig. 4.
Electron microscopy of the F proteins. Shown are F-GCNt (A), F-GCNt trypsin digested (B), F-GCNt heated to 50°C for 30 min (C), F-GCNt digested with trypsin and then heated to 50°C for 30 min (D), F-GCNt heated to 50°C for 30 min and then digested with trypsin (E), hPIV3 solF0 (F), and hPIV3 solF0 digested with trypsin (G). (Scale bar = 50 nm.)
When uncleaved F-GCNt was heated to 50°C, the trimers converted to a “golf tee”-like shape (Fig. 4C), ≈16 nm in length and their shape resembled that of the atomic structure of uncleaved postfusion hPIV3 F (18). The data indicate that, as for soluble hPIV3 F, PIV5 F-GCNt can refold into a postfusion conformation in the absence of cleavage. When F-GCNt was treated with trypsin and then heated to 50°C (Fig. 4D), the trimers adopted a golf tee-like shape and organized into rosettes with the wide ends of the golf tees oriented on the outside. These data suggest that the cleaved postfusion F molecules aggregate through interactions of the fusion peptides. When uncleaved F-GCNt was heated to 50°C, cooled, and then treated with trypsin (Fig. 4E), the trimers also formed rosettes. Thus, heating uncleaved F-GCNt most likely resulted in fusion peptide exposure on the outside of the trimer but not release.
For comparison, the morphology of soluble hPIV3 F was also examined by electron microscopy. Uncleaved hPIV3 F exhibited a golf tee-like shape (Fig. 4F) very similar to the PIV5 F-GCNt postfusion form (Fig. 4C). When uncleaved hPIV3 F was treated with trypsin to cleave F, rosettes formed with the head of the golf tee outwards (Fig. 4G). The shape of the hPIV3 F and the PIV5 F-GCNt rosettes was indistinguishable (Fig. 4, compare E and G).
Conformational Changes in F-GCNt Can Be Detected by Using mAbs.
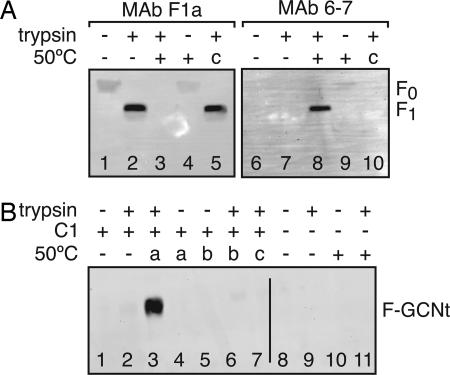
Although cleavage alone does not trigger fusion peptide release for F-GCNt, cleavage of the native F protein can affect antibody recognition (24). To determine whether the conformation of F-GCNt is affected by cleavage, F-GCNt was cleaved and/or heated and subjected to immunoprecipitation by using the mAbs F1a (25) and 6-7 (26). As shown previously for the native F protein, F1a recognized F-GCNt better after cleavage (Fig. 5A, lane 2). When cleaved F-GCNt was heated, F1a reactivity was lost (Fig. 5A, lane 3). The F1a epitope may be occluded when F-GCNt aggregates into rosettes, which would be consistent with the ability of F1a to inhibit liposome association.
Fig. 5.
mAb and C1 peptide binding to heat-triggered F-GCNt conformations. (A) Uncleaved or trypsin-cleaved F-GCNt was incubated at 25°C or 50°C for 30 min. Two samples were heated before trypsin cleavage (c, lanes 5 and 10). F-GCNt was immunoprecipitated with mAbs F1a or 6-7 and analyzed by reducing SDS/PAGE and immunoblot. (B) Uncleaved or trypsin-cleaved F-GCNt was incubated with HA-tagged C1 peptide for 30 min. Samples were heated to 50°C for 30 min at different points either during incubation with the C1 peptide (a), before adding peptide (b), or before trypsin cleavage (c). Samples in lanes 8–11 represent controls lacking C1 peptide. Peptide was immunoprecipitated by using 12CA5 mAb. The samples were analyzed by nonreducing SDS/PAGE and immunoblot.
mAb 6-7 was shown previously to react more strongly with the native F protein after heating (26, 27). Similarly, 6-7 recognized heated and cleaved F-GCNt better than F-GCNt held at 25°C (Fig. 5A, lane 8). 6-7 mAb only poorly recognized heated uncleaved F-GCNt, indicating that despite the fact that heating in the absence of cleavage can convert F-GCNt into a postfusion conformation, subtle differences remain between the postfusion conformations of cleaved versus uncleaved F-GCNt.
Despite the fact that F-GCNt can form rosettes when it is cleaved after heating, F1a and 6-7 mAbs had different reactivity for F-GCNt that was cleaved after heating (Fig. 5A, lanes 5 and 10) versus F-GCNt that was cleaved before heating, indicating that the conformation of F-GCNt after these treatments is subtly different.
C1 Peptide Binds to an F-GCNt Intermediate Conformation.
During fusion, the native PIV5 F refolds through distinct intermediates. Previous data indicate that a peptide corresponding to the amino acid residues of the F protein HRB region (C1 peptide) can block fusion by trapping the F protein at a prehairpin intermediate, a conformation that exists after fusion peptide insertion but before 6HB formation (14, 16).
To determine whether F-GCNt refolds through the same prehairpin intermediate, coimmunoprecipitation of F-GCNt with an HA-tagged C1 peptide was assessed under various conditions. When C1 peptide was added to cleaved F-GCNt and heated to 50°C, the peptide bound to F-GCNt (Fig. 5B, lane 3), suggesting that F-GCNt can adopt the prehairpin conformation. C1 peptide did not bind to cleaved F-GCNt that was heated before the addition of peptide (Fig. 5B, lane 6), consistent with the prehairpin intermediate of F-GCNt existing only transiently during heating. C1 peptide added to uncleaved F-GCNt during heating also did not bind (Fig. 5B, lane 4), confirming that cleaved and uncleaved F-GCNt have subtly different intermediate conformations.
Consistent with the model that C1 peptide binds to the prehairpin intermediate, C1 peptide did not inhibit liposome association (data not shown). The addition of excess C1 peptide to cleaved and heated F-GCNt did not alter the mAb binding (data not shown), suggesting that 6-7 mAb can recognize the prehairpin intermediate and F1a mAb cannot.
Discussion
For paramyxovirus fusion, the natural trigger to activate the F protein is the receptor binding protein, HN (or H or G), interacting with a cell surface-expressed receptor molecule. HN coassociates with F, and receptor binding is proposed to bring about a conformational change in HN that in turn activates F (7, 8, 28). One suggestion is that HN provides activating energy to F, to raise F over the energy barrier to allow protein refolding and membrane merger (14, 21). The alternative viewpoint is that in an HN/F complex HN clamps F, holding it in the prefusion form (29, 30).
It was of considerable interest to investigate whether soluble F-GCNt could be triggered to undergo conformational changes and be converted to the postfusion form. To date, incubation of sialic acid with F-GCNt and soluble tetrameric HN ectodomain that includes the stalk (31) [a domain that is considered to interact with F (32–34)] has not caused triggering of F (S.A.C. and Ping Yuan, unpublished observations). For other viral fusion proteins, such as influenza virus HA (35, 36), avian sarcoma and leukosis virus Env (37, 38), or severe acute respiratory syndrome coronavirus S (39), heat or urea have been shown to act as a surrogate trigger for protein refolding. However, for Semliki forest virus E1 protein and tick-borne encephalitis virus E protein, heat or urea cannot supplant low pH as a trigger (40, 41). For F-GCNt, heat caused conformational changes in F, and the available data indicate that F-GCNt followed a comparable pathway of protein refolding as fusion-active membrane-bound F. The GCNt trimerization domain may affect F activation properties, but addition of GCNt to the F protein was necessary to obtain PIV5 F in its prefusion state.
At 50°C cleaved F-GCNt was activated, and it bound liposomes, consistent with release of the fusion peptide. Triggered F protein could bind the C1 peptide, and conformational changes in F could be detected by mAbs. Triggered F protein was found to aggregate by rate zonal centrifugation analysis and was observed to form rosettes in the electron microscope, data consistent with release of the hydrophobic fusion peptide (Fig. 6).
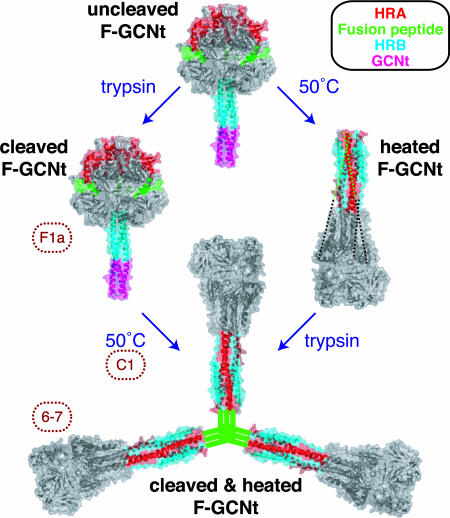
Fig. 6.
Progression from the prefusion to the postfusion conformation. The unheated and heated proteins are depicted by the PIV5 F-GCNt (Protein Data Bank ID code 2B9B) and hPIV3 solF0 (Protein Data Bank ID code 1ZTM) structures, respectively. The HRA (red), fusion peptide (green), HRB (blue), and GCNt (magenta) regions are colored. When F-GCNt is cleaved with trypsin (left side), the protein does not refold to its postfusion conformation, but a gain of mAb F1a reactivity indicates a subtle change. When cleaved F-GCNt is heated, the protein converts to the postfusion golf tee-like conformation, aggregates into rosettes through its fusion peptide, and gains mAb 6-7 reactivity. If C1 peptide is added during the heating of cleaved F-GCNt, it can bind to the protein and most likely trap the prehairpin intermediate. When F-GCNt is heated without cleavage (right side), the protein refolds into its uncleaved postfusion conformation. The 47 residues for which there is no interpretable density in the hPIV3 electron density map, including the residues encoding the cleavage site and fusion peptide, have been added as dotted lines. When heated F-GCNt is cleaved, the protein aggregates into rosettes. It is anticipated that the TM domain or GCNt domain would be adjacent to the fusion peptide in rosettes, but for clarity this has been omitted.
The soluble ectodomain of hPIV3 F (solF0) folded spontaneously into its postfusion form, despite the fact that the protein was uncleaved (19). Similarly, PIV5 F-GCNt can convert to a postfusion form after heat treatment in the absence of cleavage (Fig. 6). The uncleaved postfusion form of F-GCNt differs subtly from the cleaved postfusion F-GCNt, as determined by mAb reactivity and C1 peptide binding. This is not unexpected as the residues that contain the cleavage site, the fusion peptide, and part of HRA likely are stretched across the surface of the uncleaved postfusion F-GCNt protein (18). Thus, access of the C1 peptide and accessibility of mAb epitopes may be restricted. The epitopes on F for mAbs F1a and 6-7 have not been mapped accurately.
Previously, the morphology of a secreted soluble form of the paramyxovirus RSV F protein has been examined (42, 43). It was observed that cleavage of RSV F converted the protein from discrete cone-shaped trimers into rosettes of lollipop-shaped trimers. This transition was postulated to be a conversion from the prefusion to the postfusion form of the protein. It was determined that the thermostability of the two protein forms was equivalent (44), and thus it was considered that cleavage per se was the RSV activation event. In light of the evidence that PIV5 and hPIV3 F proteins can convert to the postfusion form in the absence of cleavage, the interpretation of the RSV F morphologic data is open to an alternative explanation. Assuming the RSV F structure is closely related to the PIV5 and hPIV3 prefusion and postfusion structures, it seems possible that the uncleaved cone-shaped RSV F may represent a postfusion conformation that folded spontaneously in the absence of a TM domain. Cleavage may trigger rosette formation simply by releasing the fusion peptide and the transition from a cone shape to a lollipop shape may reflect the movement after cleavage of the fusion peptide and some of the HRA residues from the side of the molecule to the tip.
The precise role of HN in triggering F remains to be understood but the emerging picture indicates a regulated complex biological machine. HN could exert its effects, for example, by influencing the stability of the F prefusion stalk in a receptor-dependent manner. It is postulated that one of the first steps in fusion after HN binding sialic acid is the dissociation of the HRB 3HB in prefusion F that makes available the binding site for the HRA peptide (14, 19). A model for Newcastle disease virus F-mediated fusion has been proposed in which HN binding to sialic acid alters the oligomeric arrangement of HN, creates a second sialic acid binding site and in the process activates F and tethers HN to the membrane (45, 46). However, the lack of the second sialic acid binding site in PIV5 and hPIV3 HN (31, 47) makes it unlikely to be a general model of paramyxovirus HN activation of F. For PIV5, an alternative model for HN involvement in membrane fusion was proposed that involves ligand-dependent changes in the HN oligomer that are driven by subunit-subunit interactions (31). In this model, the HN dimer of dimers forms in the absence of ligand and can interact with the F protein, potentially through lateral interactions on two sides of the tetramer. Binding of cell surface receptors could trigger the partial disassembly of the HN tetramer, driven by the energy of receptor engagement, and could lead to changes in both the HN stalk region and the interaction with F, thus activating F for membrane fusion (31).
Materials and Methods
Proteins and Peptides.
The soluble proteins PIV5 F-GCNt and hPIV3 solF0 were expressed in High Five cells by using a recombinant baculovirus as described previously (18, 19). Both proteins carry altered cleavage sites such that addition of exogenous trypsin is required for cleavage. Secreted F proteins were purified by Ni2+-chelating chromatography (18). Protein concentrations were determined by using the BCA protein assay (Pierce, Rockford, IL). The HAt-C1 peptide (14) contains the amino acid residues of the PIV5 W3A F protein HRB and extended chain region, with an HA tag (YPYDVPDYASL) at its N terminus. C1 peptide was expressed in Escherichia coli and purified as described previously (48).
Antibodies.
Ascites fluid containing PIV5 F-specific mAb F1a (25) and hybridoma supernatant containing mAb 6-7 (26), a mAb raised against the PIV5 F hyperfusogenic mutant S443P, were used. Hybridoma supernatant containing 12CA5, a mAb specific for the HA tag, was also used. IgG from F1a and 14C2 (49), a mAb specific for influenza virus M2 protein, was purified by using AffinityPak protein A columns (Pierce), and its concentration was determined by OD280. Polyclonal antibody PAb245 was raised against a PIV5 peptide (residues 388–402) (24).
Preparation of Liposomes.
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, and cholesterol in chloroform (Avanti Polar Lipids, Alabaster, AL) were mixed at an 8:2:5 molar ratio, and the chloroform was evaporated under argon. The resulting lipid films were dried under vacuum overnight and resuspended in PBS at 40 mM total lipid. After five freeze–thaw cycles, the lipids were vortexed and extruded 21 times through two 100-μm filters by using a miniextruder (Avanti Polar Lipids).
Liposome Association Assay.
For each sample, 2.2 μg of F-GCNt was cleaved with 25 milliunits of l-[tosylamido-2-phenyl] ethyl chloromethyl ketone-treated trypsin (Worthington Biochemical, Lakewood, NJ) in 100 mM phosphate buffer (pH 7.1) for 30 min at 25°C. After cleavage, 47.5 pg of soybean trypsin inhibitor (Worthington Biochemical) was added to each sample. When indicated, samples were pretreated at 60°C for 30 min or incubated with IgG (4 μg of F1a or 14C2) or C1 peptide (30 μM) at 25°C for 15 min. Liposomes (40 μl per sample) and PBS were added (80 μl final volume), and the samples were incubated at the specified temperatures for 30 min. When indicated, 80 μl of 12 M urea or carbonate (pH 11) was added at 25°C for 10 min. Sucrose was added to a final concentration of 50% (500 μl final volume). The samples were overlaid with 500 μl each of 40% sucrose, 25% sucrose, and PBS and were spun in a TLS55 rotor at 49,000 rpm for 3 h at 25°C. Fractions (500 μl) were collected from the top of the gradients. Proteins were solubilized in 0.5% Triton X-100 and precipitated by using 12.5% vol/vol trichloroacetic acid. Polypeptides were separated by SDS/PAGE and transferred to PVDF membranes (50). Blots were probed with PAb245 serum (1:2,000) and Alexa Fluor 680-conjugated goat anti-rabbit Ab (Invitrogen, Carlsbad, CA).
Rate Zonal Sucrose Sedimentation.
For each sample, 1.8 μg of F-GCNt was cleaved with trypsin as above. Trypsin inhibitor was added, and samples were incubated at 25°C or 50°C for 30 min. Gradients consisting of 500 μl of 60% sucrose, 1 ml of 50% sucrose, 1 ml of 40% sucrose, 1 ml of 30% sucrose, and 500 μl of 20% sucrose were prepared and allowed to diffuse at room temperature for 4 h. Samples were overlaid onto the gradients, and proteins were analyzed by ultracentrifugation in an SW60 rotor at 42,000 rpm for 20 h at 20°C. Gradients were fractionated, and proteins were concentrated by TCA precipitation and analyzed as above.
Electron Microscopy.
Electron microscopy was performed as described (51). Samples were absorbed onto freshly glow-discharged carbon-coated grids, stained with 2% sodium phosphotungstate (pH 6.6), and examined in a JEOL 1230 transmission electron microscope (JEOL, Peabody, MA) operating at 100 kV. The electron microscope was capable of resolving the lattice plane spacing of catalase crystals (6.85 and 8.75 nm).
Immunoprecipitation and C1 Peptide Binding.
For each sample, 2.2 μg of F-GCNt was cleaved as above and inhibitor was added. The samples were incubated at 25°C or 50°C for 30 min. C1 peptide (3 μM) was added to some samples. When indicated, the samples were heated before cleavage, before peptide addition, or during the peptide incubation. Antibodies were added (10 μl of 12CA5, 30 μl of 6-7, or 5 μl of F1a), and samples were incubated at 4°C for 2 h. A 1:1 mixture of protein A and protein G Sepharose was added in binding buffer (10 mM Tris, pH 8/100 mM NaCl/0.1% Nonidet P-40/0.05% BSA/0.05% chicken egg albumin). After 1 h at 4°C, beads were washed three times with binding buffer containing 500 mM NaCl. Polypeptides were analyzed by SDS/PAGE and immunoblotting with PAb245. To reduce cross-reactivity with the precipitating mAb, PAb245 was detected by using IRDye800-conjugated protein G (Rockland, Gilbertsville, PA).
Acknowledgments
We thank Anneli Asikainen for assistance in the production and purification of F-GCNt protein. The JEOL 1230 electron microscope used in the study is part of the Biological Imaging Facility at Northwestern University. S.A.C. and H.-S.Y. are Associates and R.A.L. is an Investigator of the Howard Hughes Medical Institute. This research was supported in part by National Institutes of Health Research Grants AI-23173 (to R.A.L.) and GM-61050 (to T.S.J.).
Glossary
Abbreviations
- RSV
respiratory syncytial virus
- 6HB
six-helix bundle
- TM
transmembrane.
Footnotes
The authors declare no conflict of interest.
References
- 1.Lamb RA, Kolakofsky D. In: Fields Virology. 4th Ed. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1305–1340. [Google Scholar]
- 2.Dorig RE, Marcil A, Chopra A, Richardson CD. Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 3.Yanagi Y, Ono N, Tatsuo H, Hashimoto K, Minagawa H. Virology. 2002;299:155–161. doi: 10.1006/viro.2002.1471. [DOI] [PubMed] [Google Scholar]
- 4.Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang LF, Eaton BT, Broder CC. Proc Natl Acad Sci USA. 2005;102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, Lee B. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 6.Feldman SA, Hendry RM, Beeler JA. J Virol. 1999;73:6610–6617. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamb RA. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 8.Lamb RA, Paterson RG, Jardetzky TS. Virology. 2006;344:30–37. doi: 10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skehel JJ, Wiley DC. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 10.Earp LJ, Delos SE, Park HE, White JM. Curr Top Microbiol Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert DM, Kim PS. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 13.Weber T, Paesold G, Galli C, Mischler R, Semenza G, Brunner J. J Biol Chem. 1994;269:18353–18358. [PubMed] [Google Scholar]
- 14.Russell CJ, Jardetzky TS, Lamb RA. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 16.Russell CJ, Kantor KL, Jardetzky TS, Lamb RA. J Cell Biol. 2003;163:363–374. doi: 10.1083/jcb.200305130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. J Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin H-S, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito M, Nishio M, Kawano M, Kusagawa S, Komada H, Ito Y, Tsurudome M. J Virol. 1997;71:9855–9858. doi: 10.1128/jvi.71.12.9855-9858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paterson RG, Russell CJ, Lamb RA. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Gorman JJ, McKimm-Breschkin J, Lawrence LJ, Tulloch PA, Smith BJ, Colman PM, Lawrence MC. Structure (London) 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 23.Connolly SA, Lamb RA. Virology. 2006 doi: 10.1016/j.virol.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 24.Dutch RE, Hagglund RN, Nagel MA, Paterson RG, Lamb RA. Virology. 2001;281:138–150. doi: 10.1006/viro.2000.0817. [DOI] [PubMed] [Google Scholar]
- 25.Randall RE, Young DF, Goswami KKA, Russell WC. J Gen Virol. 1987;68:2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- 26.Tsurudome M, Ito M, Nishio M, Kawano M, Komada H, Ito Y. J Virol. 2001;75:8999–9009. doi: 10.1128/JVI.75.19.8999-9009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waning DL, Russell CJ, Jardetzky TS, Lamb RA. Proc Natl Acad Sci USA. 2004;101:9217–9222. doi: 10.1073/pnas.0403339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison TG. Biochim Biophys Acta. 2003;1614:73–84. doi: 10.1016/s0005-2736(03)00164-0. [DOI] [PubMed] [Google Scholar]
- 29.McGinnes LW, Morrison TG. J Virol. 2006;80:2894–2903. doi: 10.1128/JVI.80.6.2894-2903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takimoto T, Taylor GL, Connaris HC, Crennell SJ, Portner A. J Virol. 2002;76:13028–13033. doi: 10.1128/JVI.76.24.13028-13033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan P, Thompson T, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structure (London) 2005;13:1–13. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 32.Deng R, Wang Z, Mahon PJ, Marinello M, Mirza A, Iorio RM. Virology. 1999;253:43–54. doi: 10.1006/viro.1998.9501. [DOI] [PubMed] [Google Scholar]
- 33.Tanabayashi K, Compans RW. J Virol. 1996;70:6112–6118. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melanson VR, Iorio RM. J Virol. 2006;80:623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruigrok RWH, Martin SR, Wharton SA, Skehel JJ, Bayley PM, Wiley DC. Virology. 1986;155:484–497. doi: 10.1016/0042-6822(86)90210-2. [DOI] [PubMed] [Google Scholar]
- 36.Carr CM, Chaudhry C, Kim PS. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith JG, Mothes W, Blacklow SC, Cunningham JM. J Virol. 2004;78:1403–1410. doi: 10.1128/JVI.78.3.1403-1410.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuyama S, Delos SE, White JM. J Virol. 2004;78:8201–8209. doi: 10.1128/JVI.78.15.8201-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F, Berardi M, Li W, Farzan M, Dormitzer PR, Harrison SC. J Virol. 2006;80:6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibbons DL, Ahn A, Chatterjee PK, Kielian M. J Virol. 2000;74:7772–7780. doi: 10.1128/jvi.74.17.7772-7780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiasny K, Allison SL, Mandl CW, Heinz FX. J Virol. 2001;75:7392–7398. doi: 10.1128/JVI.75.16.7392-7398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calder LJ, Gonzalez-Reyes L, Garcia-Barreno B, Wharton SA, Skehel JJ, Wiley DC, Melero JA. Virology. 2000;271:122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- 43.González-Reyes L, Ruiz-Argüello MB, García-Barreno B, Calder L, López JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. Proc Natl Acad Sci USA. 2001;98:9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz-Arguello MB, Martin D, Wharton SA, Calder LJ, Martin SR, Cano O, Calero M, Garcia-Barreno B, Skehel JJ, Melero JA. J Gen Virol. 2004;85:3677–3687. doi: 10.1099/vir.0.80318-0. [DOI] [PubMed] [Google Scholar]
- 45.Crennell S, Takimoto T, Portner A, Taylor G. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 46.Zaitsev V, Von Itzsteine M, Groves D, Kiefel M, Takimoto T, Portner A, Taylor G. J Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 48.Joshi SB, Dutch RE, Lamb RA. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 49.Zebedee SL, Lamb RA. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paterson RG, Lamb RA. In: Molecular Virology: A Practical Approach. Davidson A, Elliott RM, editors. Oxford: IRL Oxford Univ Press; 1993. pp. 35–73. [Google Scholar]
- 51.Wrigley NG, Brown E, Chillingworth RK. J Microsc. 1982;130:225–232. doi: 10.1111/j.1365-2818.1983.tb04220.x. [DOI] [PubMed] [Google Scholar]