Abstract
Chargaff's second parity rules for mononucleotides and oligonucleotides (CIImono and CIIoligo rules) state that a sufficiently long (>100 kb) strand of genomic DNA that contains N copies of a mono- or oligonucleotide, also contains N copies of its reverse complementary mono- or oligonucleotide on the same strand. There is very strong support in the literature for the validity of the rules in coding and noncoding regions, especially for the CIImono rule. Because the experimental support for the CIIoligo rule is much less complete, the present article, focusing on the special case of trinucleotides (triplets), examined several gigabases of genome sequences from a wide range of species and kingdoms including organelles such as mitochondria and chloroplasts. I found that all genomes, with the only exception of certain mitochondria, complied with the CIItriplet rule at a very high level of accuracy in coding and noncoding regions alike. Based on the growing evidence that genomes may contain up to millions of copies of interspersed repetitive elements, I propose in this article a quantitative formulation of the hypothesis that inversions and inverted transposition could be a major contributing if not dominant factor in the almost universal validity of the rules.
Keywords: chloroplasts, genomics, mitochondria, base composition, oligonucleotide composition
Chargaff's first parity rule, called here the CImono rule, states that “the numbers of A's and T's and the numbers of C's and G's match exactly in every DNA duplex. It is well known to be an immediate consequence of base pairing (1). Of course, not only single bases, but the oligonucleotides of each strand are paired with their reverse complements on the other, and, therefore, their numbers match exactly as well, which is called the CIoligo rule.
In contrast, Chargaff's second parity rules (denoted as CIImono and CIIoligo rules in the following), which essentially make the same claim for each single strand of a duplex (2–6), have no generally accepted explanation. Discovered almost 40 years ago (7, 8), before any sequence data were available, the rules continue to stimulate the search for their unknown underlying mechanism (6, 9–13). Obviously, base pairing does not provide one because the nucleotides of each single strand of a duplex are already paired with the nucleotides on their opposite strands and need not pair with any other on their own strand. Most puzzling, perhaps, there are no known selective advantages for genomes or organisms to comply with the rules. Yet they apply to coding and noncoding regions of the genomes equally well.
Is it possible to consider these rules as trivial? Statistically speaking, it would certainly be trivial to find a reverse complement for each oligonucleotide of length L on the same strand. If the bases are well shuffled, the next complement of any base is, on average, only 4 bases away. Likewise, on average, the nearest reverse complement on the same strand of any dinucleotide is only 42 = 16 of any trinucleotide only 43 = 64 bases away. However, this does not prove that their numbers are the same. For example, the nearest complement of three copies of TGC at positions x, x + 22, and x + 46 may be one and the same triplet GCA at position x + 61! Therefore, Chargaff's second parity rules are not trivial, and have the remarkable implication that some unknown mechanism seems to “count” and adjust the numbers of oligonucleotides and their complements to equal values on each of both strands.
For many years only the simpler CIImono rule was known, which claimed that bases and complementary bases exist in equal numbers on the same strand. Chargaff discovered it in 1968 after separating the genome of Bacillus subtilis into separate strands and analyzing the nucleotide contents of each single-strand preparation (7, 8). Since then scientists have gathered very strong evidence for its general validity (8). Nevertheless, there are exceptions to the rule (7, 9, 12, 14). However, as reported here, there seem to be none if the genome size exceeds 100 kb.
The first case of the more generalized CIItriplet rule was discovered in 1999 by Prabhu (4). It was subsequently confirmed and expanded into the general CIIoligo rule (3).
A number of scientists have tackled this enigmatic property of genomes (2, 4, 8–13). For example, Fickett et al. (2) remarked that the symmetry of the base composition between the two strands of a duplex might be explained by inversions. Also, Baisnée et al. (3) pointed, among other possibilities, to inversions as possible mechanisms and concluded that only a multiplicity of mechanisms could explain the various manifestations of the rules. Forsdyke and Bell (12) suggested stem-loop mechanisms as explanations. Lobry (13) argued that the CIImono rule might result from many single base substitutions during the course of evolution.
Lobry's hypothesis about the CIImono rule still awaits a generalization for the CIIoligo rule because the validity of the former does not automatically imply the validity of the latter. In addition, Forsdyke's stem-loop hypothesis, drawing on the stem-loops of RNA transcripts, applies only to transcribed regions, which are predominantly the coding regions.
It appears, therefore, that additional hypotheses may be needed to explain the surprisingly universal validity of Chargaff's second parity rules. Ideally, these hypotheses should (i) explain the CIIoligo rule, which, in turn, would automatically validate the CIImono rule (see supporting information, which is published on the PNAS web site); (ii) be formulated in a testable, quantitative way; and (iii) be blind to any difference between coding and noncoding regions.
The present article offers such a hypothesis. It is based on the growing evidence of large numbers of Alu, SINE, LINE, and other such dispersed, repetitive sequences in the coding and noncoding regions of the genomes of many species (15–18). These findings have increased considerably the likelihood that the earlier remarks by Fickett et al. (2) and Baisnée et al. (3) were correctly pointing to inversions as an explanation of the validity of the rules. However, these suggestions did not consider transpositions and have never been formulated and tested quantitatively. Therefore, the present article offers the quantitative formulation and simulations of the hypothesis that numerous, undirected transposition/inversions in the course of evolutionary time may have contributed substantially, if not predominantly, to the validity of CIImono and CIIoligo rules in coding and noncoding regions alike.
Results
Special Case of Triplets.
For the sake of simplicity the article focuses specifically on the example of trinucleotides (triplets) and, consequently, on the validity of the specific case of the CIItriplet rule. The generalization of the arguments to other oligonucleotides will be self-evident.
Triplet frequency distributions will simply be called “triplet profiles” in the following and written symbolically as f(△), where the variable △ represents all 64 triplets. Reverse complementary triplets of a triplet △ will be denoted by the symbol ▾. For example, if △ = GCT then ▾ = AGC.
The article will use only “running” triplet profiles (i.e., the triplets of each strand were read by frame shifts of 1) to make them invariant against frame shifts. As illustration consider the case of a hypothetical, short-nucleotide sequence of a Watson strand, 5′-ATTACGCTAGGCTA-3′. To obtain its running triplet profile one would begin at the 5′ end and extract sequentially the series of triplets △1 = ATT, △2 = TTA, △3 = TAC, △4 = ACG, …,△12 = CTA.
According to the definition of the CIIoligo rule, any test of the compliance of a genome with the CIItriplet rule requires determining how often each of the 64 possible triplets occurs along its (say) Watson strand. Next, one has to compare the frequency of every triplet in this profile with the frequency of its reverse complement to determine whether they are the same. The somewhat tedious task can be facilitated by using the following, logically equivalent formulation of the CIItriplet rule (see supporting information): If a sufficiently long (>100 kb) single strand of genomic, duplex DNA contains N copies of a triplet, then the opposite strand contains N copies of the same triplet, as well. (It is assumed that both strands are read from their 5′ to their 3′ ends.) I will use this “equivalent formulation of the CIItriplet rule” for most of the article.
According to the latter formulation the above example sequence does not comply with the CIItriplet rule, because the numbers of certain triplets are not the same on both strands. The running profile of the Crick strand 5′-TAGCCTAGCGTAAT-3′ yields the triplets △1 = TAG, △2 = AGC, △3 = GCC, △4 = CCT, …, △12 = AAT. The reader can easily verify that only the Watson strand, but not the Crick strand, contains the triplets ATT, TTA, and ACG. Therefore, the example demonstrates that, in general, arbitrary nucleotide sequences do not comply with the CIItriplet rule.
Despite this violation, the example complies with the CIImono rule, because it contains the same number of A's as T's, namely four, and the same number of C's as G's, namely three. Therefore, a sequence complying with the CIImono rule does not necessarily have to comply with the CIIoligo rule. Only the opposite is true, namely that every genome that complies with the CIIoligo rule must also comply with the CIImono rule (see supporting information). Of course, it is easy to construct much larger genomes than the above example that prove this point.
Validity and Accuracy of the CIItriplet Rule.
With very few exceptions, most of the evidence in the literature for the validity of Chargaff's second parity rule demonstrates only the validity of the CIImono rule. However, as mentioned above, the validity of the CIImono rule does not imply the validity of the CIItriplet rule. Therefore, before presenting our hypothesis of Chargaff's second parity rules, it is important to demonstrate the almost universal and uncanny accuracy of the CIItriplet rule across species and kingdom boundaries, as well.
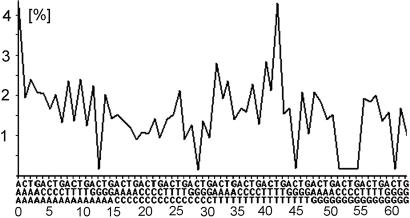
Fig. 1 shows an example of a typical triplet profile of an actual genome plotted as a function of all 64 possible triplets. According to the equivalent formulation, a test of the compliance with the CIItriplet rule needs to determine whether the triplet profiles of both strands of a DNA duplex are the same. This can be done conveniently by a correlation plot that compares the profile of the Watson strand with that of the Crick strand (Fig. 2). The more similar the two distributions are, the closer the data points fall on the diagonal line of the correlation plot, the more the tested duplex complies with the rule. One can use the correlation coefficient cWC between the two profiles as a quantitative measure of their similarity, and thus of their degree of compliance. If cWC = 1, the two profiles are identical and the tested DNA duplex complies ideally with the rule. If cWC = 0, the two profiles are unrelated and the DNA duplex violates it.
Fig. 1.
Typical triplet profile (chimpanzee chromosome 14, position 32 Mb to 40 Mb). The abscissa shows all possible triplets (to be read vertically from top to bottom). Numbers indicate the canonical numbers (see supporting information). The ordinate shows the frequency of triplets (%).
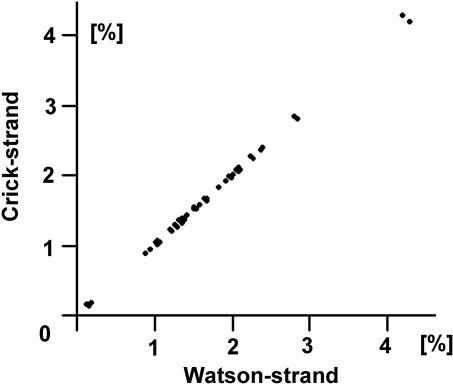
Fig. 2.
Method of testing the validity of the CIItriplet rule by using a correlation plot between the triplet frequencies of the Watson (abscissa) and Crick (ordinate) strands of the same sequence as shown in Fig. 1. If a sequence complies completely, the plot generates a straight diagonal line with a correlation coefficient of cWC = 1.0. In the above case the genome complies quite well because its correlation coefficient cWC = 0.9994.
I tested genomes whose size was <8 Mb in this way by direct analysis. If a genome was larger, it was cut into sizes of 8 Mb and their triplet profiles were measured individually. The choice of 8 Mb as the testing unit was dictated by our analysis computer program.
Based on the analysis of >500 genome segments of 8 Mb size or smaller, the triplet frequencies of their Watson and Crick strands were virtually identical. Only a subset of mitochondrial genomes violated this identity (see below). In all other cases the standard deviation of the differences between the all values of fWatson(△) and fCrick(△) was <2%. Correspondingly, the correlation coefficients between the Watson and Crick strands cWC were found to be close to unity (in the example of Fig. 2, cWC = 0.9996).
The high degree of compliance is not a matter of randomness of the genome sequences tested. Most random sequences would not even comply with the CIImono rule, let alone with the CIItriplet rule, because they would not fulfill the condition that the base frequencies f(A) = f(T) and f(C) = f(G). However, if a random sequence happened to fulfill that f(A) = f(T) and f(C) = f(G), the frequencies of all permutations of any given triplet and their reverse complements would necessarily be the same. Therefore, the correlation plots of such sequences would degenerate into a set of one to four isolated points on the diagonal. The triplet profiles of all 500+ tested genome segments were markedly different from such a profile, demonstrating that none of the naturally occurring genomes were random sequences.
Validity of the CIItriplet rule for the entire human genome and a wide range of organisms.
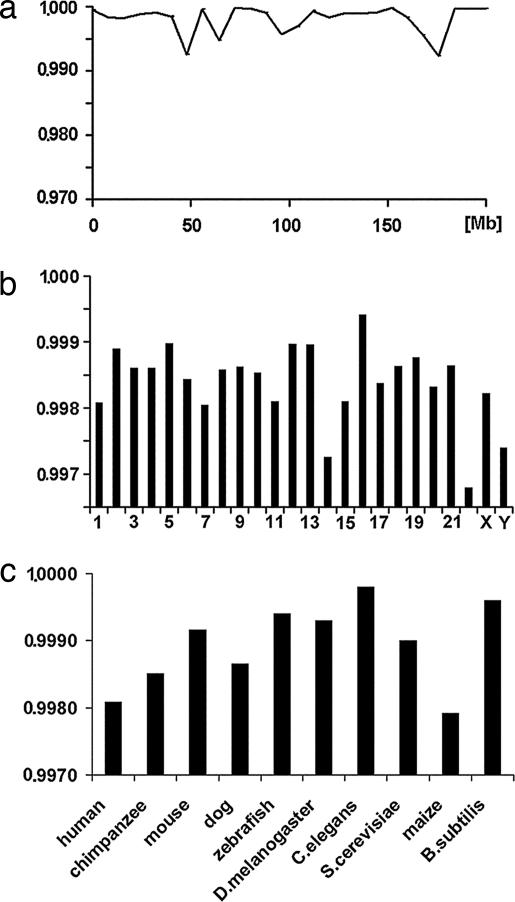
More specifically, the correlation coefficients cWC for each 8-Mb large segment of the entire human chromosome 1 were close to a value of 1.0 (Fig. 3a), although in certain locations one or several “spikes” of the correlation coefficient appeared to drop as low as 0.994.
Fig. 3.
Almost universal validity of Chargaff's second parity rules as applied to triplets. The correlation coefficient cWC is shown to vary only on the third decimal point. The ordinate shows correlation coefficient cWC, and the abscissa shows the location along the chromosome. (a) The correlation coefficients for each of the 8-Mb large segments along the entire length of human chromosome 1. (b) Average correlation coefficients cWC for all human chromosome averaged over 8 Mb segments along their entire length. (c) The correlation coefficients of arbitrarily selected entire chromosomes of various species ranging from primates to bacteria.
Similarly, I tested each human chromosome individually and found that each complied with the CIItriplet rule along its entire length (Fig. 3b). Individual chromosomes of other organisms including chimpanzee, dog, mouse, zebrafish, Drosophila melanogaster, Caenorhabditis elegans, maize, yeast (Saccharomyces cerevisiae), and B. subtilis showed similar results (Fig. 3c).
Compliance with the CIItriplet rule as a function of sequence length.
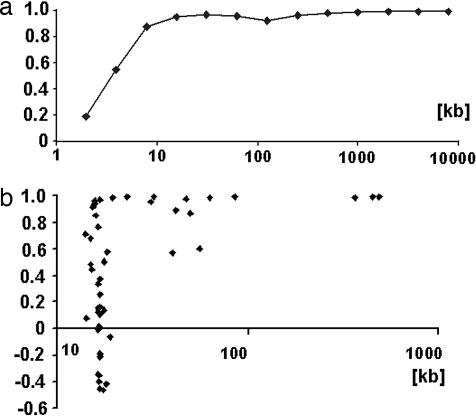
The shorter the genome segment was, the more the correlation coefficient cWC deviated from the ideal value of 1.0000. In the case of human chromosome 1 the correlation coefficient cWC = 0.995 was constant for sequences ranging in size from 10 Mb to 1 Mb. Between 1 Mb and 100 kb cWC decreased to a value of 0.93. Between 100 kb and 10 kb cWC fluctuated considerably, and at sizes below 10 kb the value of cWC decreased quite rapidly (Fig. 4a).
Fig. 4.
Role of genome size in the validity of the Chargaff second parity rules. The abscissa shows correlation coefficients cWC, and the ordinate shows genome size. (a) Correlation coefficients cWC of different size segments that include the 5′ end of human chromosome 1. (b) Lack of a size correlation between the correlation coefficients cWC of 51 mitochondrial genomes and their genome sizes.
Test of the validity of the CIItriplet rule for mitochondrial genomes.
In the course of the above tests it appeared that human mitochondrial genomes violated the CIItriplet rule. To test to what degree the same was true for all mitochondria I tested 51 mitochondrial genomes that belonged to a wide range of organisms. They included fungi, amoebae, invertebrates, insects, plants, slime mold, arthropods, and vertebrates such as amphibians, reptiles, marsupials, and mammals. They ranged in size between 14 kb (Limulus polyphemus) and 490 kb [Oryza sativa (rice)].
Seventeen mitochondrial genomes were found to comply accurately with Chargaff's second parity rule. Similar to the human mitochondrial genomes, however, 34 other mitochondrial genomes were found to violate Chargaff's second parity rule to various degrees (Fig. 4b).
The reason for the violation was not the small genome size for the following reasons.
Many of the short mitochondrial genomes were compliant at high levels. For example, the mitochondrial genomes of Chlamydomonas reinhardtii (size: 15.7 kb), Apis mellifera (honey bee) (size: 16.7 kb), and D. melanogaster (size: 19.5 kb) complied with the rules at high levels of compliance cWC = 0.94, 0.97, and 0.99, respectively, despite their small genome size.
Judging by the example of human chromosome 1 (Fig. 4a), the low compliance levels of the “violators,” which assumed even negative values (Fig. 4b), were far too low for their genome sizes of ≈16 kb.
The size–compliance relationship of mitochondrial genomes as suggested by Fig. 4b showed no gradual transition between size and compliance.
However, there is possibly an evolutionary explanation for the violation by several mitochondrial genomes, because most of the violators belonged to recent vertebrates. Examples are the mitochondrial genomes of Alligator mississippiensis, Anguilla anguilla (eel), Balaenoptera borealis (whale), Boa constrictor, Bos taurus, Canis familiaris (dog), Ciconia ciconia (stork), Equus caballus (horse), Falco peregrinus (falcon), Felis catus (cat), Gallus gallus (chicken), Gorilla gorilla, human (Japan), human (Sweden), Kaloula pulchra (bullfrog), Macaca mulatta (rhesus monkey), Mus musculus (mouse), Rattus norvegicus (rat), Sus scrofa (pig), Testudo graeca (turtle), and Macropus robustus (wallaroo). The violation of the CIItriplet rule by these mitochondrial genomes is possibly related to large number of mitochondrial genes that were transferred to the host cell genome by horizontal gene transfer, leaving behind a fragmented mitochondrial genome (19).
Test of the validity of the CIItriplet rule for chloroplast genomes.
Did some of the mitochondrial genomes violate the CIItriplet rule because mitochondria are not autonomous organisms? To examine this question, I also evaluated 42 chloroplast genomes that are not autonomous organisms, as well. The examples included those of seed plants as examples of the highest evolved plants and of nonseed plants such as protists, algae, mosses, and ferns, ranging in size between 105 kb and 201 kb (average = 150 kb, SD = 21 kb). Despite their dependence on host cells, all 42 chloroplast genomes complied quite accurately with Chargaff's second parity rule. Their average degree of compliance was cWC = 0.990 (SD = 0.017), which was considerably better than a value of cWC = 0.93 that one would expect based on their average size of 150 kb.
Transposition/Inversion Hypothesis of the CIImono and CIItriplet Rules.
In agreement with the authors of earlier hypotheses about the rules, the present article assumes that all genomes initially violated the rules because they contained arbitrary numbers of single nucleotides and triplets on their (say) Watson strands. Only their subsequent evolution rendered them increasingly compliant with the CIImono and CIItriplet rules by a number of different mechanisms. Specifically, it will propose a mechanism that is based on inversions and inverted transpositions. These genome variations insert sections of a chromosome in reverse order in their original location (inversions) or somewhere else (inverted transpositions).
To be sure, the inversion of the base sequence itself would have no significance for validity of the rules if it were not for the necessity to swap strands. In other words, the particular strand of such an inversion that was part of a Watson strand before its excision has to be inserted into the Crick strand and vice versa. As will be shown below, this action must equalize in an asymptotic fashion the base composition and oligonucleotide composition of the genome in question.
Of course, the individual steps involved in the actual mechanism of inversions and transpositions (20) are much more complex. For example, retroposons such as L1 elements involve even RNA intermediates (21). Yet these complexities will be ignored because only net changes of the genome sequence matter for present consideration. Among these, however, the article will ignore small direct repeats and other short variations generated by inversions and inverted transpositions. They are usually <10 bases long and thus contribute very little to the overall nucleotide statistics compared with the often many 100-kb large inversions and 1- to 3-kb and larger transposons (15, 16).
Qualitative description of the transposition/inversion model in the case of the CIImono rule.
Assume, e.g., that initially the number of G's is much larger than the number of C's on a Watson strand. Therefore, because of base pairing the Crick strand contains correspondingly more C's than G's. Because of its strand-swapping effect, every randomly located transposition/inversion must carry some of the supernumerary G's from the Watson strand to the Crick strand while, at the same time, it carries some of the supernumerary C's from the Crick strand to the Watson strand. The result is an ongoing equalization of the numbers of G's with C's on both strands. In a similar way, the mechanism equalizes the numbers of A's and T's on each strand. In contrast, it does not equalize the numbers of G's with A's, G's with T's, etc., because they are not paired with each other in the inverted segments.
The process is effectively irreversible because the equalization caused by a certain transposition/inversion can be undone only by reversing it exactly immediately afterward. Such an exact reversion, however, is extremely unlikely to occur in the random fashion in which the transposition/inversions are assumed to happen.
The process is also self-stabilizing, because once a genome complies with Chargaff's second parity rules, the described mechanism maintains the compliance forever. In this case both strands of the inverted segment have, on average, equal numbers of complementary nucleotides, and thus it brings as many nucleotides into a strand as it takes away from it. Thus, compliance is a stable end state of genomes that are subjected to the process described by the transposition/inversion hypothesis.
Quantitative description of the transposition/inversion model in the case of the CIImono rule.
As shown in the supporting information, the described mechanism would lead to an exponential equalization of the numbers of complementary nucleotides on the (say) Watson strand described by the following equations.
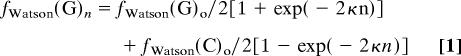 |
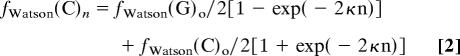 |
where the symbols mean the following: n, number of rounds of transposition/inversions; fWatson(G)o and fWatson(C)o, initial numbers of G's and C's on the Watson strand; fWatson(G)n and fWatson(C)n, final numbers of G's and C's on the Watson strand after n rounds of transposition/inversions; κ = λ/L, the rate of change; λ, average size of transposons/inversions (bases); L, total genome size (bases).
Hence, with increasing numbers n of transposition/inversions the numbers of G's and C's converge to the same value.
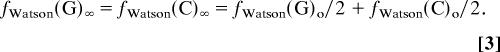 |
Thus, the resulting genome sequence eventually complies completely with the CIImono rule. The speed v of this convergence [i.e., v = (number of iterations needed to reach one-half of the final value)−1] is given by
Similar equations apply to the change of the numbers of A's and T's.
Computer simulation of the transposition/inversion model in the case of the CIImono rule.
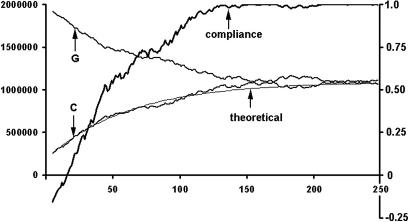
A typical simulation of the process is shown in Fig. 5. It was assumed that the initial number of G's was much larger than the number of C's and that the size of the average inverted segment is 50 kb in a genome of a size of 6 Mb. This size is unrealistically large for transposons but smaller than many inversions. Based on these parameters one can calculate the theoretical change of nucleotide numbers according to Eqs. 1 and 2 (thin line in Fig. 5). The figure shows that the theoretical curve is in excellent agreement with the simulation. The simulation also measured the changing degree of compliance of the increasingly changed genome sequence with the CIIoligo rule (thick line in Fig. 5 with the corresponding right hand ordinate). Based on the above parameters the initially noncompliant genome sequence (cWC = −0.16) became fully compliant (cWC = 0.99) after as little as 130 rounds of transposition/inversions.
Fig. 5.
Simulation of the convergence of a noncompliant genome to a compliant one by a recursive series of transposition/inversions. The abscissa shows the number of rounds of transposition/inversions, the left ordinate shows the number of G's or C's on the resulting Watson strand, and the right ordinate shows the degree of compliance of the resulting genome with the CIItriplet rule expressed as correlation coefficient cWC. The thick line labeled “compliance” depicts the simulated genome's degree of compliance with the CIItriplet rule as a function of rounds of transposition/inversions. The thinner lines labeled G and C depict the convergence of the numbers of the corresponding nucleotides during the same process. The thin line labeled “theoretical” depicts the theoretical curve of convergence calculated by Eq. 2. Note that this curve is not fitted to the simulation but merely uses the same value of (segment size)/(genome size). For the sake of graphic presentation the simulation assumed a large ratio of (size of average inverted segment)/(size of whole genome) of 0.008. It appears that the theoretical description matches quite accurately the exponential convergence of a noncompliant genome to a compliant one.
Extension of the transposition/inversion model to the CIItriplet rule.
As shown in supporting information, the almost identical arguments apply to reverse complementary triplets as applied to single bases. Again, each pair of initially very unequal numbers of reverse complementary triplets converged to a common number on each strand. The same rate of change and speed applies to each such pair as applied to each pair of reverse complementary nucleotides.
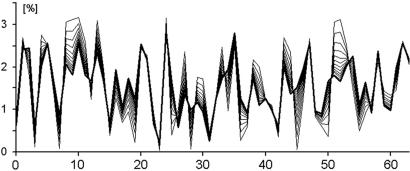
A simulation of this convergence from an arbitrary triplet profile (cWC = 0.09) to a fully compliant one (cWC = 0.993) is shown in Fig. 6. To accelerate the rate of the simulated conversion, the simulation assumed a value of κ = 0.1, which generated a compliant profile in only 12 rounds of transposition/inversion.
Fig. 6.
Simulation of the convergence of a noncompliant triplet profile to a compliant one by a recursive series of transposition/inversions. The abscissa shows all possible triplets encoded by their canonical numbers (see supporting information), and the ordinate shows the frequency of triplets (%). The figure plots into the same graph the converging series of triplet profiles starting with an initially arbitrary, noncompliant, simulated genome (cWC = 0.01) that converges to a compliant one (cWC = 0.994) during 12 recursive rounds of transposition/inversions. The final stage is marked by a thick line. For the same reasons as in Fig. 5, the simulation assumes a relatively large ratio of (inverted segment size)/(genome size) of 0.1. It appears that a recursive series of transposition/inversions as described quantitatively in supporting information is able to turn an initially noncompliant triplet profile into a compliant one.
The simulations demonstrated that any arbitrary initial triplet profile can be made compliant with the CIItriplet rule by the described transposition/inversion mechanism and, most importantly, that each initial triplet profile leads to a different final one. Expecting that different genomes had different evolutionary beginnings, one would expect that the compliant triplet profiles of the modern genomes were very different from each other. However, contrary to this expectation, most of the compliant genomes turned out to have very similar triplet profiles regardless of species and kingdom (unpublished data).
Discussion
It seems safe to assume that the evolution of genomes subjected them to many transposition/inversions. The very nature of transposable elements suggests the geometric growth of their numbers over time. Indeed, in some cases such as Alu, LINE, and SINE sequences, millions of copies were found in human, mouse, and other genomes, and they were found in coding and noncoding regions alike (15–18). It is not known exactly how many of these transposons were inverted, but the tacit assumption in the field seems to be that they are on par with the noninverted ones. Likewise, it is not known how many inversions any particular genome experienced. Yet it seems reasonable to conclude that most sufficiently large inversions transferred some coding sequences from the Watson strand to the Crick strand and vice versa. Because most genomes contain coding regions on both strands, one may infer that they also experienced a large number of inversions in their past, even if they are no longer recognizable today. In other words, it seems plausible that there were sufficiently many transposition/inversions to satisfy Eq. 4. As a consequence, the transposition/inversion hypothesis suggests that all genomes must have moved inevitably toward a stable state in which they complied with Chargaff's second parity rules.
Thus, the compliance with Chargaff's second parity rules may be interpreted as an inevitable, asymptotic product of (among other causes) numerous inversions and inverted transpositions that occurred in the course of evolution. The conversion of every initially noncompliant genome to a compliant one began presumably with relative small genomes like bacterial genomes, which gradually grew in size, while at the same time the described mechanism and the additional mechanisms described by earlier hypotheses improved their degree of compliance with the rules. As the inevitable consequence of transposition/inversions, the above mechanism changes all genomes indiscriminately. Therefore, the compliance of a genome with the rules seems to present no constraint and offers no selective advantage over less compliant ones.
The literature contains several examples of violators of the rules, notably certain mitochondria (our data), but also many viruses (e.g., ref. 6). As argued above in the case of mitochondria, small genome size or lack of genomic autonomy (i.e., dependence on a host cell genome) does not seem to explain the violations. Possible explanations for the violations may include the loss of genome material through horizontal gene transfer. Based on the hypothesis presented here, another explanation for violation may be the scarcity of transpositions/inversions in the violating mitochondrial and viral genomes.
The mathematical description (Eqs. 1 and 2) made the simplifying assumption that the mononucleotide and oligonucleotide composition of each inverted DNA segment of each transposition/inversion was that of the average of the whole genome. Consequently, the degree of compliance of all genomes with the rules increased monotonously with the number of transposition/inversions. In contrast, the simulation (Fig. 5) showed numerous jitters, indicating small temporary and local decreases of compliance. They are explained by the fact that many inverted segments must have originated in areas of the genome that were locally still less compliant than the average genome. In this way they decreased the overall level of compliance temporarily. Inevitably, though, as a genome becomes increasingly compliant, the amplitude of such jitters has to decrease steadily.
Because the described process is asymptotic in nature, no genome can ever become perfectly compliant by it. Nevertheless, as a genome experiences more and more transposition/inversions, their equalizing effect covers the entire length of the genome more and more completely. Thus, the areas where a genome still violates the rules must decrease steadily in size. In other words, consistent with the above results, the smaller a segment of a present-day genome the more likely it may still violate the rules to some degree.
Materials and Methods
The genomes used in this article included the entire human genome and several other genomes that were selected to cover a large range of species. If they exceeded 8 MB in size, the analysis program cut large chromosome sequences into 8-Mb segments. Therefore, a description like chimpanzee chr14 seg4 means that the sequence used was from chimpanzee chromosome 14 from 32 Mb to 40 Mb. The published sequences were considered Watson strands, and their complemented, inverted sequences were considered Crick strands. Because the present article constructed and evaluated in each case the complementary strand and evaluated both, our results are not affected by this problem. The individual organismal and oraganellar genomes used here are listed in supporting information. Before use, the published sequences were routinely reformatted by turning small and capital letters of nucleotides uniformly into the numbers 0,…,3. In addition, all N′s, spaces, and coordinate markers were deleted.
The investigative computer program dnaorg.exe was written by G.A.-B. using Visual C++ (Microsoft, Redmond, WA) and will be provided upon request.
Supplementary Material
Acknowledgments
I am very grateful to my wife, Dr. Veena Prahlad (Northwestern University), and my friends and colleagues, Drs. James Bartles and Richard Scarpulla (Northwestern University), for their patient criticism. I am also grateful for valuable comments from Drs. Howard Green (Harvard Medical School, Boston, MA) and Martin Zand (University of Rochester, Rochester, NY).
Footnotes
The author declares no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Watson JD, Crick FHC. Nature. 1953;177:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Fickett JW, Torney DC, Wolf DR. Genomics. 1992;13:1056–1064. doi: 10.1016/0888-7543(92)90019-o. [DOI] [PubMed] [Google Scholar]
- 3.Baisnée PF, Hampson S, Baldi P. Bioinformatics. 2002;188:1021–1033. doi: 10.1093/bioinformatics/18.8.1021. [DOI] [PubMed] [Google Scholar]
- 4.Prabhu VV. Nucleic Acids Res. 1993;21:2797–2800. doi: 10.1093/nar/21.12.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez J, Jose MV. Biochem Biophys Res Commun. 2002;299:126–134. doi: 10.1016/s0006-291x(02)02583-4. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell D, Bridge R. Biochem Biophys Res Commun. 2006;340:90–94. doi: 10.1016/j.bbrc.2005.11.160. [DOI] [PubMed] [Google Scholar]
- 7.Rudner R, Karkas JD, Chargaff E. Proc Natl Acad Sci USA. 1968;603:921–922. doi: 10.1073/pnas.60.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudner R, Karkas JD, Chargaff E. Proc Natl Acad Sci USA. 1968;603:915–920. doi: 10.1073/pnas.60.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell SJ, Forsdyke DR. J Theor Biol. 1999;197:63–76. doi: 10.1006/jtbi.1998.0858. [DOI] [PubMed] [Google Scholar]
- 10.Bell SJ, Forsdyke DR. J Theor Biol. 1999;1971:51–61. doi: 10.1006/jtbi.1998.0857. [DOI] [PubMed] [Google Scholar]
- 11.Forsdyke DR. J Mol Evol. 1995;41:573–581. doi: 10.1007/BF00175815. [DOI] [PubMed] [Google Scholar]
- 12.Forsdyke DR, Bell SJ. Appl Bioinformatics. 2004;31:3–8. doi: 10.2165/00822942-200403010-00002. [DOI] [PubMed] [Google Scholar]
- 13.Lobry JR. J Mol Evol. 1999;166:719–723. doi: 10.1093/oxfordjournals.molbev.a026156. [DOI] [PubMed] [Google Scholar]
- 14.Dang KD, Dutt PB, Forsdyke DR. Biochem Cell Biol. 1998;76:129–137. doi: 10.1139/o97-095. [DOI] [PubMed] [Google Scholar]
- 15.Simons C, Pheasant M, Makunin IV, Mattick JS. Genome Res. 2006;16:164–172. doi: 10.1101/gr.4624306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert N, Labuda D. Proc Natl Acad Sci USA. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mighell AJ, Markham AF, Robinson PA. FEBS Lett. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 19.Lang BF, Gray MW, Burger G. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 20.McClintock B. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 21.Martin SL, Li W-LP, Furano AV, Boissinot S. Cytogenet Genome Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.