Abstract
The tet(W) gene is associated with tetracycline resistance in a wide range of bacterial species, including obligately anaerobic rumen bacteria and isolates from the human gut and oral mucosa. However, little is known about how this gene is disseminated and the types of genetic elements it is carried on. We examined tetracycline-resistant isolates of the animal commensal and opportunistic pathogen Arcanobacterium pyogenes, all of which carried tet(W), and identified three genetic elements designated ATE-1, ATE-2, and ATE-3. These elements were found in 25%, 35%, and 60% of tetracycline-resistant isolates, respectively, with some strains carrying both ATE-2 and ATE-3. ATE-1 shows characteristics of a mobilizable transposon, and the tet(W) genes from strains carrying this element can be transferred at low frequencies between A. pyogenes strains. ATE-2 has characteristics of a simple transposon, carrying only the resistance gene and a transposase, while in ATE-3, the tet(W) gene is associated with a streptomycin resistance gene that is 100% identical at the DNA level with the aadE gene from the Campylobacter jejuni plasmid pCG8245. Both ATE-2 and ATE-3 show evidence of being carried on larger genetic elements, but conjugation to other strains was not observed under the conditions tested. ATE-1 was preferentially associated with A. pyogenes strains of bovine origin, while ATE-2 and ATE-3 elements were primarily found in porcine isolates, suggesting that these elements may circulate in different environments. In addition, four alleles of the tet(W) gene, primarily associated with different elements, were detected among A. pyogenes isolates.
Bacterial tetracycline resistance is widespread in nature and is particularly prominent in bacterial isolates from animals (8). The latter is a likely consequence of the use of tetracycline and its derivatives as feed additives for livestock, either for the prevention of disease or as growth promotants (34, 35). While several mechanisms of bacterial tetracycline resistance exist, the most common is that conferred by tet genes of the ribosomal protection class. Among the commonly found genes in this class is tet(M), which is thought to be prevalent because of its association with transposable elements, particularly those of the Tn916 family (8). More recently, Tet W has emerged as a widespread determinant, particularly among bacterial isolates of mucosal surfaces. This determinant was originally described for Butyrivibrio fibrisolvens and several other anaerobes from human and animal gastrointestinal tracts (4, 28, 29, 31) but has since been described for a number of bacterial species isolated from the human oral mucosa (36).
Arcanobacterium pyogenes is a common inhabitant of the mucous membranes of the upper respiratory, gastrointestinal, and urogenital tracts of a number of domestic animal species (13). It is also an important opportunistic pathogen, particularly in cattle, capable of causing a number of suppurative infections of the joints and visceral organs (7). As a mucosal commensal, A. pyogenes is exposed to antimicrobial feed additives used for growth promotion and disease prevention in animals. Therefore, it is no surprise that nearly 42% of A. pyogenes isolates are resistant to tetracycline and the derivatives commonly used in agriculture, chlortetracycline and oxytetracycline (33). We have previously demonstrated that all tetracycline-resistant A. pyogenes isolates (MIC ≥ 4 μg/ml) carry a tet(W) gene (6). A second resistance determinant, Tet 33, has been detected in some isolates, but the resistance it confers results in a tetracycline MIC of only 1 μg/ml; thus, the strains are not considered resistant (14).
Of 20 tetracycline-resistant A. pyogenes isolates, five carried tet(W) on a genetic element which could be transferred at low frequencies between A. pyogenes strains (6), and we have designated this element ATE-1 (Arcanobacterium tetracycline resistance element-1). For ATE-1, tet(W) is associated with a functional mob gene and an origin of transfer, oriT, which are presumably responsible for its ability to transfer between strains (6). Despite the widespread nature of tet(W), little is known about the genetic elements on which it is carried. In B. fibrisolvens, the tet(W) gene is associated with the conjugative transposons TnB123 and TnB1230, which confer high-frequency transfer of tet(W) to recipient strains (4, 28, 29). Nucleotide sequence from TnB1230 indicates that this transposon shares transfer genes similar to those of the Enterococcus faecalis conjugative transposon Tn1549 (21). A recent study suggests that, in addition to the B. fibrisolvens elements, a Bifidobacterium longum element in which tet(W) is associated with a transposase can be transferred to other bacterial strains (17). In four other gastrointestinal bacterial species, tet(W) is associated with orfY, a gene which is found on a number of mobile elements. However, these species were unable to transfer tet(W) in the laboratory (17). In this paper we report the presence of tet(W) on at least three genetic elements in A. pyogenes and carriage of distinct tet(W) alleles.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli DH5αMCR (Gibco-BRL) derivatives used in this study were grown either on LB agar or in LB broth (Difco) at 37°C. A. pyogenes isolates used in this study were obtained from veterinary diagnostic laboratories or personal culture collections in North America and include 20 tetracycline-resistant isolates (7 of bovine origin, 12 of porcine origin, and 1 from a macaw) and 10 tetracycline-susceptible isolates (6 of bovine origin and 4 of porcine origin). All of these are independent isolates and likely represent different strains based on U.S. state and animal host of isolation and the presence or absence of virulence genes such as cbpA (9) and nanP (15). A. pyogenes strains (Table 1) were grown on brain heart infusion (BHI) agar (Difco), supplemented with 5% citrated bovine blood, at 38°C and 5% CO2 in a humidified incubator. Liquid cultures of A. pyogenes were grown in BHI broth supplemented with 5% fetal bovine serum (Omega Scientific Inc.) at 37°C aerobically with shaking. Media were supplemented when appropriate with antibiotics at the following concentrations: for E. coli, chloramphenicol at 30 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 10 μg/ml; for A. pyogenes, erythromycin at 15 μg/ml and tetracycline at 5 μg/ml.
TABLE 1.
A. pyogenes strains
| Strain | Relevant characteristics | Source or referencea |
|---|---|---|
| BBR1 | Bovine isolate, Tetr, ATE-1 | 5 |
| OX4 | Porcine isolate, Tetr, ATE-2 | Oxford Laboratories, Worthington, MN |
| OX9 | Porcine isolate, Tetr, ATE-2 and ATE-3 | Oxford Laboratories, Worthington, MN |
| 52785-99 | Porcine isolate, Tetr, ATE-3 | Rollins ADDL, Raleigh, NC |
| 3274 | Bovine isolate, Tets | CSU, Fort Collins, CO |
| JGS610 | nanH::erm(X) derivative of strain 3274, Ermr | 6 |
ADDL, Animal Disease Diagnostic Laboratory; CSU, Colorado State University.
DNA techniques.
Procedures for E. coli transformation and plasmid extraction, DNA restriction, ligation, agarose gel electrophoresis, Southern blotting, and DNA dot blotting were performed essentially as described previously (2). Genomic DNA was prepared from A. pyogenes strains by the method of Pospiech and Neumann (24). Preparation of DNA probes, DNA hybridization, and probe detection were performed using a digoxigenin DNA labeling and detection kit (Roche Molecular Biochemicals). Primers for construction of an ATE-2 tnp-specific gene probe for dot blot analysis and a tet(W)-specific gene probe for Southern blot analysis are shown in Table 2. PCR DNA amplification was performed using Taq DNA polymerase (Promega) for 35 cycles consisting of 1 min at 94°C (DNA denaturation), 1 min at 55°C (primer annealing), and 1 min/kb at 72°C (DNA synthesis). Primers for delineation of the ATE-1 element, and linkages between tet(W) and the ATE-2 tnp gene or the ATE-3 aadE gene, are shown in Table 2. PCR amplification for nucleotide sequencing over the region covering the frameshift in the OX9 aadE gene was performed with primers tetw45 and tetw46 (Table 2). A 1,771-bp fragment of the tet(W) gene from all 20 tetracycline-resistant A. pyogenes strains was sequenced from PCR products amplified with primers tetw18 and tetw28 (Table 2).
TABLE 2.
PCR primers used in this study
| Purpose | Primer | Sequence | Region covered (bases) | Amplicon (bp) |
|---|---|---|---|---|
| Delineation of ATE-1 (A)a | tetw5 | 5′-CCCGAGGCCAAACACTGCGAGC-3′ | 511-532 of guaA | 489 |
| tetw82 | 5′-CGTGCCCTGCACGAGAAACTTG-3′ | 978-999 of guaA | ||
| Delineation of ATE-1 (B) | tetw21 | 5′-TGAGTACCGCAGAGTGTCGCAG-3′ | 38-59 3′ of guaA | 782 |
| tetw44 | 5′-ACTACGAGACGGATTTCAGGAC-3′ | 383-404 of orf171 | ||
| Delineation of ATE-1 (C) | tetw33 | 5′-GCAGGAAGGTAACGAAGATGGG-3′ | 464-485 of int | 534 |
| ATE1-1 | 5′-CGATACTTTCATGTCCCACCCG-3′ | 976-997 of int | ||
| Delineation of ATE-1 (D) | tetw22 | 5′-TGCCTGGCAGCGTCCGTCCGTG-3′ | 260-281 3′ of tet(W)-3 | 522 |
| tetw4 | 5′-AGGGCCAAGACCGCCGAGTTCC-3′ | 97-118 of orf110 | ||
| Delineation of ATE-1 (E) | tetw24 | 5′-ATAGCCGAGCTCTCCGAGTCTG-3′ | 28-49 of orf87 | 303 |
| tetw36 | 5′-ATGTGCATTGAGTGCGAGGCTG-3′ | 309-330 of orf209 | ||
| Delineation of ATE-1 (F) | tetw76 | 5′-TCGCCCAGCCCTACCAGACCTT-3′ | 116-137 of doc | 212 |
| tetw83 | 5′-TCAGCGACACCGAGCATAGCTGAG-3′ | 304-327 of doc | ||
| tnp-specific gene probe | tnp1 | 5′-GACGAGCTGTCTGGTAAACTTC-3′ | 15-36 of tnp | 822 |
| tnp2 | 5′-TAAATCTCAATGTCCTGCCTGC-3′ | 815-836 of tnp | ||
| tet(W)-specific gene probe | tetw18 | 5′-GACAACGAGAACGGACACTATG-3′ | 117-138 of tet(W) | 1,246 |
| tetw3 | 5′-AAGCGGGAGCGGCGTAACAGAC-3′ | 1,341-1,362 of tet(W) | ||
| tet(W)-tnp linkage | ATE2-3 | 5′-GTCGGCAAGCCAGTCATCCAGC-3′ | 1,840-1,861 of tet(W) | 998 |
| tnp2 | 5′-TAAATCTCAATGTCCTGCCTGC-3′ | 815-836 of tnp | ||
| tet(W)-aadE linkage | tetw10 | 5′-GTGTGCCTGACGGAACTGAAAG-3′ | 1,804-1,825 of tet(W) | 1,398 |
| tetw59 | 5′-ACTACCACTTGCAGAAGCCTAC-3′ | 415-436 of aadE | ||
| aadE sequencing | tetw45 | 5′-AGCCCGAAGATGTTAATGTGCC-3′ | 215-236 5′ of aadE | 500 |
| tetw46 | 5′-ATGAAAATCCCTTTTCTACAGC-3′ | 243-264 of aadE | ||
| tet(W) sequencing | tetw18 | 5′-GACAACGAGAACGGACACTATG-3′ | 117-138 of tet(W) | 1,843 |
| tetw28 | 5′-CGCAATAGCCAGCAATGAACGC-3′ | 18-39 3′ of tet(W) |
Letters in parentheses refer to the bars indicating the fragments amplified in Fig. 1.
Cloning of A. pyogenes tet(W) genes.
The tet(W) gene of a bovine A. pyogenes isolate, BBR1, was previously cloned in the plasmid pJGS279, a pBC KS (Stratagene) derivative containing a 12.1-kb insert (6). The tet(W) genes from A. pyogenes strains OX4, OX9, and 52785-99 were cloned from libraries of SacI-digested genomic DNA from the respective strain constructed in the vector pHSS20, pHSS21 (22), or pBC KS. The ligation mixtures were introduced into E. coli strain DH5αMCR by electroporation, and tetracycline-resistant transformants were selected on LB agar containing 10 μg/ml tetracycline. Recombinant plasmids containing A. pyogenes tet(W) genes were designated pJGS338 (OX4), pJGS464 (ATE-2, OX9) and pJGS468 (ATE-3, OX9), and pJGS529 (52785-99).
Nucleotide sequence determination.
Nucleotide sequencing of tetracycline resistance determinants was performed on a 377A DNA sequencer (Applied Biosystems Inc.) at the Genomic Analysis and Technology Core at the University of Arizona. Sequence was determined from both strands, crossing all restriction sites.
Computer analysis.
Nucleotide sequence data was compiled using the Sequencher 4.5 program (GeneCodes). Database searches were performed using the BlastN, BlastX, BlastP, and CD-search algorithms (1, 20). Sequence analysis was performed using the suite of programs available through the Genetics Computer Group, Inc. (Accelyrs). Similarity was determined from optimized sequence alignments by use of the CLUSTAL W program (32).
A. pyogenes filter matings.
Tetracycline-resistant A. pyogenes isolates were used in filter matings with the erythromycin-resistant, tetracycline-sensitive A. pyogenes recipient strain JGS610, as previously described (6). Briefly, donor and recipient strains were grown under appropriate selection conditions to an optical density at 600 nanometers (OD600) of 1.0 (∼109 bacterial cells/ml) in a Beckman DU-64 spectrophotometer with a 1 cm cell path length. The two cultures (0.5 ml of each) were mixed and filtered using a 0.45 μm filter. Following overnight incubation on a BHI-5% blood agar plate, the cells were resuspended from the filter in BHI broth. Serial dilutions were plated onto BHI-5% blood agar supplemented with 5 μg/ml tetracycline and 15 μg/ml erythromycin to select for transconjugants. Bacterial viable counts were obtained from identically treated filters containing either the donor or recipient strain alone plated on the appropriate media. Conjugation frequencies were expressed as transconjugants per donor cell recovered following overnight incubation (∼2 × 109 bacteria) and determined as averages of the results of at least three independent experiments.
Nucleotide sequence accession numbers.
The nucleotide sequences of the ATE-1 element from A. pyogenes BBR1, the ATE-2 element from A. pyogenes OX4, and the ATE-3 elements from A. pyogenes OX9 and 52785-99 have been deposited in the GenBank nucleotide sequence database under the accession numbers AY049983, DQ517519, DQ519394, and DQ519395, respectively.
RESULTS AND DISCUSSION
Nucleotide sequence of the ATE-1 element.
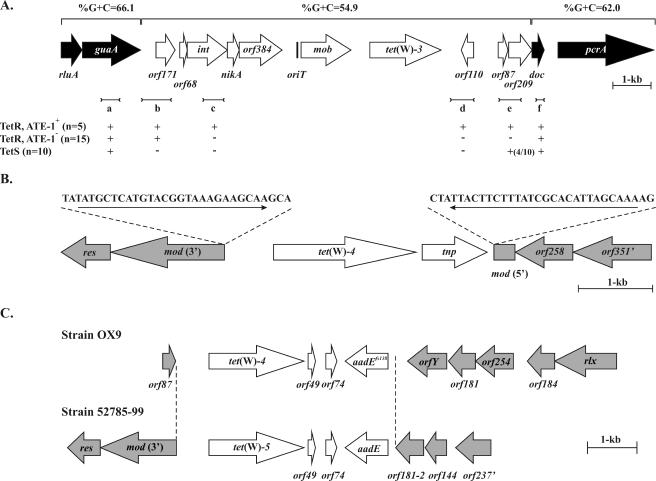
A gene map of the nucleotide sequence of the A. pyogenes BBR1 ATE-1 element is shown in Fig. 1A. This sequence was determined from pJGS279, and an adjacent 4 kb of DNA sequence, identified in a draft genome sequence of strain BBR1 generated at 454 Life Sciences (S. J. Billington and B. H. Jost, unpublished data), was confirmed by sequencing of overlapping 1-kb PCR products. As previously described, immediately upstream of tet(W) are a mob gene and its concomitant oriT site (6). In the sequence 5′ of oriT were seven open reading frames (ORFs), all oriented in the same direction as mob and tet(W) (Fig. 1A). At the left end of the sequence were two putative housekeeping genes, rluA and guaA, encoding homologues of pseudouridine synthase (conserved domain match PSRA_1; E value = 1 × 10−23) and a bifunctional GMP synthase/glutamine amidotransferase (conserved domain matches GMP_synthase_C [E value = 4 × 10−123] and GATase1_GMP_synthase [E value = 5 × 10−63]), respectively. Given their housekeeping nature, these two genes are likely part of the A. pyogenes genome flanking the left boundary of ATE-1.
FIG. 1.
Genetic organization of the ATE-1, ATE-2, and ATE-3 elements of A. pyogenes. (A) Gene map of the BBR1 ATE-1 element. ORFs and their orientations are represented by the arrows. ORFs designated by white arrows are located in the proposed ATE-1 sequence, while black ORFs at the left and right ends of the sequence are likely found in all A. pyogenes strains. Above the gene map the percentages of G+C content of the proposed ATE-1 sequence and the flanking sequences are shown. The results of PCRs (a to f) using primers (Table 2) designed to delineate ATE-1 are shown below the gene map. (B) Gene map of the OX4 ATE-2 element. White ORFs are located in the ATE-2 sequence, while shaded ORFs are present in flanking sequences. The sequences of the 26-bp imperfect inverted repeats which delineate ATE-2 are shown above the gene map. The mod ORF is disrupted by ATE-2, and both the 5′ and 3′ ends of the ORF are shown. (C) Gene maps of the OX9 and 52785-99 ATE-3 elements. The extent of ATE-3 sequence, as determined by comparison between the OX9 and 52785-99 sequences, is indicated by the dotted lines. The mod ORF is truncated at the 5′ end by ATE-3. In each figure the scale is indicated by the 1-kb bar.
Downstream of guaA were two ORFs, orf171 and orf68. The latter ORF encodes a 68-amino-acid helix-turn-helix motif protein with 23.5% to 32.4% identity to a group of small proteins, including Gp27 from the Clostridium perfringens bacteriophage φ3626 (38) and hypothetical proteins from Enterococcus faecalis (GenBank accession no. AAO82256) and Haemophilus somnus (GenBank accession no. ZP_00122065). int, which overlapped orf68, encodes a protein with similarity to DNA integrases, including those from bacteriophage φ3626 (43.9% identity, 65.4% similarity), the C. perfringens CW459 tet(M) element (28.0% identity, 51.1% similarity), and the Staphylococcus aureus pathogenicity island SaPIbov (23.5% identity, 48.1% similarity) (10, 26, 38). These elements use the 3′ end of guaA as an integration site, a site now recognized as a hot spot for site-specific recombination (18), which is consistent with the location of ATE-1 downstream of guaA.
Between int and mob were two ORFs, nikA and orf384 (Fig. 1A). NikA showed similarity to the putative Listeria monocytogenes strain 4b H7858 NikA protein (44.3% identity, 66.0% similarity) and MobC (23.8% identity, 42.6% similarity) from the Lactococcus lactis plasmid pSK11A (30), suggesting that its function may be in mobilization. Orf384 shared low-level similarity with DNA replication and repair proteins. orf384 is separated from mob by a 510-bp intergenic sequence that contains the oriT of ATE-1 (6). The 5.7 kb downstream of tet(W) contained five ORFs, including pcrA, a likely housekeeping gene, which encoded a protein with similarity to homologues of the PcrA helicases, most notably from Streptomyces coelicolor (GenBank accession no. CAB92660; 52.1% identity and 68.5% similarity). The doc gene encoded a protein with similarity to “death on curing” prophage addiction proteins, including that from Clostridium tetani (GenBank accession no. AAO36443; 33.9% identity and 48.0% similarity), while orf110, orf87, and orf209 have no known homologues.
Delineation of ATE-1 sequences.
The G+C contents of rluA and guaA (left end) and doc and pcrA (right end) were consistent with those previously observed for A. pyogenes housekeeping genes (27). However, a decreased G+C content of 54.9% from orf171 through orf209 (Fig. 1A) was consistent with the integration of ATE-1 at the 3′ end of guaA. PCR analysis was performed on the 20 tetracycline-resistant isolates and 10 tetracycline-susceptible isolates with a set of primers designed to cover the putative ATE-1 sequences. While guaA-specific primers amplified a product from all 30 strains, primers specific for int, or a fragment extending from downstream of tet(W) into orf110, amplified a product only from the 5 ATE-1 strains (Fig. 1A). doc-specific primers amplified a product from all 30 strains, suggesting that this gene is not part of ATE-1 (Fig. 1A). These results, in addition to the hybridization of a mob-specific probe only to ATE-1 strains (6), suggest that ATE-1 extends at least from int to orf110. Primers designed to amplify a fragment extending from immediately downstream of guaA into orf171 amplified this fragment in all tetracycline-resistant isolates, not just ATE-1 strains, but not in tetracycline-susceptible isolates (Fig. 1A). Conversely, all ATE-1 strains gave positive results in a PCR designed to amplify an orf87-orf209 fragment (Fig. 1A), and all results for tetracycline-resistant, non-ATE-1 isolates were negative, but the results for 4 of 10 tetracycline-susceptible isolates were positive. These results confound the delineation of the element, and while it is likely that the 3′ end of guaA represents the insertion site of ATE-1, remnants of ATE-1 or similar elements may exist in some non-ATE-1 A. pyogenes isolates.
The presence of genes involved in site-specific integration and mobilization, but not conjugation, in ATE-1 suggests that it represents a mobilizable transposon, which may explain its low frequency of transfer (6).
Cloning of the tet(W) gene from a tetracycline-resistant, ATE-1-negative A. pyogenes isolate.
As only 25% of tetracycline-resistant A. pyogenes isolates appear to contain ATE-1 (6), the tet(W) gene was cloned from the tetracycline-resistant, but ATE-1-negative, porcine isolate OX4. Nucleotide sequence of the OX4 tet(W) gene indicated that it was more similar (99.9% DNA identity) to the tet(W) gene of Bifidobacterium sp. strain ISO3519 (GenBank accession number AF202986) than it was to that of BBR1 (91.9% DNA identity). This divergence was also reflected at the protein level, with the OX4 Tet(W) protein having 99.8% identity to that of Bifidobacterium sp. strain ISO3519 but only 89.7% identity to the BBR1 Tet(W) protein. tet(W)-1 and tet(W)-2 alleles have been designated for Megasphaera elsdenii (31) and are substantially different from the two A. pyogenes alleles. Therefore, we have designated the BBR1 allele tet(W)-3 and the OX4 allele tet(W)-4. The differences between tet(W)-3 and tet(W)-4 are evenly distributed over the entire length of tet(W), and there is no evidence of a mosaic structure to these alleles. Unlike that of ATE-1, the sequence upstream of tet(W) in OX4 shares considerable (96.3%) DNA identity with the core tet(W) upstream region previously described for a number of gastrointestinal isolates (17).
Identification of ATE-2.
Nucleotide sequencing of an 8-kb SacI fragment containing the OX4 tet(W)-4 allele indicated that this gene was arranged in an operon with tnp (Fig. 1B). Tnp shared similarity with site-specific recombinases, in particular, the central regions of the large clostridial resolvases, TnpX from C. perfringens Tn4451 (31.3% identity and 54.6% similarity over amino acids 250 to 537 of TnpX) (3) and TndX from C. difficile Tn5397 (30.1% identity and 53.4% similarity over amino acids 252 to 530 of TndX) (37). However, this region of TnpX is required for dimerization and DNA binding but not for its resolvase activity (19).
tet(W) and tnp form a genetic element, designated ATE-2, which was bound by 25-bp imperfect inverted repeats (Fig. 1B). ATE-2 was inserted within an ORF, mod, which would encode a protein similar to the modification enzymes of type III restriction-modification systems, including that of Clostridium thermocellum (GenBank accession no. ZP_00504330; 44.2% identity, 64.0% similarity), and is immediately followed by the gene for its cognate restriction enzyme, res. The mod and res genes and other sequences which flank ATE-2 were not identified in the draft genome sequence of the ATE-1-carrying strain, BBR1, and their G+C contents are similar to that of ATE-2 (49.9%), which is much lower than the genome average. Therefore, while ATE-2 has characteristics of a simple transposon, it is possible that ATE-2 is part of a larger genetic element.
Distribution of ATE-2.
Genomic DNAs from the 20 tetracycline-resistant A. pyogenes isolates and 10 tetracycline-susceptible isolates were screened by DNA dot blotting using a tnp-specific gene probe (Table 2) as a marker for ATE-2 (data not shown). None of the five ATE-1 strains or the tetracycline-susceptible isolates hybridized to the tnp-specific probe. Seven of the remaining 15 tetracycline-resistant isolates hybridized, indicating that 35% (7/20) of tetracycline-resistant A. pyogenes strains carried the ATE-2 element. Linkage of tet(W)-4 and tnp in these strains was confirmed by PCR (Table 2 and data not shown).
Some ATE-2 strains carry two tet(W) genes.
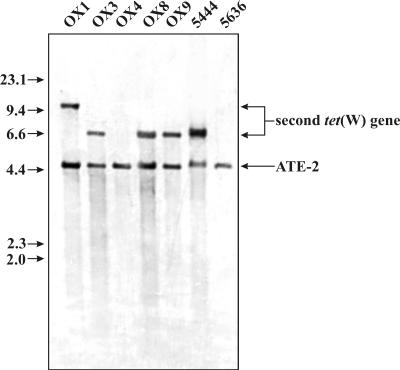
EcoRI-digested genomic DNA from each of the seven ATE-2 strains was subjected to Southern blotting with a tet(W)-specific probe (Fig. 2). All seven strains showed a hybridizing band of 5 kb consistent with the presence of the ATE-2 element. However, five of the seven strains also showed an additional band of either 7 kb or 11 kb, indicating the presence of a second copy of tet(W).
FIG. 2.
Some ATE-2 positive A. pyogenes isolates carry a second tet(W) gene. The results of Southern blot analysis of EcoRI-digested A. pyogenes genomic DNA from ATE-2-positive isolates with a digoxigenin-labeled tet(W) gene probe are shown. The A. pyogenes strains are indicated above the lanes. DNA size standards in kilobases are shown to the left of the blot, while the positions of bands corresponding to the ATE-2 element and the second tet(W) gene are indicated on the right.
Identification of ATE-3.
To investigate the additional tet(W) genes in ATE-2 strains, the tet(W) genes of the porcine strain OX9 were cloned. Two types of recombinant plasmid were obtained. Nucleotide sequencing of one plasmid type indicated that it contained ATE-2 inserted at a position identical to that found in OX4 (data not shown). In addition, the OX9 ATE-2-associated tet(W)-4 allele differed by only a single base pair from that of OX4. The second plasmid type contained a tet(W)-4 allele identical to that of ATE-2 in this strain. However, this second tet(W)-4 was associated with a defective streptomycin resistance gene, aadEfs138, in an element designated ATE-3 (Fig. 1C). This ATE-3 element may be derived from ATE-2, as the sequences from 661 bp upstream of tet(W)-4 to 99 bp downstream of tet(W)-4 are 99.9% identical between the two elements. The potential product of the aadEfs138 would be 100% identical to AadE encoded by the Campylobacter jejuni plasmid pCG8245 (23), but a frameshift at nucleotide 138 puts the remainder of the coding sequence out of frame. Interestingly, strain OX9 is resistant to streptomycin (MIC > 256 μg/ml), and sequencing of a PCR product that flanked the frameshift gave a sequence consistent with both the frame shift and a functional sequence, suggesting that there is a second functional aadE gene in OX9. This suggestion was supported by PCR sequencing of other strains carrying this gene, which gave only sequences consistent with a functional gene. The region of the ATE-3 element, from 150 bp upstream of aadE through the end of the gene, is also 100% identical to pCG8245 at the DNA level (with the exception of the aadE frameshift base deletion). These data indicate recent horizontal transfer of this aadE gene between gram-negative and gram-positive bacteria. In ATE-3, the 3′ ends of tet(W) and aadE are separated by orf49 and orf74. The genes flanking the tet(W)-aadE region are of unknown function. However, orfY, encoding a putative methyltransferase, has previously been identified in A. pyogenes strain OX7 as a gene into which an erm(B) element has inserted (16). In OX7, ATE-3 is located at the same position as in OX9 and is thus clustered with the erm(B) element (16). orfY, with near DNA identity (but designated mte), is also present as part of the Bacteroides uniformis conjugative transposon CTnBST, where it is interrupted by a Tn10-like element (12). Proteins with approximately 28% similarity to OrfY are encoded adjacent to the tet(W) genes of several animal and human gastrointestinal bacterial species. These tet(W)/orfY sequences are presumably mobile, given their high sequence identity between several species (17). orf181 has an unannotated homologue with 100% DNA identity in CTnBST (GenBank accession no. AY345595) encoded upstream of mte and three homologues on pCG8245 (23). These data, along with the lack of ATE-3 flanking genes in the BBR1 draft genome sequence and their relatively low G+C contents, suggests that ATE-3 may be part of a larger element. This suggestion is given some credence by the presence of a putative mobilization gene, rlx, ∼3 kb downstream of ATE-3 (Fig. 1C). Rlx has a conserved domain associated with relaxases (conserved domain match pfam03432 relaxases; E value = 1 × 10−9) and low similarity to a number of Mob and Rlx proteins, including that of the S. aureus plasmid pS194 (25).
Distribution of ATE-3.
An ATE-3-specific PCR, linking tet(W) and aadE, was performed with each of the 20 tetracycline-resistant isolates (data not shown). Products were obtained from each of the five ATE-2 isolates which contained a second tet(W) gene and from seven of eight strains which lacked both ATE-1 and ATE-2. Products were not obtained from any ATE-1-containing strain. The clustering of tetracycline and streptomycin resistances on ATE-3, along with the frequency of ATE-3 among tetracycline-resistant A. pyogenes isolates (60%), may explain the high correlation of tetracycline and streptomycin resistance previously observed (11).
ATE-3 can be found at multiple genetic loci.
To examine the characteristics of an ATE-3 element from a non-ATE-2 strain, the ATE-3 element was cloned from strain 52785-99. Nucleotide sequencing indicated that the organization of ATE-3 in 52785-99 was identical to that in OX9 (Fig. 1C), with similarity between the two sequences extending 4,433 bp from 663 bp 5′ to tet(W) to 154 bp 5′ to aadE, with only 51 bp of divergence between the two sequences. One of these base changes included correction of the aadE frameshift present in the OX9 ATE-3 element. The 52785-99 tet(W) gene differed nearly 1% in DNA sequence from the tet(W)-4 alleles of strains OX4 and OX9 and was thus designated tet(W)-5. In strain 52785-99, the ATE-3 element is flanked by the 3′ end of mod, as is ATE-2 in OX4. However, the 5′ end of mod is not located at the other end of ATE-3, suggesting some rearrangement, perhaps associated with ATE-3 insertion. The other end of ATE-3 is flanked by a homologue of orf181 which is 89.2% identical to that in strain OX9. Sequences flanking the 52785-99 ATE-3 element also have G+C contents of <50% and are not present in the BBR1 draft genome sequence, suggesting that ATE-3 may be carried by more than one larger genetic element.
Transferability of ATE-2 and ATE-3 elements.
To assess the transferability of ATE-2 and ATE-3, strains OX4 (ATE-2), OX9 (ATE-2 and ATE-3), and 52785-99 (ATE-3) were used as donors in mating experiments with the recipient strain, JGS610 (Table 1). Tetracycline-resistant, erythromycin-resistant transconjugants were not detected from any of these mating experiments, suggesting that ATE-2 and ATE-3 are not capable of conjugative transfer under the conditions used or that the transfer frequency was <5 × 10−10 transconjugants/donor cell. Transconjugants were detected at a frequency of 9 × 10−9 transconjugants/donor cell from matings between the ATE-1-positive strain BBR1 and JGS610.
Relationships between ATEs and tet(W).
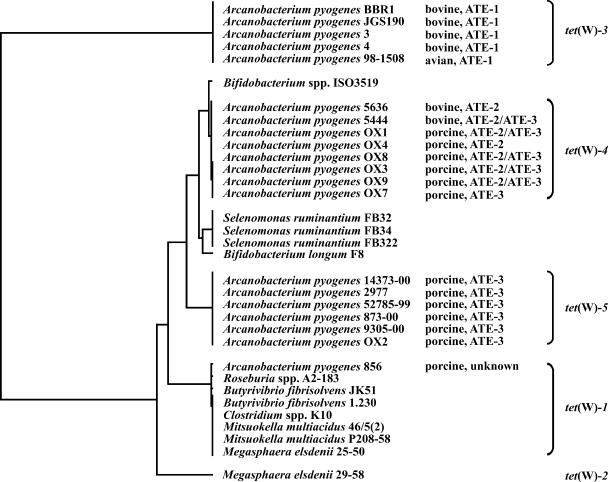
Nucleotide sequence variations in the tet(W) genes from different tetracycline-resistant A. pyogenes strains led to the identification of three tet(W) alleles. The tet(W) gene was amplified and sequenced from each of the 20 tetracycline-resistant A. pyogenes isolates, and a comparison of these sequences is shown as a dendrogram in Fig. 3. In general, particular tet(W) alleles were associated with particular ATEs. All strains which carried ATE-1 had tet(W)-3, while all strains which carried ATE-2 had tet(W)-4. Strains which carried both ATE-2 and ATE-3 gave a single unambiguous tet(W)-4 sequence, suggesting that both the ATE-2 and ATE-3 elements in these strains carried this allele. With the exception of strain OX7, ATE-3 strains consistently carried tet(W)-5. One A. pyogenes tetracycline-resistant isolate, 856, which did not carry ATE-1, ATE-2, or ATE-3, carried a tet(W) gene which differed by a single base from the M. elsdenii 25-50 tet(W)-1 allele (31). No obvious differences in levels of resistance between strains carrying different alleles, including those that carry two copies of tet(W), have been observed (33).
FIG. 3.
Phylogenetic analysis of a 1,771-bp fragment of tet(W), showing the relationships between the tet(W) genes of A. pyogenes isolates and those from other bacterial species. The figure shows an unrooted dendrogram of tet(W) sequences from A. pyogenes strains determined in this study, Bifidobacterium sp. strain ISO3519 (GenBank accession number AF202986), Bifidobacterium longum F8 (GenBank accession number DQ294299), Butyrivibrio fibrisolvens strains JK51 (GenBank accession number AJ427421) and 1.230 (GenBank accession number AJ222769), Clostridium sp. strain K10 (GenBank accession number AY601650), Megasphaera elsdenii strains 25-50 (GenBank accession AY485125) and 29-55 (GenBank accession number AY485124), Mitsuokella multiacidus strains 46/5(2) (GenBank accession number AJ427422) and P208-58 (GenBank accession number AY603069), Roseburia sp. strain A2-183 (GenBank accession number AJ421625), and Selenomonas ruminantium strains FB32 (GenBank accession number DQ294296), FB34 (GenBank accession number DQ294297), and FB322 (GenBank accession number DQ294295) compiled using PAUPsearch and Genetics Computer Group software. The host of origin of each A. pyogenes strain is indicated along with the associated ATE elements. Groups of strains carrying each of the tet(W) alleles are also indicated.
Relationship between ATEs and the host source of the isolate.
A general association exists between the host species of tetracycline-resistant A. pyogenes isolates and the type of ATE present (Fig. 3). ATE-1 was present in four of seven bovine isolates but in no porcine isolates. ATE-2 was present in 5 of 12 porcine isolates and 2 of 7 bovine isolates, while ATE-3 was present in 11 of 12 porcine isolates but only 1 of 7 bovine isolates. ATE-2 and ATE-3 were found in combination in 4 of 12 porcine isolates and 1 of 7 bovine isolates. Of the seven bovine isolates examined, four contained ATE-1, one contained ATE-2 alone, and one contained ATE-2 in combination with ATE-3. Of the 12 porcine isolates, 1 contained ATE-2 alone and 7 contained ATE-3 alone, with the remaining 4 possessing both ATE-2 and ATE-3. The single avian isolate contained ATE-1.
Conclusions.
tet(W) has recently been recognized as a widely distributed gene conferring tetracycline resistance to a number of bacterial species (4, 28, 29, 31, 36). However, little information is available about the types of genetic elements involved in its dissemination (17, 21). We have identified three genetic elements which carry tet(W) in A. pyogenes isolates. These elements are transferable, or show genetic signs of mobility, but differ from those elements that carry tet(W) in other species and carry tet(W) alleles not identified in other bacteria. These data suggest that there may be some constraints on the transfer of these elements to other bacteria. Indeed, there appears to be considerable host species preference for A. pyogenes isolates to carry certain ATE elements, suggesting that these elements may be circulating predominantly in certain environments.
Acknowledgments
This work was supported in part by National Research Initiative Competitive Grants Program/U.S. Department of Agriculture award 99-35204-7818.
We thank Ryan Knoper for performing PCR analysis to delineate ATE-1 and Maricela Pier for excellent technical assistance. We are grateful to Kasia Kazimierczak and Karen Scott (Rowett Research Institute, Aberdeen, United Kingdom) for providing sequence information and data prior to publication. Part of this work was performed in the laboratory of Glenn Songer.
Footnotes
Published ahead of print on 11 September 2006.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology, vol. 1. Greene Publishing Associates and John Wiley and Sons, Inc., New York, N.Y.
- 3.Bannam, T. L., P. K. Crellin, and J. I. Rood. 1995. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site specific recombinase excises a circular transposon molecule. Mol. Microbiol. 16:535-551. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 5.Billington, S. J., B. H. Jost, W. A. Cuevas, K. R. Bright, and J. G. Songer. 1997. The Arcanobacterium (Actinomyces) pyogenes hemolysin, pyolysin, is a novel member of the thiol-activated cytolysin family. J. Bacteriol. 179:6100-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billington, S. J., J. G. Songer, and B. H. Jost. 2002. Widespread distribution of a Tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 46:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, G. R., and M. M. Chengappa. 1991. Essentials of veterinary bacteriology and mycology, 4th ed. Lea and Febiger, Philadelphia, PA.
- 8.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esmay, P. A., S. J. Billington, M. A. Link, J. G. Songer, and B. H. Jost. 2003. The Arcanobacterium pyogenes collagen binding protein, CbpA, promotes adhesion to host cells. Infect. Immun. 71:4368-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, J. R., S. R. Monday, T. J. Foster, G. A. Bohach, P. J. Hartigan, W. J. Meaney, and C. J. Smyth. 2001. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J. Bacteriol. 183:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guérin-Faublée, V., J. P. Flandrois, E. Broye, F. Tupin, and Y. Richard. 1993. Actinomyces pyogenes: susceptibility of 103 clinical animal isolates to 22 antimicrobial agents. Vet. Res. 24:251-259. [PubMed] [Google Scholar]
- 12.Gupta, A., H. Vlamakis, N. Shoemaker, and A. A. Salyers. 2003. A new Bacteroides conjugative transposon that carries an ermB gene. Appl. Environ. Microbiol. 69:6455-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jost, B. H., and S. J. Billington. 2005. Arcanobacterium pyogenes: molecular pathogenesis of an animal opportunist. Antonie Leeuwenhoek 88:87-102. [DOI] [PubMed] [Google Scholar]
- 14.Jost, B. H., A. C. Field, H. T. Trinh, J. G. Songer, and S. J. Billington. 2003. Tylosin resistance in Arcanobacterium pyogenes is encoded by an Erm X determinant. Antimicrob. Agents Chemother. 47:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jost, B. H., J. G. Songer, and S. J. Billington. 2002. Identification of a second Arcanobacterium pyogenes neuraminidase and involvement of neuraminidase activity in host cell adhesion. Infect. Immun. 70:1106-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jost, B. H., H. T. Trinh, J. G. Songer, and S. J. Billington. 2004. A second tylosin resistance determinant, Erm B, in Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 48:721-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazimierczak, K. A., H. J. Flint, T. M. Barbosa, and K. P. Scott. 2006. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob. Agents Chemother. 50:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavigne, J. P., A. C. Vergunst, G. Bourg, and D. O'Callaghan. 2005. The IncP island in the genome of Brucella suis 1330 was acquired by site-specific integration. Infect. Immun. 73:7779-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucet, I. S., F. E. Tynan, V. Adams, J. Rossjohn, D. Lyras, and J. I. Rood. 2005. Identification of the structural and functional domains of the large serine recombinase TnpX from Clostridium perfringens. J. Biol. Chem. 280:2503-2511. [DOI] [PubMed] [Google Scholar]
- 20.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melville, C. M., R. Brunel, H. J. Flint, and K. P. Scott. 2004. The Butyrivibrio fibrisolvens tet(W) gene is carried on the novel conjugative transposon TnB1230, which contains duplicated nitroreductase coding sequences. J. Bacteriol. 186:3656-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickoloff, J. A., and R. J. Reynolds. 1991. Subcloning with new ampicillin- and kanamycin-resistant analogs of pUC19. BioTechniques 10:469-472. [PubMed] [Google Scholar]
- 23.Nirdnoy, W., C. J. Mason, and P. Guerry. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob. Agents Chemother. 49:2454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 25.Projan, S. J., S. Moghazeh, and R. P. Novick. 1988. Nucleotide sequence of pS194, a streptomycin-resistance plasmid from Staphylococcus aureus. Nucleic Acids Res. 16:2179-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts, A. P., P. A. Johanesen, D. Lyras, P. Mullany, and J. I. Rood. 2001. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology 147:1243-1251. [DOI] [PubMed] [Google Scholar]
- 27.Rudnick, S. T., B. H. Jost, J. G. Songer, and S. J. Billington. 2003. The gene encoding pyolysin, the pore-forming toxin of Arcanobacterium pyogenes, resides within a genomic islet flanked by essential genes. FEMS Microbiol. Lett. 225:241-247. [DOI] [PubMed] [Google Scholar]
- 28.Scott, K. P., T. M. Barbosa, K. J. Forbes, and H. J. Flint. 1997. High-frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 63:3405-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siezen, R. J., B. Renckens, I. van Swam, S. Peters, R. van Kranenburg, M. Kleerebezem, and W. M. de Vos. 2005. Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl. Environ. Microbiol. 71:8371-8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanton, T. B., J. S. McDowall, and M. A. Rasmussen. 2004. Diverse tetracycline resistance genotypes of Megasphaera elsdenii strains selectively cultured from swine feces. Appl. Environ. Microbiol. 70:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trinh, H. T., S. J. Billington, A. C. Field, J. G. Songer, and B. H. Jost. 2002. Susceptibility of Arcanobacterium pyogenes to tetracyclines, macrolides and lincosamides. Vet. Microbiol. 85:353-359. [DOI] [PubMed] [Google Scholar]
- 34.U.S. Department of Agriculture. 2000. Feedlot '99 part III: health management and biosecurity in U.S. feedlots, 1999. USDA:APHIS:VS, CEAH, no. N336.1200. National Animal Health Monitoring System, U.S. Department of Agriculture, Fort Collins, CO.
- 35.U.S. Department of Agriculture. 1996. Swine '95 part II: reference of 1995 U.S. grower/finisher health & management practices. USDA:APHIS:VS, CEAH. National Animal Health Monitoring System, U.S. Department of Agriculture, Fort Collins, CO.
- 36.Villedieu, A., M. L. Diaz-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, H., A. P. Roberts, D. Lyras, J. I. Rood, M. Wilks, and P. Mullany. 2000. Characterization of the ends and target sites of the novel conjugative transposon Tn5397 from Clostridium difficile: excision and circularization is mediated by the large resolvase, TndX. J. Bacteriol. 182:3775-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmer, M., S. Scherer, and M. J. Loessner. 2002. Genomic analysis of Clostridium perfringens bacteriophage φ3626, which integrates into guaA and possibly affects sporulation. J. Bacteriol. 184:4359-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]