Abstract
Resistance to macrolides and ketolides occurs mainly via alterations in RNA moieties of the drug-binding site. Using an A2058G mutant of Mycobacterium smegmatis, additional telithromycin resistance was acquired via deletion of 15 residues from protein L22. Molecular modeling, based on the crystal structure of the large ribosomal subunit from Deinococcus radiodurans complexed with telithromycin, shows that the telithromycin carbamate group is located in the proximity of the tip of the L22 hairpin-loop, allowing for weak interactions between them. These weak interactions may become more important once the loss of A2058 interactions destabilizes drug binding, presumably resulting in a shift of the drug toward the other side of the tunnel, namely, to the vicinity of L22. Hence, the deletion of 15 residues from L22 may further destabilize telithromycin binding and confer telithromycin resistance. Such deletions may also lead to notable differences in the tunnel outline, as well as to an increase of its diameter to a size, allowing the progression of the nascent chain.
Ribosomes provide a target for several antibiotic families, among which is the macrolide-ketolide group. High-resolution crystal structures showed that macrolides and their derivatives bind to a specific pocket of the nascent protein exit tunnel (2, 3, 7, 16, 20, 21, 25, 29), the universal feature of the large ribosomal subunit through which nascent proteins emerge. The same pocket is exploited by all members of the macrolide family, and effective inhibitory action is achieved when the drug consumes a significant portion of the tunnel cross-section (1, 31, 32). Typically, resistance to macrolides is acquired through either efflux or target-based alteration (methylation or mutation of nucleotides involved in drug binding [for reviews, see references 11 and 28]).
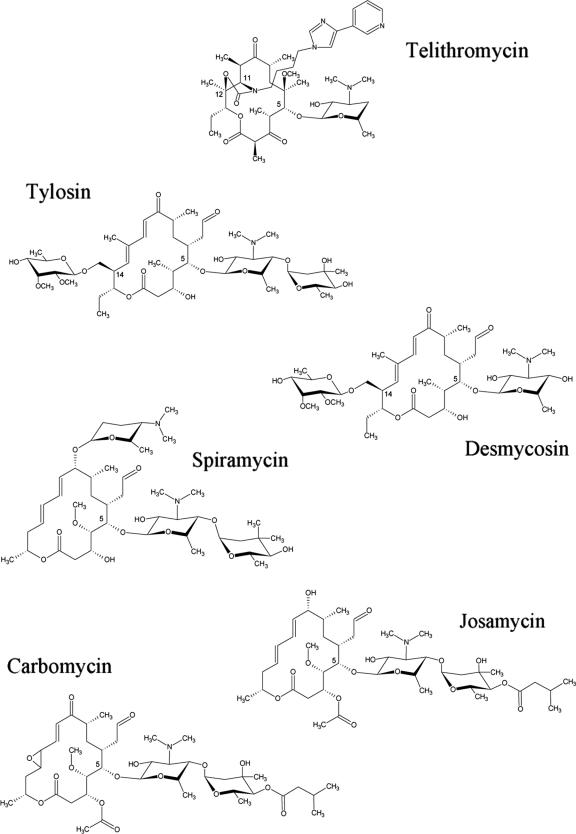
Ketolides are an advanced generation of the macrolide antibiotics, which, in part, provide activity against macrolide-resistant pathogens. They are semisynthetic derivatives of erythromycin, the first macrolide in use. Like erythromycin, ketolides are composed of a 14-membered macrolactone ring (Fig. 1). However, their macrolactone ring lacks a cladinose sugar and possesses a keto group at position 3, a cyclic carbamate, and an extended arm. Ketolides and macrolides share a similar, albeit not identical inhibitory mechanism. Owing to their more elaborated chemistry, in addition to their binding to the macrolide pocket, the ketolides extend further into the tunnel and interact with rather remote sites (2, 20, 29). Among ketolides, telithromycin carries an aryl-alkyl extension bound to its cyclic carbamate (Fig. 1). The crystal structure of the large ribosomal subunit from Deinococcus radiodurans (called D50S) in complex with telithromycin indicates that telithromycin interacts with 23S RNA domain II nucleotides (2, 29). This finding is consistent with a number of previous biochemical and mutagenesis studies showing that, in addition to nucleotides in domain V, domain II nucleotide A752 (Escherichia coli numbering is used throughout, unless mentioned otherwise) is protected by telithromycin from dimethylsulfate in footprinting studies and that a deletion in helix H35 of domain II strongly influences drug resistance (4, 13, 30).
FIG. 1.
Chemical structures of the ketolides and 16-membered macrolides used in the present study.
Telithromycin can exert its action on most macrolide-resistant Streptococcus pneumoniae strains (6). Nevertheless, resistance to telithromycin, though still not fully understood, has been observed in bacterial pathogens. In particular, telithromycin resistance mutations of the 23S RNA were identified in domains II and V (4, 24, 30). To a lesser extent, mutations in the ribosomal proteins L4 and L22 have been associated with ketolide resistance (4, 24, 27).
In the present study, we analyzed the occurrence of mutations related to telithromycin resistance in Mycobacterium smegmatis by using the crystal structure of its complex with the large ribosomal subunit of Deinococcus radiodurans. The choice of these organisms is justified by the high similarity between their ribosomes to those of bacterial pathogens. We report mutations in domain V of the 23S RNA and a deletion of 15 amino acid residues in the ribosomal L22. The latter is the largest deletion in protein L22 thus far identified that is associated with resistance to macrolides or ketolides.
MATERIALS AND METHODS
Bacterial strains.
The single rRNA M. smegmatis allelic mc2155 SMR5 rrnB minus mutant strain (18) and its derivative the M. smegmatis mc2155 SMR5 rrnB A2058G mutant strain (16) were used to select for spontaneous telithromycin-resistant mutants. The strains were cultivated in LB medium containing 0.05% Tween 80. For the selection of telithromycin-resistant mutants LB medium was supplemented with 1.5% agar and telithromycin (32 μg ml−1 for M. smegmatis mc2155 SMR5 rrnB and 512 μg ml−1 for the M. smegmatis mc2155 SMR5 rrnB A2058G mutant). Drug-resistant mutants were colony purified on LB medium supplemented with 1.5% agar. The strains studied are listed in Table 1.
TABLE 1.
Strains used in this study
| Mutant straina | Source or reference |
|---|---|
| mc2155 SMR5 rrnB* | 16 |
| mc2155 SMR5 rrnB 2058G† | 16 |
| mc2155 SMR5 rrnB 2058C† | 16 |
| mc2155 SMR5 rrnB 2058G/2059G‡ | 16 |
| mc2155 SMR5 rrnB 2058G/rplV ΔIle85-Arg99‡ | This study |
*, Parental strain; †, recombinant mutants of M. smegmatis mc2155 SMR5 rrnB; ‡, spontaneous mutants of M. smegmatis mc2155 SMR5 rrnB 2058G mutant.
DNA techniques.
The primers used for the amplification of domains II and V of 23S rRNA and for the amplification of rplD (coding for ribosomal protein L4) and rplV (coding for ribosomal protein L22) are given in Table 2. Nucleic acid sequencing was done with fluorescence-labeled nucleotides and the Taq cycle sequencing system of Applied Biosystems. rplD was wild type in all strains, and mutational alterations were limited to rplV and 23S rRNA as specified in the text.
TABLE 2.
Primers used in this study
| Primera | Sequence (5′ to 3′) | Nucleotide position
|
|
|---|---|---|---|
| Location | Positions | ||
| 23S rRNA (lrs)* | |||
| #625 | GGC GTC TGG GGG GAA CGC GG | 23S rRNA (lrs) | 168-187 |
| #86 | GGA GGT AGA GCT ACT GGA TGG | 23S rRNA (lrs) | 855-874 |
| #696 | CCA TCC AGT AGC TCT ACC TCC | 23S rRNA (lrs) | 874-855 |
| #85 | TAC GGC TAC CTT CCT GCG TC | 23S rRNA (lrs) | 1444-1425 |
| #601 | GTA GCG AAA TTC CTT GTC GGG TA | 23S rRNA (lrs) | 1929-1951 |
| #603 | GGT GGG TAG TTT AAC TGG GG | 23S rRNA (lrs) | 2233-2252 |
| #604 | CGC GCG GCG GAT AGA GAC CG | 23S rRNA (lrs) | 2625-2606 |
| Downstream of 5S rRNA* | |||
| #630A | TCT CAC GGG TTA TGG GGG CGG CCT CG | Downstream of rrnA | 155-130 |
| L22 (rplV)† | |||
| #737 | CGC ACG TTC AAG GGT CAC AT | Upstream of rplV | 47-28 |
| #738 | TAC TGC TTG TCG GCG TAC CA | Downstream of rplV | 82-63 |
| L4 (rplD)† | |||
| #775 | CGG CAA GAC GGA CGG TTC TG | rplD | 33-52 |
| #739 | TGT TCA AGG GCA CCC GCA TG | Upstream of rplD | 207-187 |
| #740 | CCG TAC GAC TTC TCC GAG AT | Downstream of rplD | 64-45 |
*, E. coli numbering (with exception of downstream region of rrn operon A which is numbered according to contig:3563:m_smegmatis, positions 5023050 to 5025075 from www.tigr.org); †, numbered according to contig:3563:m_smegmatis, positions 1537075 to 1541332 from www.tigr.org).
Determination of the MICs.
MIC tests were done in a 200-μl plate format using liquid LB medium supplemented with 0.05% Tween 80 (16). The antibiotics to be tested were added in a twofold series of dilutions ranging from 1,024 to 0.25 μg/ml. The drugs were obtained from Pfizer (carbomycin), Sigma (josamycin, spiramycin, tylosin), Eli Lilly (desmycosin), and Aventis Pharma (telithromycin). The MIC is defined as the drug concentration at which the growth of the cultures is completely inhibited after 72 h of incubation at 37°C, corresponding to 24 generations.
Molecular modeling.
The effect of the mutations identified on the ribosome structure was studied by molecular modeling using the program “O” (10). In the case of the Ile85-Arg99 deletion, the structure of L22 from M. smegmatis was modeled by using that of L22 from D. radiodurans as a template. This allowed the identification of nucleotides neighboring the Ile85-Arg99 region in M. smegmatis. The Ile85-Arg99 deletion was then modeled, followed by a stereochemical regularization of the protein structure. To compare drug locations, superposition of the large subunits D50S and H50S was performed by using the program LSQMAN (12).
RESULTS AND DISCUSSION
Isolation of mutants.
M. smegmatis mc2155 SMR5 rrnB (18) was used to select for mutants resistant to more than 64 μg of telithromycin/ml (for the chemical structure of telithromycin compared to the classical macrolides, see Fig. 1); mutants were obtained with a frequency of 4 × 10−9. M. smegmatis mc2155 SMR5 rrnB A2058G mutant was used to select for mutants resistant to more than 512 μg of telithromycin/ml; mutants were obtained with a frequency of 3 × 10−7.
Genetic characterization of the mutants.
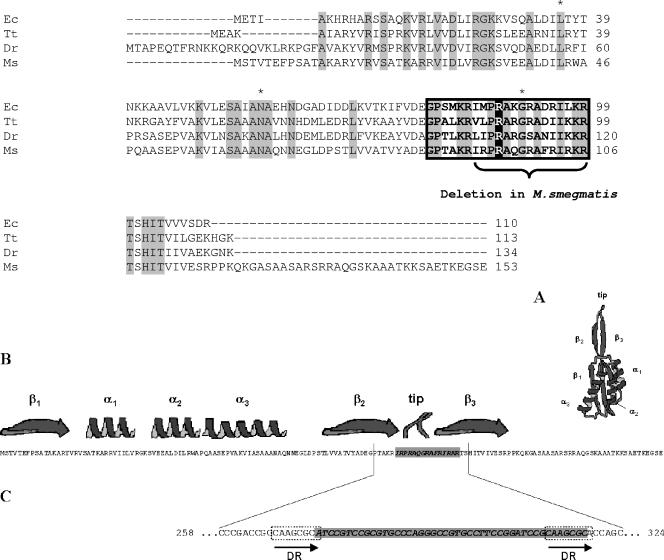
Based on the available crystal structures of D50S complexed with ketolide antibiotics (2, 20), we assumed that the resistance mutations should localize to 23S rRNA domains II and V. Gene amplification of corresponding regions of the 23S rRNA gene and subsequent sequence determination revealed that the telithromycin resistance phenotype was associated with a single point mutation in domain V in M. smegmatis rrnB, i.e., either 2058C (six of nine isolates) or 2058G (three of nine isolates). Thirteen of the fifteen telithromycin-resistant isolates obtained from M. smegmatis rrnB A2058G mutant demonstrated the double mutation 2058G/2059G. In two of fifteen of the analyzed mutants, no additional sequence alteration within 23S rRNA domains II or V other than the parental 2058A→G alteration was found, but we observed a deletion in rplV that resulted in the loss of 15 amino acids from protein L22 (Ile85-Arg99) (Fig. 2). Inspection of the nucleotides around the deleted DNA region revealed the presence of flanking direct repeats (Fig. 2), suggesting that either homologous recombination or slipped stranded mispairing (23) resulted in the loss of residues between Ile85 and Arg99.
FIG. 2.
(Top) Multiple sequence alignment of protein L22 of Escherichia coli (Ec; GenBank accession no. X02613), Thermus thermophilus (Tt; GenBank accession no. X84708), Deinococcus radiodurans (Dr; GenBank accession no. AE001892), and Mycobacterium smegmatis (Ms; GenBank accession no. Y13227). The sequences were aligned by using the CLUSTAL W algorithm (17). Amino acids identical to the consensus sequence provided by the CCD (accession no. pfam00237.11) are highlighted with gray boxes (14). Residues forming the conserved β-hairpin (positions 79 to 99 according to E. coli numbering) are shown within the black box with the tip of the hairpin highlighted in black. The region deleted in the telithromycin-resistant M. smegmatis strains is indicated by a brace. (Bottom panel) (A) Ribbon presentation of the overall fold of L22 from M. smegmatis (in black) based on its structure within the large subunit of D. radiodurans (in gray) (9). (B) The secondary structure elements are indicated above the amino acid sequence of the L22 protein of M. smegmatis (GenBank accession no. Y13227). The amino acids deleted in the telithromycin-resistant mc2155 SMR5 rrnB 2058G/rplV ΔIle85-Arg99 mutant are highlighted by a gray box. (C) DNA sequence of M. smegmatis around the location of the deletion (gray box). The deleted nucleotides are flanked by direct repeats (DR, open boxes).
Physiological investigations.
We next determined MICs to telithromycin and several 16-membered-ring macrolides (Table 3). As previously shown (16), A2058C confers high-level resistance to telithromycin (relative resistance [RR] > 4,096) and results in a significant resistance to spiramycin, josamycin, and carbomycin but has only a small effect on the interactions of tylosin and desmycosin with the ribosome. The A2058G mutation marginally alters the susceptibility of the ribosome to 16-membered macrolides but confers significant resistance to telithromycin (RR = 512). The double mutation 2058G/2059G results in high-level resistance to all ketolides and 16-membered macrolides. For M. smegmatis A2058G/rplV ΔIle85-Arg99, a fourfold increase in resistance to telithromycin, tylosin, and desmycosin is found compared to M. smegmatis A2058G. In contrast, the deletion in rplV did not affect the MICs for josamycin, carbomycin, and spiramycin. We also found that the generation time of the 23SrRNA A2058G/rplV ΔIle85-Arg99 double mutant is similar to that of strain M. smegmatis A2058G (data not shown), indicating that the partial rplV deletion does not grossly affect protein biosynthesis.
TABLE 3.
Drug susceptibility of M. smegmatis mutants
| Antibiotic | MIC (μg/ml) and RRa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| WT MIC |
M. smegmatis mc2155
|
M. smegmatis mc2155 2058G
|
|||||||
| 2058Cb
|
2058Gb
|
2059Gb
|
ΔL22
|
||||||
| MIC | RR | MIC | RR | MIC | RR | MIC | RR | ||
| Telithromycin | 0.25 | >1,024 | >4,096 | 128 | 512 | >1,024 | >4,096 | 512-1,024 | 2,048-4,096 |
| Tylosin | 16 | 64 | 4 | 32 | 2 | >1,024 | >64 | 128 | 8 |
| Desmycosin | 16 | 256 | 16 | 128 | 8 | >1,024 | >64 | 512 | 32 |
| Spiramycin | 32 | >1,024 | >32 | 256 | 8 | >1,024 | >32 | 256 | 8 |
| Josamycin | 4 | 1,024 | 256 | 32 | 8 | >1,024 | >256 | 32 | 8 |
| Carbomycin | 2 | 512 | 256 | 64 | 32 | >512 | >256 | 64 | 32 |
WT, wild type. The RR was calculated by dividing MIC for the mutant by the MIC for the wild type.
These data have been reported previously (16) and were experimentally confirmed in the present study. They have been included for comparison.
Structural basis for telithromycin resistance.
Molecular modeling, based on the crystal structure of telithromycin complexed with the ribosomal large subunit of D. radiodurans, provides a feasible structural basis for the telithromycin resistance mechanisms conferred by the alterations isolated. Thus, both mutations of nucleotide A2058 observed in the M. smegmatis rrnB mutant, namely, A2058G or A2058C, disturb the interactions of telithromycin with the nucleotide at position 2058, either by steric hindrance (A→G mutation) or by placing the antibiotic molecule distant from position 2058, thus hampering optimal interactions (A→C mutation). Both mutations are therefore likely to decrease the binding affinity of telithromycin (Table 3). Nucleotide 2058 was found to play a key role in the binding of 14-membered macrolides and ketolides (19). Consistently, crystal structures of complexes of macrolides or ketolides, with eubacterial ribosomes possessing an adenosine at position 2058, show that A2058 is implicated in hydrogen bonding with the drug (2, 3, 16, 20, 21, 29).
As shown in Table 3, the A2058G mutation alone provokes significant resistance to telithromycin (MIC of 128 μg/ml). When M. smegmatis A2058G was subjected to selection with 512 μg of telithromycin/ml, resistance was accompanied by additional mutations. One of the additional mutations was found to be 2059A→G (16): consistent with its contribution to macrolides or ketolides binding observed in complexes of the eubacterial large ribosomal subunit (2, 3, 20, 21), the additional A2059G should result in a steric hindrance and binding destabilization. These results, as well as those described below, suggest the existence of multiple steps alongside several types of mutations that can yield telithromycin resistance.
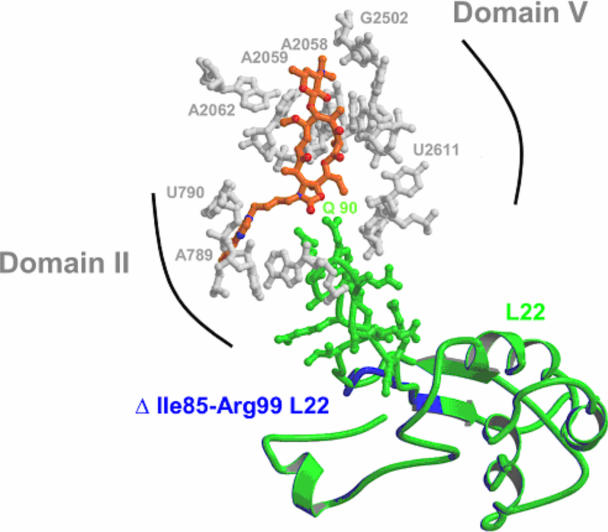
The other mechanism for additional resistance involves the deletion of 15 residues in the protein L22 (Ile85-Arg99), most of which line the D50S tunnel (9), in a location that is rather close to the ketolides long arm (Fig. 3) (2, 21). The A→G mutation of 2058, the key nucleotide for macrolide/ketolide binding, which leads to additional space consumption, should cause a shift in telithromycin position, thus decreasing telithromycin affinity to the 2058 region of the macrolide binding pocket and may, at the same time, facilitate the formation of new drug contacts. The most likely direction of this shift is toward the other side of the tunnel, proximal to the L22 hairpin tip. Consequently, telithromycin may interact with L22, and these interactions may become rather critical for its binding to eubacterial A2058G mutated ribosomes.
FIG. 3.
Modeled telithromycin (in orange)-binding site in M. smegmatis. Note the tight packing of the desosamine sugar against the rRNA bases A2058 and A2059 (in gray). The modeled mutated (blue) and intact (green) ribosomal forms of protein L22 are superimposed.
We analyzed the resistance mechanism mediated by the deletion in ribosomal protein L22, by modeling its M. smegmatis structure using the structure of L22 in D. radiodurans as a template. Protein L22 is composed of a globular domain, a long N-terminal extension, and a long β-hairpin, which is a constituent of the ribosome exit tunnel. The Ile85-Arg99 stretch belongs to the tip of L22 β-hairpin (Fig. 2 and 3) and is involved in intensive interactions with the 23S rRNA (for a list of nucleotides located within 4 Å of the region Ile85-Arg99, see Table 4). The modeled L22 protein from M. smegmatis shows that a few direct interactions between telithromycin and the L22 β-hairpin are possible and that the L22 residue closest to telithromycin is Gln90, whose side chain is located 4 Å from the drug (Fig. 3). The L22 region Ile85-Arg99 is positively charged; as such, it may contribute to stabilize the local RNA architecture via coulomb effects. As shown in Table 4, in the crystal structure of the telithromycin/D50S complex, the region Ala89-Gly91 interacts with domain II nucleotides U747, G748, A750, and A751 (2, 29). This region was also found to play a role in telithromycin binding by biochemistry, site-directed mutagenesis, and studies of clinical isolates with acquired drug resistance (4, 5, 8, 15, 26, 27, 30). Besides broadening the tunnel cross-section, a deletion of Ile85-Arg99 in L22 may cause a structural rearrangement of these nucleotides, with a resulting decrease in telithromycin binding affinity.
TABLE 4.
Nucleotides located within 4 Å from the region L22 Ile85-Arg99
| L22 residue | 23S RNA nucleotide(s)a |
|---|---|
| Ile85 | A1630 (A1614) |
| Arg86 | A1281 (A1268), G1338 (U1325), U1994 (U2011), G1995 (G2012) |
| Pro87 | A1630 (A1614), C1631 (C1615) |
| Arg88 | U760 (U747), G761 (G748), A1281 (A1268) |
| Ala89 | U760 (U747), G761 (G748), A763 (A750) |
| Gln90 | A764 (A751) |
| Gly91 | A764 (A751) |
| Arg92 | A1630 (A1614), A1997 (A2014), A1998, (A2015) |
| Phe94 | A1996 (A2013), A1997 (A2014) |
| Arg95 | A1996 (A2013) |
| Ile96 | G1995 (G2012), A1996 (A2013) |
| Arg97 | G1995 (G2012), A1996 (A2013), A1997 (A2014) |
| Lys98 | G1336 (C1323), G1337 (G1324), G1995 (G2012) |
| Arg99 | G1995 (G2012), A1996 (A2013) |
According to D. radiodurans numbering. The corresponding E. coli numbers are shown in parentheses.
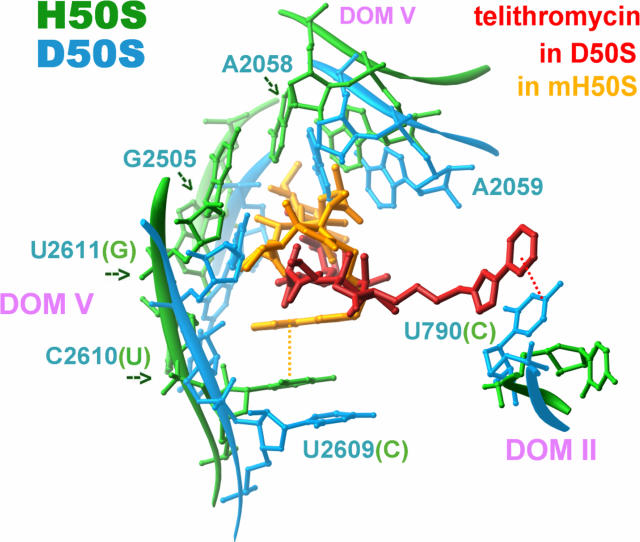
The only structural study that challenges the interaction of telithromycin with the 23S RNA domain II, in the proximity of L22 hairpin tip, is based on the crystal structure of telithromycin in complex with the G2058A mutant of the large ribosomal subunit from the archaeon Haloarcula marismortui, called here mH50S (25). Since interactions with domain II nucleotides have neither been observed in the crystal structure of this complex nor been identified by footprinting experiments (A. Mankin, unpublished observations), this structure cannot provide a structural explanation as to the L22 deletion. It is conceivable, however, that the lack of telithromycin contacts with domain II is ascribed, partially, to the different conformations of domain II nucleotides involved in telithromycin binding in eubacteria and archaea (Fig. 4). Thus, in the D50S complex nucleotide 790 forms stacking interactions with the aryl-alkyl arm of telithromycin, thus stabilizing its conformation. A similar conformation of the drug's aryl-alkyl arm would not benefit from this stabilizing interaction in the mH50S complex, since the mH50S equivalent to nucleotide 790 is flipped away from the position that could facilitate interactions with the telithromycin aryl-alkyl arm (Fig. 4). An additional reason for the inconsistency between the large volume of biochemical and crystallographic studies may be linked to the high salinity essential for H. marismortui optimal growth and integrity (22), which may mask several potential ribosomal entities that could have interacted with the drug.
FIG. 4.
Comparison of telithromycin-binding modes in D50S (binding pocket in light blue; telithromycin in red) and in mH50S (binding pocket in green; telithromycin in dark yellow). The nucleotide (U, C, and G) in parentheses corresponds to the H. marismortui sequence in case it is not identical to that of D. radiodurans. Note the different stacking interactions of telithromycin's aryl-alkyl arm in the two structures.
Structural basis for resistance to 16-membered lactone-ring macrolides.
All 16-membered lactone ring macrolides share a larger lactone ring compared to the 14-membered lactone ring. The additional two atoms seem to allow for higher conformational flexibility and also provide more potential interactions with the ribosome. Both properties should increase binding to altered ribosomes, since the higher flexibility and the increased number of interactions with the binding pocket can stabilize the drug binding and thus compensate for the loss of contacts with a mutated or methylated nucleotide at position 2058.
Among the 16-membered macrolides, tylosin and desmycosin carry a mycinose sugar at position 14 of their macrolactone rings, whereas spiramycin, carbomycin, and josamycin lack this sugar (Fig. 1). Structurally, it was shown that tylosin binds to D50S (29) and to the large ribosomal subunit of H. marismortui, H50S (7), in a similar fashion, with its mycinose sugar involved in interactions both with L22 and with domain II nucleotides. Hence, both binding modes are consistent with the resistance conferred by the Ile85-Arg99 L22 deletion (Table 3).
Our results show that the 16-membered ring macrolides that lack the mycinose sugar (i.e., spiramycin, josamycin, and carbomycin) are more susceptible to the A2058C mutation compared to tylosin and desmycosin, indicating that the mycinose sugar can serve as an additional interacting moiety. These findings are in accord with the structures of their complexes with H50S (7). The finding that the Ile85-Arg99 L22 deletion did not confer resistance to spiramycin, josamycin, and carbomycin, is also in line with this interpretation.
Conclusions.
Based on the findings reported here, supported by a large volume of biochemical and genetic evidence, as well as by crystallographic observations, we conclude that domain II and protein L22 can stabilize the binding of telithromycin and 16-membered lactone rings containing a mycinose sugar. Consistently, these interactions may become imperative as a result of the A2058G mutation and thus can play a role in resistance to these antibiotics. Our results suggest that the L22 deletion causes alterations in the tunnel cross-section and eliminates possible direct drug-ribosome interactions, thus portraying the way that the L22 deletion exerts its effects in the presence of a mutated 2058A.
Acknowledgments
Funds were provided by the Swiss National Science Foundation (E.C.B.), L.R.5 Regione Campania (R.B.), the University of Zurich (E.C.B.), the U.S. National Institutes of Health (GM34360 [A.Y.]), and the Kimmelman Center for Macromolecular Assembly (A.Y.). A.Y. holds the Helen and Martin S. Kimmel Professorial Chair.
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Auerbach, T., A. Bashan, and A. Yonath. 2004. Ribosomal antibiotics: structural basis for resistance, synergism, and selectivity. Trends Biotechnol. 22:570-576. [DOI] [PubMed] [Google Scholar]
- 2.Berisio, R., J. Harms, F. Schluenzen, R. Zarivach, H. A. Hansen, P. Fucini, and A. Yonath. 2003. Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 185:4276-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berisio, R., F. Schluenzen, J. Harms, A. Bashan, T. Auerbach, D. Baram, and A. Yonath. 2003. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 10:366-370. [DOI] [PubMed] [Google Scholar]
- 4.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183-193. [DOI] [PubMed] [Google Scholar]
- 6.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham. 2004. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 48:3169-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 8.Hansen, L. H., P. Mauvais, and S. Douthwaite. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S rRNA. Mol. Microbiol. 31:623-631. [DOI] [PubMed] [Google Scholar]
- 9.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High-resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 10.Jones, T. A., M. Bergdoll, and M. Kjeldgaard. 1990. A macromolecular modeling environment, p. 189-195. In C. E. Bugg and S. E. Ealick (ed.), Crystallographic and modeling methods in molecular design. Springer-Verlag, New York, N.Y.
- 11.Katz, L., and G. W. Ashley. 2005. Translation and protein synthesis: macrolides. Chem. Rev. 105:499-528. [DOI] [PubMed] [Google Scholar]
- 12.Kleywegt, G. J., and T. A. Jones. 1996. Efficient rebuilding of protein structures. Acta Crystallogr. D 52:829-832. [DOI] [PubMed] [Google Scholar]
- 13.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfister, P., N. Corti, S. Hobbie, C. Bruell, R. Zarivach, A. Yonath, and E. C. Bottger. 2005. 23S rRNA base pair 2057-2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A→G. Proc. Natl. Acad. Sci. USA 102:5180-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfister, P., S. Jenni, J. Poehlsgaard, A. Thomas, S. Douthwaite, N. Ban, and E. C. Bottger. 2004. The structural basis of macrolide-ribosome binding assessed using mutagenesis of 23S rRNA positions 2058 and 2059. J. Mol. Biol. 342:1569-1581. [DOI] [PubMed] [Google Scholar]
- 17.Rick, S. W., and R. E. Cachau. 2000. The nonplanarity of the peptide group: molecular dynamics simulations with a polarizable two-state model for the peptide bond. J. Chem. Phys. 112:5230-5241. [Google Scholar]
- 18.Sander, P., T. Prammananan, and E. C. Bottger. 1996. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 22:841-848. [DOI] [PubMed] [Google Scholar]
- 19.Sander, P., T. Prammananan, A. Meier, K. Frischkorn, and E. C. Bottger. 1997. The role of rRNAs in macrolide resistance. Mol. Microbiol. 26:469-480. [DOI] [PubMed] [Google Scholar]
- 20.Schluenzen, F., J. M. Harms, F. Franceschi, H. A. Hansen, H. Bartels, R. Zarivach, and A. Yonath. 2003. Structural basis for the antibiotic activity of ketolides and azalides. Structure 11:329-338. [DOI] [PubMed] [Google Scholar]
- 21.Schluenzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 22.Shevack, A., H. S. Gewitz, B. Hennemann, A. Yonath, and H. G. Wittmann. 1985. Characterization and crystallization of ribosomal particles from Halobacterium marismortui. FEBS Lett. 184:68-71. [Google Scholar]
- 23.Springer, B., P. Sander, L. Sedlacek, W. D. Hardt, V. Mizrahi, P. Schar, and E. C. Bottger. 2004. Lack of mismatch correction facilitates genome evolution in mycobacteria. Mol. Microbiol. 53:1601-1609. [DOI] [PubMed] [Google Scholar]
- 24.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257-270. [DOI] [PubMed] [Google Scholar]
- 26.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh, F., J. Willcock, and S. Amyes. 2003. High-level telithromycin resistance in laboratory-generated mutants of Streptococcus pneumoniae. J. Antimicrob. Chemother. 52:345-353. [DOI] [PubMed] [Google Scholar]
- 28.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, D. N., J. M. Harms, K. H. Nierhaus, F. Schlunzen, and P. Fucini. 2005. Species-specific antibiotic-ribosome interactions: implications for drug development. Biol. Chem. 386:1239-1252. [DOI] [PubMed] [Google Scholar]
- 30.Xiong, L., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633-639. [DOI] [PubMed] [Google Scholar]
- 31.Yonath, A. 2005. Antibiotics targeting ribosomes: resistance, selectivity, synergism, and cellular regulation. Annu. Rev. Biochem. 74:649-679. [DOI] [PubMed] [Google Scholar]
- 32.Yonath, A. 2005. Ribosomal crystallography: peptide bond formation, chaperone assistance and antibiotics activity. Mol. Cell 20:1-16. [PubMed] [Google Scholar]