Abstract
A plasmid containing the qnrS quinolone resistance determinant and the gene encoding the SHV-2 β-lactamase has been discovered from a clinical Klebsiella pneumoniae strain isolated in Taiwan. The complete 98-kb sequence of this plasmid, designated pK245, was determined by using a whole-genome shotgun approach. Transfer of pK245 conferred low-level resistance to fluoroquinolones in electroporant Escherichia coli epi300. The sequence of the immediate region surrounding qnrS in pK245 is nearly identical (>99% identity) to those of pAH0376 from Shigella flexneri and pINF5 from Salmonella enterica serovar Infantis, the two other qnrS-carrying plasmids reported to date, indicating a potential common origin. Other genes conferring resistance to aminoglycosides (aacC2, strA, and strB), chloramphenicol (catA2), sulfonamides (sul2), tetracycline (tetD), and trimethoprim (dfrA14) were also detected in pK245. The dfrA14 gene is carried on a class I integron. Several features of this plasmid, including three separate regions containing putative replicons, a partitioning-control system, and a type II restriction modification system, suggest that it may be able to replicate and adapt in a variety of hosts. Although no critical conjugative genes were detected, multiple insertion sequence elements were found scattered throughout pK245, and these may facilitate the dissemination of the antimicrobial resistance determinants. We conclude that pK245 is a chimera which acquired its multiple antimicrobial resistance determinants horizontally from different sources. The identification of pK245 plasmid expands the repertoire of the coexistence of quinolone and extended-spectrum-β-lactam resistance determinants in plasmids carried by various species of the family Enterobacteriaceae in different countries.
Plasmid-mediated quinolone resistance was first identified in a clinical Klebsiella pneumoniae strain in the United States in 1998 (29). The gene responsible for this mechanism, qnrA, was later characterized (43). Several QnrA- and QnrB-like proteins have since been identified in different species of the family Enterobacteriaceae, including Escherichia coli, Enterobacter cloacae, and K. pneumoniae from the United States, Europe, and Asia (6, 21, 22, 27, 32, 36, 37, 44, 45). Another qnr gene, qnrS, was recently found on a transferable plasmid from a clinical Shigella flexneri isolate in Japan (19) and has subsequently been found in a Salmonella enterica serovar Infantis isolate of avian origin (23). Although many qnrA- and qnrB-positive plasmids were also found to carry genes encoding extended-spectrum β-lactamases (ESBLs) (21, 32, 34), no ESBLs were found on the plasmids that carried qnrS.
We recently identified a multidrug resistance plasmid from K. pneumoniae strain NK245. NK245 was isolated in January 2002 from a patient with hospital-acquired urinary tract infection. The plasmid, designated pK245, conferred resistance to several classes of antimicrobials in the E. coli electroporant, including low-level resistance to fluoroquinolones (FQs). The complete sequence of plasmid pK245 was determined because of its broad range of antimicrobial resistance. The present report describes the genomic structure of pK245, which carried several known resistance genes, including the blaSHV-2 ESBL gene and the qnrS quinolone resistance determinant.
MATERIALS AND METHODS
Bacterial strain.
K. pneumoniae NK245 was collected from the National Cheng Kung University Hospital in Tainan, Taiwan. The isolate was subcultured onto 5% sheep blood agar and MacConkey agar plates (BBL, Becton Dickinson Microbiology Systems, Cockeysville, MD) to check for purity. Species identification was confirmed by using a Vitek Gram-Negative Plus Identification card (bioMérieux Vitek, Inc., Hazelwood, MO) and conventional biochemical methods, as needed.
Plasmid manipulations.
Plasmid pK245 was one of the plasmids that we have isolated in the course of investigating the plasmid contents of several K pneumoniae clinical strains. Initially, the plasmids were isolated from the total DNA of K. pneumoniae by electrophoresis and were purified by QIAquick gel extraction columns (QIAGEN, Valencia, CA) and the gene for kanamycin resistance (kanr) was inserted into the plasmids by using an EZ-Tn5<KAN-2> kit (Epicenter Technologies, Madison WI). After in vitro transposon tagging, the chimeric plasmid, hereafter designated pK245-km, was introduced into competent E. coli epi300 cells by electroporation (Bio-Rad Gene Pulser, Hercules, CA). The electroporants were selected on Luria-Bertani agar containing 50 μg/ml of kanamycin. Functional analysis of pK245-km in the E. coli electroporant was carried out for this study.
Antimicrobial susceptibility testing.
The MICs of various classes of antimicrobials for clinical strain K. pneumoniae NK245, electroporant KM10 containing pK245-km, and the reference E. coli epi300 strain were determined by the broth microdilution method following the guidelines of the Clinical and Laboratory Standards Institute (formerly NCCLS) (5) by using custom-designed 96-well panels (Sensititre; Trek Diagnostics, East Essex, England). The MICs of several antimicrobials were further tested, and confirmatory testing for ESBL production was performed by Etest (AB Biodisk, Solna, Sweden), following the manufacturer's instructions.
Shotgun sequencing, assembly, and analysis of plasmid pK245.
DNA sequencing of plasmid pK245 was determined as part of the process of sequencing of the genome of K. pneumoniae strain NK245 by using a whole-genome shotgun approach. Sequence gaps were closed by primer walking on linking clones and by sequencing of the PCR products from the plasmid DNA. The sequences were assembled by using the Phred/Phrap/Consed software obtained from University of Washington, Seattle (12, 13, 16). This gave a total of 490 reads to be assembled into one contig, which was subsequently circularized by sequencing of the amplicon generated on the plasmid by PCR. The accuracy, order, and orientation of the contigs were verified based on the linking information from the forward and reverse sequence ends of each clone. Open reading frames (ORFs) in the plasmid sequence were identified by using the Glimmer 2.13 (9), GeneMark 2.4, and GeneMark.hmm 2.1 (38) programs. ORFs with less than 30 translated amino acids were abandoned. The ORFs predicted by the different programs were merged if they had the same stop position and were in the same frame, and the start position was chosen from the longest length. The ORF start positions were modified according to the results of a search with BLASTP. ORFs that overlapped with each other were visually inspected and in some cases removed. The iterons and repeat sequences were identified by Repeats Finder 4.0 (3).
Nucleotide sequence accession number.
The annotated sequence of pK245 has been submitted to the GenBank nucleotide sequence database under accession number DQ449578.
RESULTS AND DISCUSSION
General features of plasmid pK245.
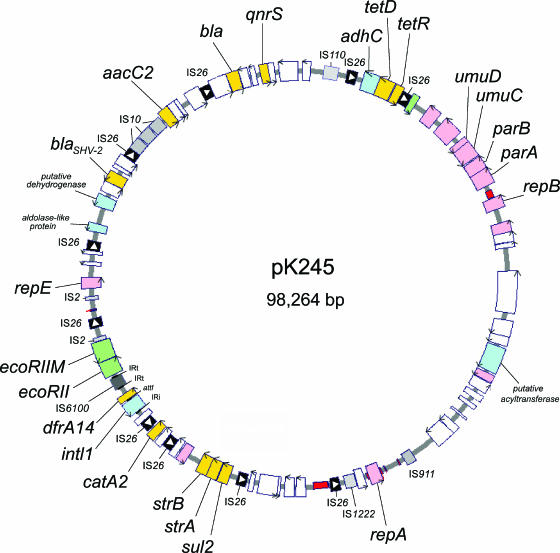
Plasmid pK245 was determined to be a 98,264-bp circular plasmid with a G+C content of 52%. From the annotated data, 90 genes were predicted, of which 12 genes encode replication, partitioning, multimer resolution, and DNA restriction/modification functions and 23 genes have possible roles in transposition and recombination. Several genes on pK245 are predicted to be involved in resistance to different classes of antimicrobials, including a blaSHV-2 gene for extended-spectrum β-lactam resistance and a qnrS gene for quinolone resistance. The genetic map of pK245 is shown in Fig. 1.
FIG. 1.
Circular map of plasmid pK245. Genes are color coded as follows: yellow, antimicrobial resistance associated; pink, plasmid replication and partitioning; black (with white triangles indicating their orientation), IS26; different shades of gray, other IS elements; green, nuclease and methylase; light blue, other functions; white, unknown functions. The arrows on the ORFs indicate the direction of transcription. Iteron regions are marked red.
Quinolone resistance gene and its organization.
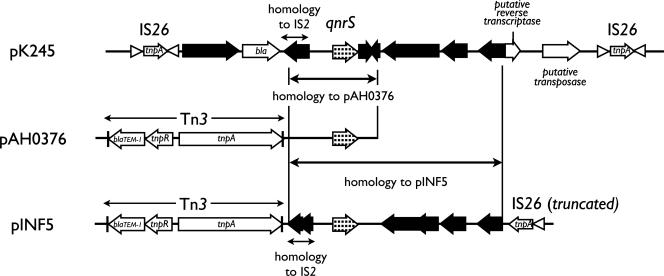
A 654-bp ORF with 100% identity in its deduced 218-amino-acid sequence to the amino acid sequence of the qnrS quinolone resistance gene was found in pK245. The qnrS-containing region of pK245 was compared to those of plasmids pAH0376 from S. flexneri and pINF5 from S. enterica serovar Infantis, the two qnrS-carrying plasmids reported to date (19, 23) (Fig. 2). Similar to pAH0376 and pINF5, we did not find any integron-related elements in the qnrS-containing region in pK245. This is different from the situation for qnrA-like genes, the majority of which were found to be associated with class I integrons (32).
FIG. 2.
Schematic diagram comparing the qnrS-containing region of pK245 with those of pAH0376 from S. flexneri and pINF5 from S. enterica serovar Infantis. The ORFs are shown as arrows, with the arrowhead indicating the direction of transcription. A 12.4-kb region of pK245, which is flanked by two copies of IS26, is illustrated on the top. The terminal inverted repeats on both sides of IS26 are shown in triangles. The Tn3 element is absent from pK245, and the deduced amino acid sequence of bla from pK245 is 61% identical and 75% similar to the TEM-1 β-lactamase. The 2.6-kb and 5.7-kb regions in pK245 that are similar to pAH0376 and pINF5, respectively, are also indicated by vertical lines. The stippled arrows indicate the qnrS gene. Black arrows are hypothetical ORFs. Although some predictions of the putative ORFs in these three plasmids are different, the nucleotide sequences of the homologous regions are more than 99.8% identical.
The immediate 2,677-bp region flanking qnrS was nearly identical (>99% identity) among the three plasmids, and another ∼3-kb region downstream was also identical between pK245 and pINF5, indicating a potential common origin (Fig. 2). The Tn3 element upstream of the qnrS region in pAH0376 and pINF5 was not found in pK245. The deduced amino acid sequence of bla upstream of qnrS in pK245 shared no more than 75% similarity with any other known β-lactamase genes. The transfer of this bla gene into E. coli HB101 conferred resistance only to ampicillin in the recipient (the MIC increased from 4 μg/ml to >256 μg/ml), and no ESBL activity was detected (data not shown).
No critical conjugation-related genes were detected in pK245, suggesting that it is not self-conjugative. We also performed conjugation test using E. coli HB101 as a recipient, with which no transconjugant could be obtained (data not shown). This is another feature of pK245 that differs from those of pAH0376 and pINF5, both of which have been reported to be conjugative (19, 23). However, the region containing qnrS and bla in pK245 appears to be a composite transposon bounded by two flanking IS26 elements in a head-to-tail arrangement which may be responsible for the mobility of qnrS (Fig. 2). The putative transposase and reverse transcriptase genes and the hypothetical insertion sequence (IS) genes at the vicinity of qnrS may also facilitate its spread.
When pK245-km was transferred into competent E. coli epi300 cells, increases in the MICs of multiple classes of antimicrobials were observed in electroporant E. coli KM10, including a >32-fold increase in the MICs of four FQs (ciprofloxacin, levofloxacin, gatifloxacin, and moxifloxacin) from ≤0.008 to 0.25 μg/ml (Table 1). Although pK245-km conferred low-level resistance to FQs and increased the nalidixic acid MIC (from 1 to 3 μg/ml) in KM10, it did not confer resistance to nalidixic acid in KM10. The same effect has also been noted in other studies of qnrA- and qnrS-containing plasmids (19, 23, 28, 34). The higher MICs of the FQs for clinical K. pneumoniae strain NK245 (2 to 6 μg/ml) compared to those for pK245-km-containing electroporant KM10 (0.125 to 0.25 μg/ml) could be due to changes in the porin or the efflux pump in NK245, because we did not find any mutations in its GyrA or ParC quinolone resistance-determining regions.
TABLE 1.
MICs of different antimicrobials for clinical strain K. pneumoniae NK245, its plasmid pK245-km electroporant KM10, and reference strain E. coli epi300
| Antimicrobial class and agenta | MIC (μg/ml)b
|
||
|---|---|---|---|
| NK245 | KM10 | E. coli epi300 | |
| Aminoglycosides | |||
| Amikacin | 12 | 1.5 | 1.5 |
| Gentamicin | 24 | 128 | 0.5 |
| Kanamycin | >64 | >64 | 4 |
| β-Lactams | |||
| Ampicillin | >256 | >256 | 4 |
| Amoxicillin-clavulanic acid | 6 | 8 | 6 |
| Ampicillin-sulbactam | 6 | 8 | 6 |
| Aztreonam | 24 | 0.75 | 0.094 |
| CTX | 4 | 1.5 | 0.064 |
| CAZ | >32 | 2 | 0.019 |
| Cefoxitin | 4 | 4 | 4 |
| Cefepime | 1.5 | 0.75 | 0.125 |
| Imipenem | 0.25 | 0.25 | 0.25 |
| Quinolone and fluoroquinolones | |||
| Nalidixic acid | >256 | 3 | 1 |
| Ciprofloxacin | 4 | 0.25 | 0.004 |
| Levofloxacin | 4 | 0.25 | 0.008 |
| Gatifloxacin | 2 | 0.125 | <0.002 |
| Moxifloxacin | 6 | 0.19 | 0.003 |
| Others | |||
| Chloramphenicol | >256 | >256 | 1.5 |
| Tetracycline | 6 | 256 | 1.5 |
| Trimethoprim-sulfamethoxazole | >32 | >32 | 0.094 |
| ESBL testingc | |||
| CTX/CTX + CLA | 4/0.032 | 1.5/0.094 | <0.25/0.064 |
| CAZ/CAZ + CLA | >32/0.19 | 2/0.38 | <0.5/<0.5 |
CTX, cefotaxime; CAZ, ceftazidime; CLA, clavulanic acid.
Based on Etest results for all agents except ampicillin and cefoxitin, which were from broth microdilution tests.
A >3 twofold concentration decrease in an MIC for either antimicrobial tested in combination with clavulanic acid versus its MIC when tested alone is considered positive for ESBL (or a ratio of >8 for either cefotaxime/cefotaxime plus clavulanic acid or ceftazidime/ceftazidime plus clavulanic acid).
ESBL and other antimicrobial resistance genes.
In pK245, an 858-bp ORF was identified to encode the SHV-2 ESBL (100% identity in 286 amino acids [aa]) (1, 20). ESBL activity was detected in KM10 (Table 1). Although the MICs of cefotaxime, ceftazidime, and aztreonam were similar to the data previously reported for SHV-2 (14), the MICs of ceftazidime and aztreonam were higher for clinical K. pneumoniae isolate NK245 than for electroporant E. coli KM10. Based on our whole-genome sequencing results (unpublished data), another copy of the blaSHV-2 gene was identified on the NK245 chromosome, which likely contributed to the additional resistance to the extended-spectrum β-lactams.
Several of the proteins encoded by the pK245 plasmid are identical to those known to confer resistance to gentamicin (aacC2), chloramphenicol (catA2), trimethoprim (dfrA14), streptomycin (strA and strB), sulfonamide (sul2), and tetracycline (tetD) in other bacterial species. The higher gentamicin MIC for electroporant KM10 (128 μg/ml) containing pK245-km compared to that for K. pneumoniae NK245 (24 μg/ml) most likely resulted from insertion of the kanamycin resistance marker into pK245. The difference in the MICs of tetracycline for electroporant E. coli KM10 (256 μg/ml) and K. pneumoniae NK245 (6 μg/ml) was unexpected. It has been reported recently that, as a transcriptional regulator, the product of tetD can activate a subset of SoxS/MarA/Rob regulon genes and confers resistance to redox-cycling compounds and antimicrobials (17). It is possible that the unexpected MIC of tetracycline was a result of the different genetic backgrounds of the two species.
The sul2 and the strA and strB genes, which encode resistance to sulfonamide and streptomycin, respectively, are localized in an approximately 3-kb region (Fig. 1), which closely resembles that of the broad-host-range plasmid RSF1010 (40), the multidrug resistance island of S. enterica DT193 (8), and the trimethoprim-sulfamethoxazole resistance-encoding transposon-like element of Vibrio cholerae (2). The homology of the 3-kb region to these known sequences is truncated by flanking IS elements. Its common occurrence suggests that the sul2-strA-strB region is a highly diffused genetic trait which can be transferred between the chromosomes and plasmids of different bacteria.
A 3.4-kb region representing a class I integron was identified. This integron region contains an intI1 integron-specific recombinase gene, an attI integration site, and a gene cassette that carries the dfrA14 dihydrofolate reductase gene. The integron is bounded by a 25-bp imperfect inverted repeat (IRi) downstream of intI1 on one end, and IS6100, which is flanked by two copies of a terminal inverted repeat (IRt), on the other end (Fig. 1). The role of gene cassettes and class I integrons in the spread and acquisition of antimicrobial resistance determinants has been well delineated (18).
There are a total of 10 IS26 elements interspersed throughout the entire plasmid, 9 of which are flanked by intact terminal inverted repeats (IRs). Four of the IS26 elements, starting with the one with a truncated terminal IR, are in a head-to-tail orientation on the complementary strand. The other six IS26 elements are also aligned in a head-to-tail orientation but on the opposite strand (Fig. 1). Since the donor of transposition requires two copies of the ISs in direct relative orientation flanking an interstitial DNA segment (26), the head-to-tail placement of these IS26 elements in pK245 may further facilitate the dissemination of its antimicrobial resistance determinants.
Other key features of pK245.
Several features of this plasmid suggest that it may be able to replicate and adapt in a variety of hosts and environments. An adhC gene with 99% similarity to the putative alcohol dehydrogenase of Pasteurella piscicida (24) and 94% similarity to the formaldehyde dehydrogenase of E. coli (25) was also found on pK245. Although the primary function of this enzyme is likely related to the metabolism of endogenous formaldehyde, it can also confer bacterial resistance to formaldehyde disinfectant (11). Thus, the adhC gene may provide an added survival advantage to the host in the hospital environment, where disinfectants are routinely used.
An umu DNA repair system locus containing two genes (umuC and umuD) was identified. The umuD and umuC genes, also known as mucA and mucB, respectively, have long been used in the Ames test to increase the rate of mutagenesis (30). Therefore, the existence of the umu DNA repair system may have exacerbated the growing problem of antimicrobial resistance by increasing the rate of target gene mutation in this bacterium.
Three genes were identified to encode replication machineries, including the replication initiator proteins found in Pantoea stewartii plasmid pSW800 repA (70% similarity in 293 aa) (46), the RepB replication protein of K. pneumoniae plasmid pGSH500 (96% similarity in 287 aa) (10), and repE of E. coli IncHI plasmid R27 (99% similarity in 251 aa) (39). Multiple iterons were found in the vicinity of the rep genes (Fig. 1). These iterons presumably play a role in plasmid replication and maintenance. A number of plasmids possess more than one replicons or replication initiation protein genes, including the K. pneumoniae plasmids pGSH500 (33) and pLVPK (4). It is plausible that, by encoding different sets of replication initiation proteins, these plasmids may have a better chance to replicate in a broader range of hosts.
Two genes, parA and parB, were identified downstream from the aforementioned repB gene in pK245. By comparison of the regions near the par genes with the known partitioning sites, features such as the repeat sequences of the binding sites and the central integration host factor-binding motif of the P1par family were also identified (7, 15). Presumably, the par genes and the nearby partitioning site are involved in the partitioning and compatibility of the plasmid.
Two adjacent ORFs that encode proteins with more than 99% identity to the EcoRII restriction endonuclease (EcoRII) and methyltransferase (M.EcoRII) of E. coli (41), respectively, were found in a convergent orientation of transcription and were positioned in between two of the repeated IS elements of the plasmid (Fig. 1). The two ORFs represent a type II restriction modification system similar to those identified in prokaryotic genomes and plasmids (35, 42). In addition to assisting with fighting foreign invaders, this restriction modification system has also been reported to contribute to the spread and maintenance of the plasmid (31).
Conclusions.
Complete nucleotide sequencing of plasmid pK245 not only has identified the molecular mechanisms contributing to multidrug resistance in K. pneumoniae isolate NK245 but also has provided insight into the means for the rapid adaptation and transmission of this plasmid within the hospital environment. This is the first report of qnrS plasmid-mediated quinolone resistance in K. pneumoniae. The identification of plasmid pK245 provides additional evidence for the association of quinolone and extended-spectrum β-lactam resistance in K. pneumoniae and expands the repertoire of the coexistence of ESBL and quinolone resistance-encoding genes in plasmids carried by various species of the family Enterobacteriaceae in different countries. Further investigations are needed to determine the prevalence of plasmid-mediated quinolone resistance and the role that it plays in facilitating coresistance to fluoroquinolones, extended-spectrum β-lactams, and other antimicrobials in different bacterial species.
Acknowledgments
This work was supported in part by a grant from the Department of Health Executive Yuan (DOH94-TD-G-111-038), and by an intramural grant from the National Health Research Institutes (CL-094-PP-01), Taiwan.
Footnotes
Published ahead of print on 28 August 2006.
REFERENCES
- 1.Barthélémy, M., J. Peduzzi, H. B. Yaghlane, and R. Labia. 1988. Single amino acid substitution between SHV-1 beta-lactamase and cefotaxime-hydrolyzing SHV-2 enzyme. FEBS Lett. 231:217-220. [DOI] [PubMed] [Google Scholar]
- 2.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y. T., H. Y. Chang, Y. C. Lai, C. C. Pan, S. F. Tsai, and H. L. Peng. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 6.Corkill, J. E., J. J. Anson, and C. A. Hart. 2005. High prevalence of the plasmid-mediated quinolone resistance determinant qnrA in multidrug-resistant Enterobacteriaceae from blood cultures in Liverpool, UK. J. Antimicrob. Chemother. 56:1115-1117. [DOI] [PubMed] [Google Scholar]
- 7.Dabrazhynetskaya, A., K. Sergueev, and S. Austin. 2005. Species and incompatibility determination within the P1par family of plasmid partition elements. J. Bacteriol. 187:5977-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly, M., L. Villa, C. Pezzella, S. Fanning, and A. Carattoli. 2005. Comparison of multidrug resistance gene regions between two geographically unrelated Salmonella serotypes. J. Antimicrob. Chemother. 55:558-561. [DOI] [PubMed] [Google Scholar]
- 9.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Silva-Tatley, F. M., and L. M. Steyn. 1993. Characterization of a replicon of the moderately promiscuous plasmid, pGSH5000, with features of both the mini-replicon of pCU1 and the ori-2 of F. Mol. Microbiol. 7:805-823. [DOI] [PubMed] [Google Scholar]
- 11.Dorsey, C. W., and L. A. Actis. 2004. Analysis of pVU3695, a plasmid encoding glutathione-dependent formaldehyde dehydrogenase activity and formaldehyde resistance in the Escherichia coli VU3695 clinical strain. Plasmid 51:116-126. [DOI] [PubMed] [Google Scholar]
- 12.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 13.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Fortineau, N., L. Poirel, and P. Nordmann. 2001. Plasmid-mediated and inducible cephalosporinase DHA-2 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 47:207-210. [DOI] [PubMed] [Google Scholar]
- 15.Funnell, B. E., and R. A. Slavcev. 2004. Partition systems of bacterial plasmids, p. 81-104. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, D.C.
- 16.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 17.Griffith, K. L., S. M. Becker, and R. E. Wolf, Jr. 2005. Characterization of TetD as a transcriptional activator of a subset of genes of the Escherichia coli SoxS/MarA/Rob regulon. Mol. Microbiol. 56:1103-1117. [DOI] [PubMed] [Google Scholar]
- 18.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 19.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49:801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacoby, G. A. 2006. Beta-lactamase nomenclature. Antimicrob. Agents Chemother. 50:1123-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonas, D., K. Biehler, D. Hartung, B. Spitzmuller, and F. D. Daschner. 2005. Plasmid-mediated quinolone resistance in isolates obtained in German intensive care units. Antimicrob. Agents Chemother. 49:773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehrenberg, C., S. Friederichs, A. de Jong, G. B. Michael, and S. Schwarz. 2006. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J. Antimicrob. Chemother. 58:18-22. [DOI] [PubMed] [Google Scholar]
- 24.Kim, E. H., and T. Aoki. 1994. The transposon-like structure of IS26-tetracycline, and kanamycin resistance determinant derived from transferable R plasmid of fish pathogen, Pasteurella piscicida. Microbiol. Immunol. 38:31-38. [DOI] [PubMed] [Google Scholar]
- 25.Kümmerle, N., H. H. Feucht, and P. M. Kaulfers. 1996. Plasmid-mediated formaldehyde resistance in Escherichia coli: characterization of resistance gene. Antimicrob. Agents Chemother. 40:2276-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mammeri, H., L. M. Van De, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Martinez, L., A. Pascual, I. Garcia, J. Tran, and G. A. Jacoby. 2003. Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 51:1037-1039. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 30.McCann, J., N. E. Spingarn, J. Kobori, and B. N. Ames. 1975. Detection of carcinogens as mutagens: bacterial tester strains with R factor plasmids. Proc. Natl. Acad. Sci. USA 72:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naito, T., K. Kusano, and I. Kobayashi. 1995. Selfish behavior of restriction-modification systems. Science 267:897-899. [DOI] [PubMed] [Google Scholar]
- 32.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463-469. [DOI] [PubMed] [Google Scholar]
- 33.Osborn, A. M., F. M. Silva Tatley, L. M. Steyn, R. W. Pickup, and J. R. Saunders. 2000. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 146:2267-2275. [DOI] [PubMed] [Google Scholar]
- 34.Poirel, L., L. M. Van De, H. Mammeri, and P. Nordmann. 2005. Association of plasmid-mediated quinolone resistance with extended-spectrum beta-lactamase VEB-1. Antimicrob. Agents Chemother. 49:3091-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts, R. J., and D. Macelis. 2001. REBASE—restriction enzymes and methylases. Nucleic Acids Res. 29:268-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robicsek, A., D. F. Sahm, J. Strahilevitz, G. A. Jacoby, and D. C. Hooper. 2005. Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob. Agents Chemother. 49:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Martinez, J. M., A. Pascual, I. Garcia, and L. Martinez-Martinez. 2003. Detection of the plasmid-mediated quinolone resistance determinant qnr among clinical isolates of Klebsiella pneumoniae producing AmpC-type beta-lactamase. J. Antimicrob. Chemother. 52:703-706. [DOI] [PubMed] [Google Scholar]
- 38.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saul, D., D. Lane, and P. L. Bergquist. 1988. A replication region of the IncHI plasmid, R27, is highly homologous with the RepFIA replicon of F. Mol. Microbiol. 2:219-225. [DOI] [PubMed] [Google Scholar]
- 40.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 41.Som, S., A. S. Bhagwat, and S. Friedman. 1987. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 15:313-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takano, T., T. Watanabe, and T. Fukasawa. 1968. Mechanism of host-controlled restriction of bacteriophage lambda by R factors in Escherichia coli K12. Virology 34:290-302. [DOI] [PubMed] [Google Scholar]
- 43.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, M., D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 48:1295-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, C. Y., J. F. Fu, and S. T. Liu. 2001. The replicon of pSW800 from Pantoea stewartii. Microbiology 147:2757-2767. [DOI] [PubMed] [Google Scholar]