Abstract
We report a new Salmonella genomic island 1 variant antibiotic resistance gene cluster called SGI1-L in a Salmonella enterica serovar Newport isolate containing a dfrA15 gene cassette conferring resistance to trimethoprim. The isolate carried another class 1 integron containing the aacC5 and aadA7 gene cassettes conferring resistance to gentamicin and streptomycin/spectinomycin, respectively.
A chromosomal genomic island called Salmonella genomic island 1 (SGI1) has been initially described in epidemic multidrug-resistant Salmonella enterica serovar Typhimurium phage type DT104 strains (2, 9). SGI1 contains an antibiotic resistance gene cluster and most often confers resistance to ampicillin, chloramphenicol/florfenicol, streptomycin/spectinomycin, sulfonamides, and tetracyclines.
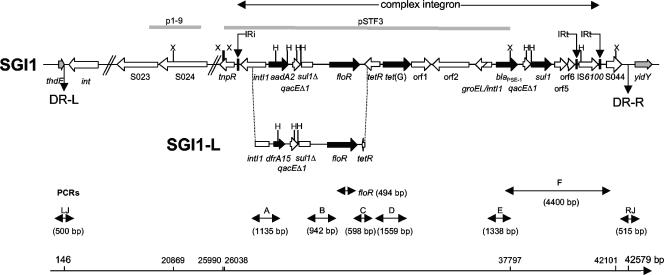
The 43-kb SGI1 is located between the thdF and int2 genes of the chromosome of S. enterica serovar Typhimurium. The int2 gene is part of a retron sequence which has been reported to date only in S. enterica serovar Typhimurium (2, 9). In other S. enterica serovars, SGI1 is located between the thdF and yidY genes (2, 9). The antibiotic resistance gene cluster is located near the 3′ end of SGI1 and constitutes a complex class 1 integron that belongs to the In4 group, which has been recently named In104 (6) (Fig. 1). At the first attI1 site of this complex integron, the cassette carries the aadA2 gene, which confers resistance to streptomycin and spectinomycin, and a 3′ conserved segment (3′-CS) with a truncated sul1 (sul1Δ) gene is present. At the second attI1 site, the cassette contains the β-lactamase gene blaPSE-1, conferring resistance to ampicillin, and the 3′-CS comprises a complete sul1 gene, conferring resistance to sulfonamides. Flanked by the two cassettes are the floR gene, which confers cross-resistance to chloramphenicol and florfenicol, and the tetracycline resistance genes tetR and tet(G). SGI1 has been recently demonstrated to be an integrative mobilizable element (4).
FIG. 1.
Genetic organization of the antibiotic resistance gene clusters of SGI1 and the new SGI1-L variant of S. enterica serovar Newport strain 00-4093. The direction of transcription of genes is indicated by arrows. Black and gray arrows correspond to SGI1 antibiotic resistance genes and chromosomal genes flanking SGI1, respectively. DR-L and DR-R are the left and right direct repeats, respectively, bracketing SGI1. IRi and IRt are 25-bp imperfect inverted repeats defining the left and right ends of complex class 1 integrons. PCRs used to assess the genetic organization of the antibiotic resistance genes (PCRs floR, A, B, C, D, E, and F) and the SGI1 junctions to the chromosome (PCRs LJ and RJ [for left and right junctions, respectively]) are indicated. Abbreviations for restriction sites: X, XbaI; and H, HindIII. Gray thick bars indicate the p1-9 and pSTF3 probes. The positions of the DR-L, DR-R, and XbaI restriction sites are indicated on the scale according to GenBank accession no. AF261825.
Variant SGI1 antibiotic resistance gene clusters have been described in a wide variety of S. enterica serovars (9). SGI1 variant antibiotic resistance gene clusters were accordingly classified in SGI1-A to SGI1-K (GenBank accession no. AY463797). These gene clusters were likely generated after chromosomal recombinational events or by antibiotic resistance gene cassette replacement at one of the attI1 sites (9).
In this study, we examined an S. enterica serovar Newport strain, named 00-4093, isolated from stools from a French patient with gastroenteritis. He had been infected by this strain while traveling to Egypt in 2000. Strain 00-4093 displayed the multidrug resistance profile typical of the SGI1 antibiotic resistance gene cluster but with additional resistance to gentamicin and trimethoprim.
Detection of SGI1, its location, and mapping of antibiotic resistance genes were performed by PCR using primers and conditions described previously (Fig. 1) (2, 4, 5). PCR results were positive for the left junction of SGI1 with the chromosomal thdF gene and for the right junction with the chromosomal yidY gene but negative for the junction with the int2 gene. Thus, these results indicated that strain 00-4093 harbors SGI1 at the same chromosomal location, i.e., between the thdF and yidY genes, as in non-Typhimurium serovars. PCR mapping of the antibiotic resistance gene cluster showed the presence of fragments B, C, D, E, F, and floR of the sizes expected from the S. enterica serovar Typhimurium DT104 control strain BN9181 harboring SGI1, indicating the presence of the floR, tetR, tet(G), and blaPSE-1 genes (Fig. 1). However, PCR A specific for the aadA2 resistance gene cassette was negative. The positive result for PCR B nevertheless indicated the presence of the 3′-CS with the truncated sul1 gene. To assess the gene cassette array at the first attI1 site of the SGI1 complex integron, a PCR with the forward primer of PCR A, targeting the intI1 gene, and the reverse primer of PCR C, targeting the tetR gene, was performed (Fig. 1). Strain 00-4093 yielded a fragment of 4.4 kb that was cloned and sequenced (GenBank accession no. DQ647028). Sequence analysis identified the trimethoprim resistance gene cassette dfrA15 at the first attI1 site (Fig. 1). This dfrA15 gene showed 99% nucleotide identity to those found in class 1 integrons from Vibrio cholerae and Vibrio fluvialis strains (GenBank accession no. AY605681, AB219235, AF221900, AB113114, and AY605692). The deduced amino acid sequence showed one amino acid change (I91T) compared with that of other DfrA15 proteins. This new variant SGI1 antibiotic resistance gene cluster was named SGI1-L according to the previously proposed nomenclature of SGI1 variants (9).
To identify the genes conferring resistance to gentamicin, streptomycin, and spectinomycin, PCR amplification of class 1 integrons using 5′-CS and 3′-CS primers was performed as described previously (10). This PCR yielded three fragments with strain 00-4093, of which two were linked to the SGI1-L complex class 1 integron. The third fragment of 1.6 kb was cloned (plasmid pKP04) and sequenced (GenBank accession no. DQ647029). Sequence analysis identified the gene cassettes aacC5, also called aac(3)-Id, and aadA7, conferring resistance to gentamicin and streptomycin/spectinomycin, respectively (1, 3, 5, 7, 8). This gene cassette array was previously identified in SGI1-H of S. enterica serovar Newport and in SGI1-K of S. enterica serovar Kentucky (GenBank accession no. AY463797) (5). The class 1 integron containing aacC5 and aadA7 in strain 00-4093 was probably located elsewhere in the chromosome, since the gentamicin and streptomycin/spectinomycin resistances were not transferable and since PCRs using the 5′-CS and 3′-CS primers on plasmid extracts were negative (data not shown). This class 1 integron was also not located in SGI1-L, since PCR using a forward primer of S026 found upstream of the SGI1 complex integron and a reverse primer of aacC5 was negative, whereas it was positive with SGI1-H carrying the S. enterica serovar Newport control strain (data not shown).
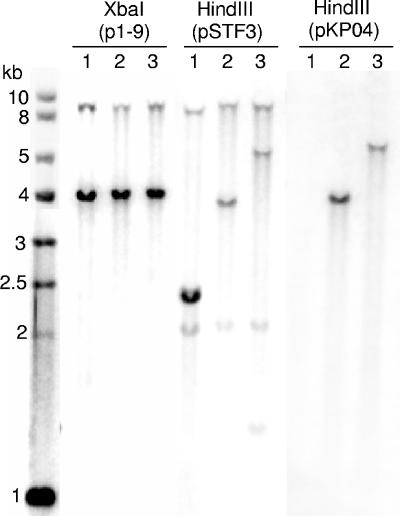
The presence of the 13-kb region found upstream of the SGI1 complex integron in strain 00-4093 was confirmed by Southern blot hybridization of XbaI-digested genomic DNA using the p1-9 probe (Fig. 1) (2, 4, 5). This probe corresponds to a 2-kb EcoRI fragment comprising parts of S023 and S024 open reading frames in the central region of SGI1 which encode putative helicase and exonuclease proteins (2). The p1-9 probe showed two XbaI fragments of the expected 4- and 9-kb sizes as in the SGI1-carrying control strains (Fig. 2). The new SGI1-L variant in strain 00-4093 was confirmed by Southern blotting of HindIII-digested genomic DNA with the pSTF3 probe containing nearly the entire SGI1 antibiotic resistance gene cluster of DT104 control strain BN9181 (Fig. 1 and 2). Probe pKP04 containing the aacC5 and aadA7 genes revealed a fragment of 6 kb in the HindIII-digested genomic DNA of strain 00-4093, suggesting as indicated above that these genes were not linked to SGI1-L (Fig. 2).
FIG. 2.
Southern blot hybridization with the p1-9, pSTF3, or pKP04 probes of XbaI- or HindIII-digested genomic DNAs of S. enterica serovar Typhimurium DT104 strain BN9181 carrying SGI1 (lanes 1), S. enterica serovar Newport strain 01-2174 carrying SGI1-H (lanes 2), and S. enterica serovar Newport strain 00-4093 carrying SGI1-L (lanes 3). The 9- and 4-kb XbaI fragments obtained with the p1-9 probe correspond to XbaI site positions 11949 and 20869 and 20869 and 24937, respectively (as shown in Fig. 1 and according to GenBank accession no. AF261825).
XbaI macrorestriction analysis by PFGE showed that strain 00-4093 was distinct from serovar Newport strains carrying SGI1-H which were also imported cases from Egypt (5; data not shown).
In conclusion, the SGI1 complex class 1 integron may contribute to the capture of a wide diversity of resistance gene cassettes in the Salmonella chromosome and thus may generate diverse antibiotic resistance gene clusters.
Footnotes
Published ahead of print on 11 September 2006.
REFERENCES
- 1.Ahmed, A. M., T. Nakagawa, E. Arakawa, T. Ramamurthy, S. Shinoda, and T. Shimamoto. 2004. New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J. Antimicrob. Chemother. 53:947-951. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabrera, R., F. Marco, J. Vila, J. Ruiz, and J. Gascon. 2006. Class 1 integrons in Salmonella strains causing traveler's diarrhea. Antimicrob. Agents Chemother. 50:1612-1613. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 5.Doublet, B., F.-X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levings, R. S., S. R. Partidge, D. Lightfoot, R. M. Hall, and S. P. Djordjevic. 2005. New integron-associated gene cassette encoding a 3-N-aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 49:1238-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michael, G. B., P. Butaye, A. Cloeckaert, and S. Schwarz. 2006. Genes and mutations conferring antimicrobial resistance in Salmonella: an update. Microbes Infect. 8:1898-1914. [DOI] [PubMed] [Google Scholar]
- 9.Mulvey, M. R., D. A. Boyd, A. B. Olson, B. Doublet, and A. Cloeckaert. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 8:1915-1922. [DOI] [PubMed] [Google Scholar]
- 10.Weill, F.-X., R. Lailler, K. Praud, A. Kérouanton, L. Fabre, A. Brisabois, P. A. D. Grimont, and A. Cloeckaert. 2004. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 42:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]