The vancomycin-resistant Enterococcus raffinosus strain GV5 was resistant to vancomycin (MIC, 1,024 μg/ml) and teicoplanin (MIC, 256 μg/ml). The species of strain GV5 was determined by sequencing of a specific PCR product for the 16S rRNA gene of E. raffinosus. GV5 was isolated from a stool specimen and from a bedsore on the necrotic inferior limb of a diabetic 73-year-old man in Japan.
DNA sequence analysis of the vanD operon was performed by sequencing the PCR products with primers specific for each gene in the vanD4 operon of E. faecium 10/96A (8); it showed that GV5 encodes a 5,654-bp vanD gene cluster consisting of vanRDSDYDHDDXD, which is homologous to the corresponding genes in the reported VanD-type strains and is located on the chromosome (accession no. AB242319) (3, 6, 7, 9). The vanD gene cluster was compared with that of the corresponding genes of the vanD4 gene cluster of E. faecium 10/96A (Fig. 1) (8). vanRD and vanXD were completely identical to the equivalent genes in 10/96A. There was one amino acid substitution in both VanHD and VanD, where Ile169 was converted to Phe and Gly121 was converted to Val, respectively. The reported VanSD contains five blocks of the conserved sequences H, N, G1, F, and G2 (2, 4, 8), which are contained in phosphate transmitters of two-component regulator systems (1, 11). Block H sequences consist of the residues L164AHDLKTPLS173, including a putative autophosphorylation site, His166 (14). The Thr170 residue in the block H sequence has been replaced by Ile in VanSD of GV5, suggesting that this mutation might result in the constitutive expression of resistance due to impaired VanSD function to dephosphorylate phosphorylated VanRD. vanYD of GV5, which has a molecular size of 1,068 bp, is completely identical to that of 10/96A with the exception of an additional adenosine insertion in vanYD of 10/96A (8). The nucleotide sequence from position 346 to position 354 of GV5 vanYD is C346AAAAAAAC354, and the sequence from position 346 to position 355 of 10/96A vanYD is C346AAAAAAAAC355. If an adenosine residue were inserted within the seven adenosines located between nucleotides 346 and 354 of GV5 vanYD, the codon sequence at positions 415 to 417 of the resulting gene would become the TGA translation stop codon as a result of the frameshift mutation, and translation would be terminated prematurely after amino acid 138, as in the VanYD protein of 10/96A (8).
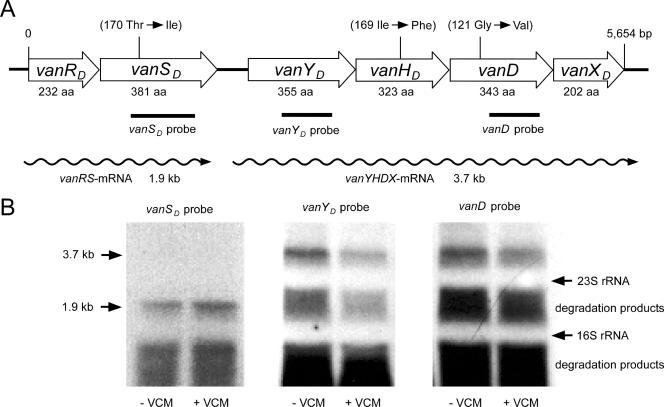
FIG. 1.
Schematic representation of the vanD gene cluster from E. raffinosus strain GV5 and Northern blot analysis of the vanD cluster. (A) Open arrows represent coding sequences and indicate the direction of transcription. The PCR fragments internal to the vanSD, vanYD, and vanD genes used in the hybridization experiments are indicated below the corresponding regions. Amino acids with arrows within parentheses indicate substitutions compared with the reported sequence of the vanD4 operon of E. faecium 10/96A (8). (B) Northern hybridization was performed according to a protocol described previously (13). RNAs were prepared from strains cultured with 6 μg/ml of vancomycin (+VCM) or without vancomycin (−VCM) for 2 h. Thirty micrograms of RNA was used in each lane. The sizes of the RNAs were determined by using the sizes of RNA molecular weight markers (Invitrogen, Inc.), and the arrows and the numbers on the left indicate the positions and sizes of the largest bands in each experiment.
In Northern hybridizations with vanYD and vanD probes, identical bands of about 3.7 kb in size, which correspond to the transcript of vanYDHDDXD (5), were observed in both the absence and the presence of vancomycin (Fig. 1). The vanSD probe detected an approximately 1.9-kb band, which corresponds to the size of the transcript of vanRDSD, in the absence and presence of vancomycin (Fig. 1). These results indicate that the vanD4 cluster in GV5 is expressed constitutively (2, 3, 6, 9, 12).
Analysis of the d-Ala:d-Ala ligase gene (ddl) on the chromosome of strain GV5 (accession no. AB242318) revealed that there are two amino acid substitutions—Asn271 is converted to Asp, and Gly319 is converted to Asp—compared to the wild-type DDL of E. raffinosus JCM8733 (accession no. AB242317), which implies that the amino acid substitutions might result in impaired function of GV5 DDL (10).
Several VanD-type vancomycin-resistant enterococci have been identified among E. faecium and E. faecalis (3, 6, 7, 9). We have described the first VanD-type E. raffinosus strain and showed evidence that there is species divergence in enterococci that encode VanD resistance as well as nucleotide divergence between the VanD determinants (3, 6, 7, 9).
Acknowledgments
This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology [Tokuteiryoiki (2) (16017216), Kiban (C) (17590384)] and from the Japanese Ministry of Health, Labor, and Welfare (H15-Shinko-9, H18-Shinko-11).
We thank Yoshiyuki Ozawa and Elizabeth Kamei for helpful advice.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Baptista, M., P. Rodrigues, F. Depardieu, P. Courvalin, and M. Arthur. 1999. Single-cell analysis of glycopeptide resistance gene expression in teicoplanin-resistant mutants of a VanB-type Enterococcus faecalis. Mol. Microbiol. 32:17-28. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D. A., J. Conly, H. Dedier, G. Peters, L. Robertson, E. Slater, and M. R. Mulvey. 2000. Molecular characterization of the vanD gene cluster and a novel insertion element in a vancomycin-resistant enterococcus isolated in Canada. J. Clin. Microbiol. 38:2392-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D. A., P. Kibsey, D. Roscoe, and M. R. Mulvey. 2004. Enterococcus faecium N03-0072 carries a new VanD-type vancomycin resistance determinant: characterization of the VanD5 operon. J. Antimicrob. Chemother. 54:680-683. [DOI] [PubMed] [Google Scholar]
- 4.Casadewall, B., and P. Courvalin. 1999. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J. Bacteriol. 181:3644-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadewall, B., P. E., Reynolds, O. H. Ambur, and P. Courvalin. 2001. Regulation of expression of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J. Bacteriol. 183:3436-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalla Costa, L. M., P. E. Reynolds, H. A. Souza, D. C. Souza, M. F. Palepou, and N. Woodford. 2000. Characterization of a divergent VanD-type resistance element from the first glycopeptide-resistant strain of Enterococcus faecium isolated in Brazil. Antimicrob. Agents Chemother. 44:3444-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Depardieu, F., M. Kolbert, H. Pruul, J. Bell, and P. Courvalin. 2004. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 48:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Depardieu, F., P. E. Reynolds, and P. Courvalin. 2003. VanD-type vancomycin-resistant Enterococcus faecium 10/96A. Antimicrob. Agents Chemother. 47:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostrowsky, B. E., N. C. Clark, C. Thauvin-Eliopoulos, L. Venkataraman, M. H. Samore, F. C. Tenover, G. M. Eliopoulos, R. C. Moellering, Jr., and H. S. Gold. 1999. A cluster of VanD vancomycin-resistant Enterococcus faecium: molecular characterization and clinical epidemiology. J. Infect. Dis. 180:1177-1185. [DOI] [PubMed] [Google Scholar]
- 10.Ozawa, Y., P. Courvalin, and M. Gaiimand. 2000. Identification of enterococci at the species level by sequencing of the genes for d-alanine:d-alanine ligases. Syst. Appl. Microbiol. 23:230-237. [DOI] [PubMed] [Google Scholar]
- 11.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 12.Perichon, B., P. Reynolds, and P. Courvalin. 1997. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother. 41:2016-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanimoto, K., and Y. Ike. 2002. Analysis of the conjugal transfer system of the pheromone-independent highly transferable enterococcus plasmid pMG1: identification of a tra gene (traA) up-regulated during conjugation. J. Bacteriol. 184:5800-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright, G., T. Holman, and C. Walsh. 1993. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 32:5057-5063. [DOI] [PubMed] [Google Scholar]