Abstract
Thrombin-induced platelet microbicidal protein 1 (tPMP-1) is a staphylocidal peptide released by activated platelets. This peptide initiates its microbicidal activity by membrane permeabilization, with ensuing inhibition of intracellular macromolecular synthesis. RP-1 is a synthetic congener modeled on the C-terminal microbicidal α-helix of tPMP-1. This study compared the staphylocidal mechanisms of RP-1 with those of tPMP-1, focusing on isogenic tPMP-1-susceptible (ISP479C) and -resistant (ISP479R) Staphylococcus aureus strains for the following quantitative evaluations: staphylocidal efficacy; comparative MIC; membrane permeabilization (MP) and depolarization; and DNA, RNA, and protein synthesis. Although the proteins had similar MICs, RP-1 caused significant killing of ISP479C (<50% survival), correlating with extensive MP (>95%) and inhibition of DNA and RNA synthesis (>90%), versus substantially reduced killing of ISP479R (>80% survival), with less MP (55%) and less inhibition of DNA or RNA synthesis (70 to 80%). Interestingly, RP-1-induced protein synthesis inhibition was equivalent in both strains. RP-1 did not depolarize the cell membrane and caused a relatively short postexposure growth inhibition. These data closely parallel those previously reported for tPMP-1 against this strain set and exemplify how synthetic molecules can be engineered to reflect structure-activity relationships of functional domains in native host defense effector molecules.
The growing problem of microbial resistance to existing antibiotics has stimulated great interest in the development of novel antimicrobial agents with new modes of action (16, 17, 21). One such strategy is based on naturally occurring cationic antimicrobial peptides (APs), which represent evolutionarily optimized effectors of innate immunity (27-29) and have been isolated from prokaryotes, eukaryotes, plants, and humans (2, 8-11, 19, 41).
Recently, a homologous group of cationic APs from rabbit and human platelets has been characterized and termed platelet microbicidal proteins (PMPs) (20, 30, 39). These peptides are released into the bloodstream following platelet activation by agonists generated at sites of endovascular damage or infection (e.g., thrombin) (6, 7). Among these, thrombin-induced PMP-1 (tPMP-1) has been most extensively studied (12-14, 22-24, 31). tPMP-1 exerts rapid and potent microbicidal activities in submicromolar concentrations against pathogens that commonly access the bloodstream, including staphylococci (22, 23, 27, 32, 33). In addition, this peptide is active against microbial pathogens in a physiologically relevant range of pH (5.5 to 8.0), as well as in plasma or serum. The latter observations suggest that the in vitro antimicrobial activities of tPMP-1 are influenced by contexts that exist in relevant settings of infection in vivo (31). Consistent with this concept, tPMP-1 appears to exert minimal host cell toxicity at concentration ranges over which it exerts antimicrobial activity (34). Of note, we have previously demonstrated that in vitro susceptibility of Staphylococcus aureus, viridans streptococci, and Candida albicans to tPMP-1 is negatively correlated with strain virulence in experimental endocarditis models (3-5, 32, 33).
The staphylocidal mechanisms of tPMP-1 appear to involve initial cytoplasmic membrane permeabilization (MP) in a voltage-dependent (12, 31) but nondepolarizing (31) manner, with subsequent inhibition of intracellular macromolecular synthesis (22, 24). Recently, Yount et al. defined the structure of tPMP-1 and evaluated its structure-activity relationships (39). Amino acid sequencing and mass spectroscopy demonstrated that tPMP-1 has the following features: (i) it is relatively small (mass of 7.95 kDa); (ii) it consists of two isoforms, featuring 72 or 73 amino acid residues; (iii) it has a cationic and α-helical C terminus; (iv) it has an extended N terminus containing a hallmark CXC chemokine motif; and (v) it shares a marked degree of sequence and three-dimensional homology with human platelet factor 4 (PF-4) (39). Collectively, these data indicate that tPMP-1 is a kinocidin (microbicidal chemokine) with distinct microbicidal and chemoattractant domains (39, 40).
To further study the structure-activity relationships of tPMP-1, Yeaman et al. designed synthetic chimeric peptides (e.g., RP-1) based on structural themes in C-terminal microbicidal α-helices of mammalian PF-4 family proteins and other kinocidins (e.g., tPMPs) (35). These synthetic APs have been previously shown to exert potent antimicrobial activities reflective of the native peptide (35) and are highly microbicidal in relevant blood and blood-derived biomatrices (35). However, it is not known whether the microbicidal mechanisms of the synthetic APs recapitulate those of native tPMP-1. Such analyses are critical to the rational design of novel antimicrobial agents with optimal efficacy against antibiotic-resistant pathogens. Thus, our present study was developed to examine and compare RP-1 with native tPMP-1 as regards their microbicidal mechanisms by using an isogenic staphylococcal strain set with differing in vitro susceptibilities to tPMP-1.
(This work was presented in part at the 105th General Meeting of the American Society for Microbiology [abstract E-008], Atlanta, Georgia, 5 to 9 June 2005.)
MATERIALS AND METHODS
Microorganisms.
ISP479C and ISP479R comprise a well-characterized isogenic methicillin-susceptible S. aureus strain pair differing principally in their in vitro tPMP-1 susceptibility profiles (1, 4, 22). Additionally, strains COL (a clinical methicillin-resistant genome-characterized S. aureus strain [1]), 67-0 (clinical methicillin-resistant S. aureus and β-lactamase-producing strain [10]), and ATCC 29213 (control MSSA strain) were tested. The latter panel of laboratory and clinical strains was used in selected studies. For studies, all organisms were cultured identically and prepared as described elsewhere (22, 23).
Antimicrobial agents.
RP-1 is an 18-amino-acid peptide (N-ALYKK5FKKKL10LKSLK15RLG-C; mass, 2.1 kDa) designed to incorporate structure-activity attributes characteristic of tPMP-1 (35). RP-1 was synthesized with a multiplex synthesizer (Rainin, Woburn, MA) and authenticated by mass spectroscopy and amino acid analysis as previously described (35). The purification of tPMP-1 by reverse-phase high-pressure liquid chromatography is described in detail elsewhere (30, 36). Novobiocin, rifampin, and tetracycline were purchased from Sigma Chemical Co. (St. Louis, MO), Marion-Merrell Dow (Kansas City, MO), and Aldrich Chemical Company, Inc. (Milwaukee, WI), respectively, and were used as prototype DNA, RNA, and protein synthesis inhibitors, respectively (22).
Radioisotopes.
Isotopes [methyl-3H]thymidine, [5-3H]uridine, and l-[4,5-3H]leucine were purchased from Amersham Life Science (Piscataway, NJ), and their specific activities were adjusted as recommended by the manufacturer (22).
MIC assays.
MIC assays were performed following Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) guidelines but modified for assessment of antimicrobial polypeptides. In brief, standard assay conditions employing cation-supplemented 100% Mueller-Hinton broth (MHB) were used in pilot studies. As previously reported (35), this assay condition impeded the antimicrobial activities of both RP-1 and tPMP-1. Therefore, minimal essential medium (MEM) containing increasing proportions (vol/vol) of MHB was used to compare the relative MICs of peptides for the panel of S. aureus laboratory and clinical strains listed above.
Staphylocidal activities.
The staphylocidal activities of RP-1 and tPMP-1 were evaluated as described previously (22, 23). In brief, log-phase S. aureus cells (107 CFU/ml) were exposed to RP-1 (25, 50, or 100 μg/ml) or tPMP-1 (2 μg/ml). At selected time points, aliquots were plated on Trypticase soy agar plates (Difco Laboratories, Detroit, MI) and incubated at 37°C (22). Staphylocidal activities of RP-1 or tPMP-1 were expressed as the mean percentage of survival (± standard deviation [SD]) versus untreated controls (set at 100% survival).
RP-1 postexposure effect.
We assessed the antistaphylococcal growth-inhibiting effects following RP-1 exposure as previously described (36). In brief, log-phase S. aureus cells (107 CFU/ml) were exposed to RP-1 (50 μg/ml) for 60 min at 37°C. Surviving cells were washed and incubated in fresh MHB (Difco Laboratories, Detroit, MI). Culture samples were removed hourly and quantitatively cultured. The post-RP-1 exposure effect duration was defined as the difference in time required for RP-1-exposed S. aureus cells versus controls (RP-1 unexposed) to achieve 1 log10 CFU/ml increase (36).
Membrane permeabilization.
S. aureus MP by RP-1 or tPMP-1 was quantified by flow cytometry (FACScalibur; Becton Dickinson, San Jose, CA) via the release of the preloaded fluorophore calcein as previously described (13, 23). In brief, log-phase S. aureus cells (109 CFU/ml) were loaded with calcein acetoxymethyl ester (2 μM; Molecular Probes, Eugene, OR) prior to peptide exposure for 2 h at 37°C. Calcein-loaded S. aureus cells (107 CFU/ml) were exposed to either RP-1 (50 or 100 μg/ml), tPMP-1 (2 μg/ml), or MEM (controls). At selected time points, samples were removed and 10,000 S. aureus cells were analyzed for MP (13, 23). Cells with fluorescence (FL-1) units of ≥100 were considered to possess intact membranes, while those with FL-1 units of ≤100 were considered to be permeabilized by peptide (see Results).
Membrane depolarization.
S. aureus membrane depolarization was examined by flow cytometry using 3,3′-dipentyloxacarbocyanine iodide (DiOC5) (Molecular Probes, Eugene, OR) as previously described (18, 31). Briefly, DiOC5-labeled S. aureus cells (107 CFU/ml) were resuspended in K+ MEM in the presence or absence of RP-1. Valinomycin (Sigma) was utilized as a positive control. Reductions in mean DiOC5 fluorescence were interpreted as membrane depolarization (31).
Macromolecular synthesis.
The effects of RP-1 on DNA, RNA, and protein synthesis in the S. aureus strain pair were determined by measuring the respective incorporations of thymidine, uridine, or leucine, respectively, into trichloroacetic acid-insoluble material, as described in detail elsewhere (22). Briefly, radioactive precursors were added to log-phase S. aureus cells (10 μCi/ml of [methyl-3H]thymidine, 5 μCi/ml of [5-3H]uridine, or 5 μCi/ml of l-[4,5-3H]leucine) in MEM only (controls) or in medium containing RP-1. Intracellular macromolecular synthesis was measured by liquid scintillation counting (LS-100; Beckman Instruments, Fullerton, CA). The impact of RP-1 on radioisotopic precursor incorporation over time was determined as [1 − (experimental counts per minute/control counts per minute)] × 100% and was expressed as the mean percentage of inhibition versus control (±SD).
Statistical analyses.
Differences in staphylocidal effects, membrane permeabilization, or membrane depolarization between control, RP-1-, or tPMP-1-treated cells were compared by the unpaired Student's t test, employing the statistical package within Sigma Plot (version 2.02). Probability values (P) of ≤0.05 were considered significant.
RESULTS
Staphylocidal activities.
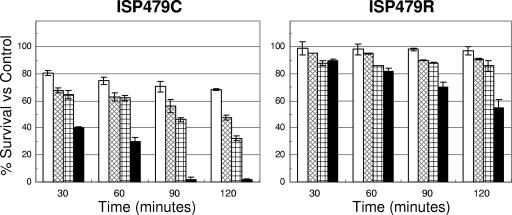
The comparative staphylocidal effects of RP-1 and tPMP-1 on strains ISP479C and ISP479R are summarized in Fig. 1. Consistent with our previously published data (13, 22), ISP479C was significantly more susceptible to tPMP-1 than ISP479R (30% versus 82% of cells survived after 60 min of incubation, respectively; P < 0.05) (Fig. 1). The staphylocidal activities caused by RP-1 occurred in a time- and concentration-dependent manner, reflecting the intrinsic tPMP-1 susceptibilities of ISP479C and ISP479R. For example, 100 μg/ml of RP-1 exerted a significant microbicidal effect against ISP479C (e.g., ∼30% survival versus control at 120 min of incubation; P < 0.05). By comparison, at the same RP-1 concentration, the rate and extent of RP-1-induced killing of ISP479R were substantially reduced (e.g., ∼90% survival versus control at 120 min of incubation). These survival data were significantly higher than those observed with RP-1 exposure of ISP479C cells (P < 0.05). There was no significant loss in the viability of cells exposed to control buffer over the 2-h period (data not shown). On the basis of these data demonstrating statistically significant differential staphylocidal efficacy versus the strain pair, a concentration of RP-1 of 50 μg/ml was selected for use in the ensuing assays (see below).
FIG. 1.
Staphylocidal activities of RP-1 and tPMP-1 on ISP479C and ISP479R at an initial inoculum of 107 CFU/ml. Logarithmic-phase S. aureus cells were exposed to either 25 (□), 50 ( ), or 100 (
) μg/ml of RP-1 or 2 μg/ml (▪) of tPMP-1. The mean percentage (±SD) of S. aureus survival versus controls was calculated from at least three independent experiments.
), or 100 (
) μg/ml of RP-1 or 2 μg/ml (▪) of tPMP-1. The mean percentage (±SD) of S. aureus survival versus controls was calculated from at least three independent experiments.
MICs.
Using an assay format modified to assess antimicrobial peptide efficacy, the relative MICs of RP-1 and tPMP-1 against a panel of S. aureus isolates were investigated in vitro, as summarized in Table 1. As documented in previous studies (35), the antistaphylococcal efficacies of RP-1 and tPMP-1 were significantly influenced by assay condition. For example, the MIC ranges for RP-1 were 8 to 32 μg/ml in MEM containing up to 20% MHB but >128 μg/ml in 100% MHB. Similarly, the MICs of tPMP-1 were >32 μg/ml when MEM contained 10 to 20% MHB.
TABLE 1.
Influence of in vitro assay conditions on antistaphylococcal MICs of RP-1 and tPMP-1
| Strain | MIC (μg/ml) ina: |
|||||||
|---|---|---|---|---|---|---|---|---|
| MEM + 10% MHB |
MEM + 20% MHB |
MEM + 50% MHB |
MEM + 100% MHB |
|||||
| RP-1b | tPMP-1c | RP-1 | tPMP-1 | RP-1 | tPMP-1 | RP-1 | tPMP-1 | |
| ISP479C | 16 | >32 | 16 | >32 | 64 | NDd | >128 | ND |
| ISP479R | 8 | >32 | 16 | >32 | 32 | ND | >128 | ND |
| COL | 16 | >32 | 32 | >32 | 64 | ND | >128 | ND |
| 67-0 | 32 | >32 | 32 | >32 | 64 | ND | >128 | ND |
| 29213 | 32 | >32 | 32 | >32 | 128 | ND | >128 | ND |
Data represent geometric means of a minimum of two studies performed independently. Modified standard CLSI guidelines were used, such that MEM with increasing proportions (vol/vol) of MHB was assessed (see Materials and Methods). Note that RP-1 and tPMP-1 yielded comparable results when tested in identical conditions. For example, in MEM + 20% MHB, the MICs of tPMP-1 were >32 μg/ml, while those for RP-1 were 32 μg/ml for strains COL, 67-0, and 29213. The limited availability of purified tPMP-1 precluded testing at levels >32 μg/ml.
The maximum RP-1 concentration tested was 128 μg/ml.
The maximum tPMP-1 concentration tested was 32 μg/ml; availability precluded further testing.
ND, not determined (see footnote c).
Post-RP-1 exposure effect.
Similar to native tPMP-1 (36), exposure to RP-1 (50 μg/ml) produced a relatively short postexposure growth inhibitory effect (∼0.5 h) for ISP479C. In contrast, there was no detectable postexposure effect for ISP479R, paralleling its tPMP-1-resistant phenotype.
Membrane permeabilization.
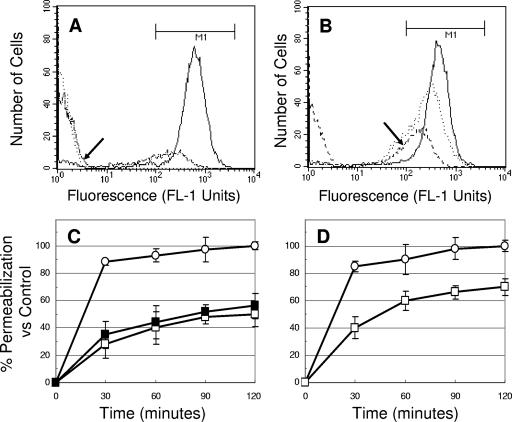
Figure 2 shows the MP caused by RP-1 or tPMP-1 in S. aureus ISP479C and ISP479R. Exposure of ISP479C to RP-1 (50 μg/ml) resulted in rapid and significant MP (Fig. 2A and C). For example, ∼99% calcein leakage from ISP479C cells was observed over the 120 min of the RP-1 exposure period (P < 0.05 versus untreated controls) (Fig. 2A, arrow, and C). In contrast, exposure of ISP479R to RP-1 at the same concentration yielded only 50% MP (Fig. 2B, arrow, and C). Interestingly, exposure of ISP479R to a higher concentration of RP-1 (100 μg/ml) did not substantially increase the extent of MP (Fig. 2C). MP by RP-1 versus that by tPMP-1 were also directly compared. Consistent with our prior published data (13), tPMP-1 (2 μg/ml) significantly permeabilized the cell membranes of both strains in a time-dependent manner (Fig. 2D), although ISP479C was more susceptible to MP than ISP479R (99% versus 70% of cells permeabilized after 120 min of incubation, respectively). There was no significant loss of calcein observed in control cells over the 2-h experimental period (data not shown).
FIG. 2.
S. aureus MP by RP-1 and tPMP-1. Panels A and B show the fluorescence distribution (FL-1 units) versus the number of S. aureus ISP479C (A) and ISP479R (B) cells treated with 50 μg/ml RP-1 over 0 (—), 60 ( · · · ), and 120 (- - - - ) minutes. Arrows in panels A and B represent the extent of fluorescence of ISP479C and ISP479R at 120 min of RP-1 (50 μg/ml) exposure, respectively. Panel C shows the percent MP by RP-1 at 50 μg/ml on ISP479C (○) and ISP479R (□) and RP-1 at 100 μg/ml on ISP479R (▪) versus controls (±SD). Panel D shows the percent MP of ISP479C (○) and ISP479R (□) caused by tPMP-1 (2 μg/ml) versus controls. Each experiment was done in triplicate.
Membrane depolarization.
Similar to native tPMP-1 (31), RP-1 did not depolarize either S. aureus strain. As anticipated, exposure to valinomycin caused significant membrane depolarization in both strains (>95%; P < 0.05 versus controls).
Inhibition of intracellular macromolecular synthesis.
RP-1- and antibiotic-induced DNA (Fig. 3), RNA, and protein (Fig. 4) synthesis inhibition in the test strains was examined.
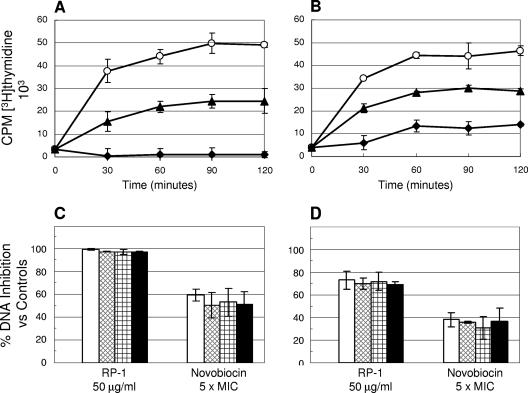
FIG. 3.
Effect of RP-1 on DNA synthesis in S. aureus ISP479C and ISP479R. Panel A (ISP479C) and panel B (ISP479R) present the actual [methyl-3H]thymidine incorporation kinetic curves (○, untreated control; ▴, novobiocin; and ⧫, RP-1). CPM, counts per minute. Panel C (ISP479C) and panel D (ISP479R) show the percent DNA synthesis inhibition versus untreated controls over 120 min of incubation (□, 30 min; ▩, 60 min; , 90 min; and ▪, 120 min). Novobiocin was used as a positive control DNA synthesis inhibitor.
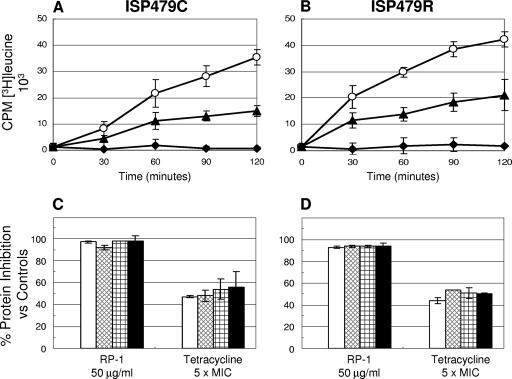
FIG. 4.
Effect of RP-1 on protein synthesis in S. aureus ISP479C and ISP479R. Panel A (ISP479C) and panel B (ISP479R) present the actual [4,5-3H]leucine incorporation kinetic curves (○, untreated control; ▴, novobiocin; and ⧫, RP-1). Panel C (ISP479C) and panel D (ISP479R) show the percent protein synthesis inhibition versus untreated controls over 120 min of exposure (□, 30 min; ▩, 60 min; , 90 min; and ▪, 120 min). Tetracycline was used as a positive control protein synthesis inhibitor.
RP-1 (50 μg/ml) caused rapid and significant inhibition of DNA synthesis in ISP479C (P < 0.05 versus untreated control) (Fig. 3A and C). In contrast, for ISP479R, the extent of DNA synthesis inhibition was substantially less (by ∼30%) than that observed for strain ISP479C (P < 0.05) (Fig. 3B and D). As expected, 5× MIC of the comparator, novobiocin, resulted in substantial inhibition of DNA synthesis in both S. aureus strains versus untreated controls.
Paralleling the data above for DNA synthesis inhibition, nearly complete inhibition of RNA synthesis was seen in ISP479C (>90% inhibition; P < 0.05 versus control). In contrast, for ISP479R, only 80 to 90% RNA synthesis inhibition was observed over the same time period. As predicted, rifampin (5× MIC) caused RNA synthesis inhibition (30% to 60% versus controls) in both strains (P < 0.05 versus control; data not shown).
In contrast to the differential inhibitory effects of RP-1 observed for DNA and RNA synthesis between the study strains, RP-1 reduced protein synthesis to equivalent extents in both strains over the 2-h period (∼90% inhibition; P < 0.05 versus controls) (Fig. 4). The control agent, tetracycline, at 5× MIC reduced protein synthesis by ∼40 to 60% over the 2-h incubation period for strains ISP479C and ISP479R (P < 0.05 versus controls) (Fig. 4). The present observations of macromolecular synthesis inhibition by RP-1 are similar to those previously generated and published for native tPMP-1 (22).
DISCUSSION
APs from a wide variety of organisms, including bacteria (e.g., polymyxins), insects (e.g., melittins and cecropins), amphibians (e.g., magainins), and mammals (e.g., defensins, lactoferricins, and cathelicidins), have been isolated and characterized (2, 8, 9, 11, 19, 27, 29, 41). PMPs are a family of small cationic APs released from mammalian platelets which have multiple structural and functional characteristics that contribute to innate host defense against microbial infections (27, 28). These functions include (i) direct interactions with microorganisms leading to cell death (22, 23, 27, 30); (ii) reductions in microbial binding to target host tissues (e.g., platelets and endothelial cells) (28); and (iii) chemokine-mediated phagocyte clearance of microbes (15, 25, 27, 37).
These distinct but complementary activities have been putatively linked to distinct structural domains within tPMP-1. For example, Yount et al. (39) found that the C-terminal domain of tPMP-1 is cation rich and α-helical, properties important to the microbicidal functions of other APs (26, 37, 38). In contrast, the N terminus contains a CXC domain characteristic of classic α-chemokine molecules. These two termini are linked by an interposing γ-core common to other disulfide-stabilized APs (39). Yet the critical structure-activity relationships governing the antimicrobial functions of PMPs, such as tPMP-1, have not been fully defined. The present study addresses the hypothesis that the activities and mechanisms of antimicrobial determinants in native PMP molecules can be precisely recapitulated using engineered peptide congeners.
Recently, Yeaman et al. engineered the synthetic congener peptide RP-1, modeled upon the C-terminal α-helices of native tPMP-1 and other mammalian PF-4 homologues (35). Notably, this synthetic peptide exerts direct, highly potent, and durable antimicrobial activity on a serum-resistant Escherichia coli isolate in human blood, plasma, and serum (35). Of great importance, they demonstrated that the antimicrobial activity of RP-1 was logarithmically amplified in such biomatrices but minimal in artificial media, such as those used for standard MIC assays. The latter observations were confirmed in the present study. These facts highlight the concept that antimicrobial host defense molecules such as microbicidal peptides and kinocidins are likely optimized to function in cognate host defense contexts (37, 40).
In the current studies, several important observations emerged. First, RP-1 essentially retained the in vitro antistaphylococcal mechanisms of native tPMP-1. Thus, killing by both molecules occurred in a concentration- and time-dependent manner. Also, similar to native tPMP-1 (22), RP-1 showed bactericidal activities that were significantly more rapid and extensive against the tPMP-1-susceptible strain than the tPMP-1-resistant strain. In addition, the former but not the latter strain exhibited a definable, albeit small, post-RP-1 exposure growth inhibition effect. Second, RP-1 and tPMP-1 caused MP in both S. aureus strains, with the extent and rapidity of effect paralleling their intrinsic tPMP-1 susceptibility profiles and the degree and kinetics of peptide-induced killing. In addition, similar to tPMP-1 (31), RP-1 did not depolarize the S. aureus membrane in either strain. This membrane effect profile (extensive MP without depolarization) is characteristic of several PMPs (31).
Equivalent to tPMP-1, RP-1 significantly inhibited relevant isotope incorporation into macromolecules, indicating direct and/or indirect inhibition of DNA, RNA, and protein synthesis in each study strain. However, the extent of DNA and RNA synthesis inhibition caused by RP-1 was substantially greater in the tPMP-1-susceptible strain than in the tPMP-1-resistant strain, precisely mirroring our previous studies with native tPMP-1 (22). As previously discussed for the native molecule (22), it is not clear at this time if such differences in RP-1-induced inhibition of DNA and RNA synthesis relate to differences in (i) the extent of intracellular RP-1 access or (ii) the extent of MP, secondarily dampening nucleic acid synthesis (e.g., due to osmoregulatory collapse or reduced cellular energetics). Interestingly, similar to results with native tPMP-1 (22), the extents of protein synthesis inhibition were equivalent for both strains. The latter observations may relate to differential inhibition thresholds for the three different intracellular targets. Thus, although the degrees of DNA and RNA synthesis inhibition due to RP-1 were substantially less in the tPMP-1-resistant strain than in the tPMP-1-susceptible cells, it is possible that the extent of inhibition in both strains may have exceeded a threshold sufficient to impair downstream protein synthesis.
It should be recognized that the extents of antistaphylococcal effects (MICs and staphylocidal kinetics) differed for tPMP-1 and RP-1 depending on assay conditions. At least two possible considerations may account for these differences. First, the antistaphylococcal efficacies of tPMP-1 and RP-1 are underestimated in artificial media. In contrast, as illustrated in our prior studies using RP-1, the antimicrobial activities of such molecules are dramatically enhanced in relevant blood and blood-derived matrices, such as plasma and serum (35). Such observations underscore the benefit of employing nonconventional assay systems, such as the biomatrix assay, to assess the relevance of the antimicrobial functions of anti-infective polypeptides. Second, as synthetic RP-1 (∼2.1 kDa [35]) and native tPMP-1 (∼8 kDa [40]) have significant differences in mass, the molar concentrations of these agents differ when equivalent masses are tested. Therefore, one or both of the above variables may contribute to the apparent differences in the antistaphylococcal potency of these molecules when evaluated using in vitro assay systems. Nonetheless, the current data indicate that despite potential differences in molar potency under certain test conditions, the net mechanisms of action of RP-1 and tPMP-1 are equivalent. These considerations reinforce the overarching rationale for engineering novel synthetic peptides for systemic efficacy based on native templates evolved to function in cognate environments in vivo.
Taken together, these data support the notion that relevant microbicidal mechanisms of selected APs can be faithfully reproduced by synthetic molecules modeled upon corresponding structural domains. It should be emphasized, however, that the collective antimicrobial effects of such APs in vivo (e.g., direct microbicidal activity and indirect leukocyte potentiation) likely benefit from the complementary effects of additional structural domains, such as those potentially conferring chemokine (39) or scaffold (e.g., γ-core) (40) functions. Nonetheless, the present findings underscore the concept that novel synthetic congeners can be designed to exert specific functions equivalent to those of cognate effector domains in native effector molecules and which may optimize multiple host defense functions.
Acknowledgments
This research was supported by grants from the National Institutes of Health (AI39108 to A.S.B. and AI39001 and AI48031 to M.R.Y.) and the American Heart Association (0265054Y and 0465142Y to Y.Q.X.).
We gratefully acknowledge Deborah Kupferwasser and Kimberly Gank for technical assistance.
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Bayer, A. S., P. McNamara, M. R. Yeaman, N. Lucindo, T. Jones, A. L. Cheung, H.-G. Sahl, and R. A. Proctor. 2006. Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1. J. Bacteriol. 188:211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechinger, B., M. Zasloff, and S. J. Opella. 1993. Structure and orientation of the antibiotic peptide magainin in membranes by solid-state nuclear magnetic resonance spectroscopy. Protein Sci. 2:2077-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhawan, V. K., M. R. Yeaman, and A. S. Bayer. 1990. Influence of in vitro susceptibility phenotype against thrombin-induced platelet microbicidal protein on treatment and prophylaxis outcomes of experimental Staphylococcus aureus endocarditis. J. Infect. Dis. 180:1561-1568. [DOI] [PubMed] [Google Scholar]
- 4.Dhawan, V. K., M. R. Yeaman, A. L. Cheung, E. Kim, P. M. Sullam, and A. S. Bayer. 1997. Phenotypic resistance to platelet microbicidal protein in vitro is correlated with enhanced virulence in experimental endocarditis due to Staphylococcus aureus. Infect. Immun. 65:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhawan, V. K., A. S. Bayer, and M. R. Yeaman. 1998. In vitro resistance to thrombin-induced platelet microbicidal protein is associated with enhanced progression and hematogenous dissemination in experimental Staphylococcus aureus infective endocarditis. Infect. Immun. 66:3476-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake, T. A., G. M. Rodgers, and M. A. Sande. 1984. Tissue factor is a major stimulus for vegetation formation in enterococcal endocarditis in rabbits. J. Clin. Investig. 73:1750-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake, T. A., and M. Pang. 1988. Staphylococcus aureus induces tissue factor expression in cultured human cardiac valve endothelium. J. Infect. Dis. 157:749-756. [DOI] [PubMed] [Google Scholar]
- 8.Gazit, E., I. R. Miller, P. C. Biggin, M. S. P. Sansom, and Y. Shai. 1996. Structure and orientation of the mammalian antibacterial peptide cecropin P1 within phospholipid membranes. J. Mol. Biol. 258:860-870. [DOI] [PubMed] [Google Scholar]
- 9.Hancock, R. E. W., T. Falla, and M. H. Brown. 1995. Cationic bactericidal peptides. Adv. Microb. Physiol. 37:135-175. [DOI] [PubMed] [Google Scholar]
- 10.Hirano, L., and A. S. Bayer. 1991. β-Lactam-β-lactamase-inhibitor combinations are active in experimental endocarditis caused by β-lactamase-producing oxacillin-resistant staphylococci. Antimicrob. Agents Chemother. 35:685-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang, P. M., N. Zhou, X. Shan, C. H. Arrowsmith, and H. J. Vogel. 1998. Three-dimensional solution structure of lactoferricin b, an antimicrobial peptide derived from bovine lactoferrin. Biochemistry 37:4288-4298. [DOI] [PubMed] [Google Scholar]
- 12.Koo, S. P., A. S. Bayer, B. L. Kagan, and M. R. Yeaman. 1999. Membrane permeabilization by thrombin-induced platelet microbicidal protein 1 is modulated by transmembrane voltage polarity and magnitude. Infect. Immun. 67:2475-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koo, S. P., A. S. Bayer, and M. R. Yeaman. 2001. Diversity in antistaphylococcal mechanisms among membrane-targeting antimicrobial peptides. Infect. Immun. 69:4916-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koo, S. P., M. R. Yeaman, C. C. Nast, and A. S. Bayer. 1997. The cytoplasmic membrane is a primary target for the staphylocidal action of thrombin-induced platelet microbicidal protein. Infect. Immun. 65:4795-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy, P. M. 1997. Neutrophil receptors for interleukin-8 and related CXC chemokines. Semin. Hematol. 34:311-318. [PubMed] [Google Scholar]
- 16.Nicolas, P., D. Vanhoye, and M. Amiche. 2003. Molecular strategies in biological evolution of antimicrobial peptides. Peptides 24:1669-1680. [DOI] [PubMed] [Google Scholar]
- 17.Shah, P. M. 2005. The need for new therapeutic agents: what is the pipeline? Clin. Microbiol. Infect. 11(Suppl. 3):36-42. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro, H. M. 1990. Cell membrane potential analysis. Methods Enzymol. 33:25-35. [DOI] [PubMed] [Google Scholar]
- 19.Storm, D. R., K. S. Rosenthal, and P. E. Swanson. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46:175-187. [DOI] [PubMed] [Google Scholar]
- 20.Tang, Y. Q., M. R. Yeaman, and M. E. Selsted. 2002. Antimicrobial peptides from human platelets. Infect. Immun. 70:6524-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toke, O. 2005. Antimicrobial peptides: new candidates in the fight against bacterial infections. Biopolymers 80:717-735. [DOI] [PubMed] [Google Scholar]
- 22.Xiong, Y. Q., A. S. Bayer, and M. R. Yeaman. 2002. Inhibition of intracellular macromolecular synthesis in Staphylococcus aureus by thrombin-induced platelet microbicidal proteins. J. Infect. Dis. 185:348-356. [DOI] [PubMed] [Google Scholar]
- 23.Xiong, Y. Q., K. Mukhopadhyay, M. R. Yeaman, J. Adler-Moore, and A. S. Bayer. 2005. Functional interrelationships between cell membrane and cell wall in antimicrobial peptide-mediated killing of Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3114-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong, Y. Q., M. R. Yeaman, and A. S. Bayer. 1999. In vitro antibacterial activities of platelet microbicidal protein and neutrophil defensin against Staphylococcus aureus are influenced by antibiotics differing in mechanism of action. Antimicrob. Agents Chemother. 43:1111-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan, Z., J. Zhang, J. C. Holt, G. J. Steward, S. Niewiarowski, and M. Poncz. 1994. Structural requirements of platelet chemokines for neutrophil activation. Blood 84:2329-2339. [PubMed] [Google Scholar]
- 26.Yang, D., Q. Chen, D. M. Hoover, P. Staley, K. D. Tucker, J. Lubkowski, and J. J. Oppenheim. 2003. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J. Leukoc. Biol. 74:448-455. [DOI] [PubMed] [Google Scholar]
- 27.Yeaman, M. R. 1997. The role of platelets in antimicrobial host defense. Clin. Infect. Dis. 25:951-970. [DOI] [PubMed] [Google Scholar]
- 28.Yeaman, M. R., and A. S. Bayer. 1999. Antimicrobial peptides from platelets. Drug Resist. Updates 2:116-126. [DOI] [PubMed] [Google Scholar]
- 29.Yeaman, M. R., and N. Y. Yount. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27-55. [DOI] [PubMed] [Google Scholar]
- 30.Yeaman, M. R., Y. Q. Tang, A. J. Shen, A. S. Bayer, and M. E. Selsted. 1997. Purification and in vitro activities of rabbit platelet microbicidal proteins. Infect. Immun. 65:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeaman, M. R., A. S. Bayer, S. P. Koo, W. Foss, and P. M. Sullam. 1998. Platelet microbicidal proteins and neutrophil defensin disrupt the Staphylococcus aureus cytoplasmic membrane by distinct mechanisms of action. J. Clin. Investig. 101:178-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeaman, M. R., D. Cheng, B. Desai, L. I. Kupferwasser, Y. Q. Xiong, K. D. Gank, J. E. Edwards, Jr., and A. S. Bayer. 2004. Susceptibility to thrombin-induced platelet microbicidal protein is associated with increased fluconazole efficacy against experimental endocarditis due to Candida albicans. Antimicrob. Agents Chemother. 48:3051-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeaman, M. R., S. S. Soldan, M. A. Ghannoum, J. E. Edwards, Jr., S. G. Filler, and A. S. Bayer. 1996. Resistance to platelet microbicidal protein results in increased severity of experimental Candida albicans endocarditis. Infect. Immun. 64:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeaman, M. R., A. S. Ibrahim, J. A. Ritchie, S. G. Filler, A. S. Bayer, J. E. Edwards, Jr., and M. A. Ghannoum. 1995. Sublethal concentrations of platelet microbicidal protein reduce Candida albicans interactions with vascular endothelial cells in vitro, abstr. 491. Abstr. IDSA Annu. Meet. 1995.
- 35.Yeaman, M. R., K. D. Gank, A. S. Bayer, and E. P. Brass. 2002. Synthetic peptides that exert antimicrobial activities in whole blood and blood-derived matrices. Antimicrob. Agents Chemother. 46:3883-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeaman, M. R., D. C. Norman, and A. S. Bayer. 1992. Platelet microbicidal protein enhances antibiotic-induced killing of and postantibiotic effect in Staphylococcus aureus. Antimicrob. Agents Chemother. 36:1665-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeaman, M. R., and N. Y. Yount. 2005. Code among chaos: immunorelativity and the AEGIS model of antimicrobial peptides. ASM News 71:21-27. [Google Scholar]
- 38.Yeaman, M. R. 2005. Modular determinants of coordinated functions in platelet kinocidins. Presented at the 105th General Meeting of the American Society for Microbiology, Atlanta, Ga., 5 to 9 June 2005.
- 39.Yount, N. Y., K. D. Gank, Y. Q. Xiong, A. S. Bayer, T. Pender, W. H. Welch, and M. R. Yeaman. 2004. Platelet microbicidal protein 1: structural themes of a multifunctional antimicrobial peptide. Antimicrob. Agents Chemother. 48:4395-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yount, N. Y., and M. R. Yeaman. 2004. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 101:7363-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanetti, M., R. Gennaro, and D. Romeo. 1995. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 374:1-5. [DOI] [PubMed] [Google Scholar]