Abstract
The small antimicrobial peptide PAF26 (Ac-RKKWFW-NH2) has been identified by a combinatorial approach and shows preferential activity toward filamentous fungi. In this work, we investigated the mode of action and inhibitory effects of PAF26 on the fungus Penicillium digitatum. The dye Sytox Green was used to demonstrate that PAF26 induced cell permeation. However, microscopic observations showed that sub-MIC concentrations of PAF26 produced both alterations of hyphal morphology (such as altered polar growth and branching) and chitin deposition in areas of no detectable permeation. Analysis of dose-response curves of inhibition and permeation suggested that growth inhibition is not solely a consequence of permeation. In order to shed light on the mode of PAF26 action, its antifungal properties were compared with those of melittin, a well-known pore-forming peptide that kills through cytolysis. While the 50% inhibitory concentrations and MICs of the two peptides against P. digitatum mycelium were comparable, they differed markedly in their fungicidal activities toward conidia and their hemolytic activities toward human red blood cells. Kinetic studies showed that melittin quickly induced Penicillium cell permeation, while PAF26-induced Sytox Green uptake was significantly slower and less efficient. Therefore, the ultimate growth inhibition and morphological alterations induced by PAF26 for P. digitatum are not likely a result of conventional pore formation. Fluorescently labeled PAF26 was used to demonstrate its specific in vivo interaction and translocation inside germ tubes and hyphal cells, at concentrations as low as 0.3 μM (20 times below the MIC), at which no inhibitory, morphological, or permeation effects were observed. Interestingly, internalized PAF26 could bind to cellular RNAs, since in vitro nonspecific RNA binding activity of PAF26 was demonstrated by electrophoretic mobility shift assays. We propose that PAF26 is a short, de novo-designed penetratin-type peptide that has multiple detrimental effects on target fungi, which ultimately result in permeation and killing.
Antimicrobial peptides (AMP) of natural and synthetic origin inhibit the growth of human and plant pathogens (2, 32, 40). Knowledge of the mode of action of AMP is critical for attempts to increase their potency and, even more challenging, their specificity. Numerous studies aimed at understanding the mechanism of AMP action by using different experimental approaches have been reported in recent years (2, 6, 12, 29, 38). A major group of AMP includes the so-called cationic antimicrobial peptides (CAMP), which usually also display amphipathic properties. Interaction of CAMP with membrane mimetics or with selected microbial cells has led to the conclusion that peptide-membrane interactions drive their antimicrobial properties and that many of them permeabilize target cells (4, 12, 29). However, it is open to discussion whether this is their primary or even unique toxic effect, and neither the mode of action of membrane-lytic AMP nor the bases for their selectivity toward specific cells are fully understood. Recent studies on natural peptides point toward the existence of additional functions and properties related to host defense that are not linked to cell permeation but could mediate microbial killing (2, 6, 12, 38).
In previous work, we identified from a peptide combinatorial library and characterized a group of hexapeptides, named PAFs, with antimicrobial activity against certain filamentous fungi, including plant pathogens (13, 15) and human dermatophytes (B. López-García et al., manuscript in preparation). They inhibit in vivo infection of selected phytopathogens. PAFs are very short CAMP with closely related sequences and distinct activity profiles, and some of them show high antimicrobial activity against fungi but lower toxicity against nontarget bacterial and yeast cells. Although these peptides were identified through a nonbiased approach, they show properties of natural AMP, together with other similarly identified synthetic peptides (5, 18, 26). We share the view, suggested previously (12), that these short peptides could be very valuable for a better understanding of the mode of action of this new class of antibiotics, since they represent a minimum core domain for biological activity and thus can be used as tools to dissect the factors involved in the microbicidal activity and specificity of CAMP.
PAF26 displayed activity against several filamentous fungi with a potency similar to that of the cytotoxic peptide melittin (Table 1), but it did not show the high toxicity of melittin toward Escherichia coli or Saccharomyces cerevisiae (15). Melittin is a natural membrane-lytic peptide of 26 amino acids isolated from honeybees that is toxic to microbes but also to human cells, since it kills by forming pores in cell membranes with poor specificity (37).
TABLE 1.
Sequence and growth inhibition properties of synthetic peptides toward Penicillium digitatum
| Peptide | Amino acid sequence | Conidia (5% PDB)
|
Mycelium (5% PDB)
|
||
|---|---|---|---|---|---|
| IC50 (μM)a | MIC (μM) | IC50 (μM)a | MIC (μM) | ||
| PAF26 | Ac-RKKWFW-NH2 | 2.2 ± 0.2 | 4.0 | 1.3 ± 0.4 | 6.0 |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ | 1.1 ± 0.5 | 4.0 | 2.9 ± 0.7 | 8.0 |
| Mel.subK7I | GIGAVLIVLTTGLPALISWIKRKRQQ | 3.2 ± 0.9 | 8.0 | ND | >15.0 |
IC50 represent averages of at least three independent experiments with standard deviations. ND, not determined.
Efforts are currently under way by our group to increase the potency and specificity of PAFs, particularly PAF26. In order to achieve this, it is important to understand the mode of action of PAFs and the bases for their selectivity. We know that PAF26 interacts electrostatically and nonchirally with the fungal envelope. Also, distinct PAF peptides interact with and insert into model lipid membranes, and their antifungal potency correlates with the strength of such interactions (14). To gain information on the mode of action of PAF26, we investigated the inhibitory effects of the peptide in relation to its ability to permeate the membrane of the fungus Penicillium digitatum in a comparison study with melittin. The interaction and morphological changes that PAF26 induces on mycelium were also evaluated in the context of its antifungal activity, translocation, and permeation properties.
MATERIALS AND METHODS
Microorganism.
The fungal isolate used in this study was Penicillium digitatum PHI-26 (13), a field isolate highly virulent to citrus fruits. It was cultured on potato dextrose agar (PDA) (Difco, Detroit, Mich.) plates for 7 to 10 days at 24°C. Conidia were collected and transferred to distilled water, filtered, titrated with a hemocytometer, adjusted to the appropriate concentration, and used in the assays.
Synthesis of peptides.
Peptides (Table 1) were purchased at >90% purity from Genscript Corporation (Piscataway, N.J.), where they were synthesized by solid-phase methods using N-(9-fluorenyl)methoxycarbonyl (Fmoc) chemistry. PAF26 was acetylated at the N terminus (Ac) and amidated at the C terminus (NH2) (Table 1). Stock solutions of each peptide were prepared at 1 mM in 5 mM 3-(N-morpholino)-propanesulfonic acid (MOPS) (pH 7) buffer and stored at −20°C. Peptide concentrations were determined by measuring the absorbance at 280 nm (ɛ280 for the Trp residue, 5,600 M−1 cm−1) and rechecked before each experiment. PAF26 was also synthesized labeled with fluorescein 5-isothiocyanate (FITC) by covalent modification of its N terminus with FITC. The concentration of FITC-labeled PAF26 was determined by measuring the absorbance at 495 nm (ɛ495 for the FITC fluorophore, 68,000 M−1 cm−1).
Fluorescence microscopy.
Conidia (90 μl, 2.5 × 104 conidia/ml) dispensed into wells of sterile 96-well microtiter plates (Nunc, Roskilde, Denmark) were grown in 1/20-diluted potato dextrose broth (5% PDB) at 24°C to an optical density (OD) at 492 nm of approximately 0.1. Subsequently, 10 μl of peptides from 10× stock solutions were added to a final concentration of 0.1 to 100 μM and allowed to grow for another 48 h in the presence of peptides. After this incubation time, a Sytox Green (SG) (Molecular Probes, Eugene, Oreg.) stock solution (4 μM) was added to a final concentration of 0.2 μM, and samples were incubated for 5 min in the dark; subsequently, 0.1% (wt/vol) calcofluor white (CFW) (Fluorescent Brightener 28; Sigma-Aldrich) was added to a final concentration of 50 μg/ml, and samples were incubated again for 5 min in the dark (24). Finally, mycelium was washed, resuspended in 20% glycerol solution, and visualized under the microscope. Fluorescence was examined and photographed with an Eclipse E 600 epifluorescence microscope (Nikon) with the filters set at an excitation wavelength of 450 to 490 nm and an emission wavelength at 515 to 565 nm for SG detection and at an excitation wavelength of 395 nm and an emission wavelength of 440 nm for CFW detection.
To visualize the interaction of PAF26 with fungal structures, conidia or mycelia were incubated in sterile water or 5% PDB with 0.3, 3, or 30 μM FITC-labeled PAF26 for different times. Samples were visualized with a Leica TCS SL confocal laser scanning microscope (Leica), with excitation at 488 nm and emission at 510 to 560 nm. Controls used free FITC (Sigma-Aldrich, St. Louis, Mo.).
In vitro antimicrobial activity assays.
Antimicrobial activities were determined using a microtiter plate assay as previously described (1, 15) on 2.5 × 104 conidia/ml. Growth was determined by the OD at 492 nm in a Multiskan Spectrum microplate spectrophotometer (Thermo Electron Corporation, Finland). The growth medium was 5% PDB containing 0.003% (wt/vol) chloramphenicol. Three replicates were prepared for each treatment. The MIC was defined as the lowest peptide concentration that showed no growth at the end of the experiment (after 4 days of incubation) in all the experiments carried out with that peptide. The IC50 of a peptide was defined as the concentration required to obtain 50% inhibition, and the value for each experiment was estimated by adjustment to a four-parameter logistic curve (SigmaPlot, version 8.02; SPSS Inc., Chicago, Ill.).
Fungicidal kinetics of peptides toward P. digitatum conidia were determined by incubating 104 conidia/ml with 30 μM of either PAF26 or melittin in distilled water at 24°C. At each time point, aliquots were removed, diluted, and plated onto peptide-free PDA plates. Plates were incubated 48 h at 24°C, and CFU were counted. Three replicas were analyzed for each time point and peptide treatment.
Sytox Green uptake quantification.
SG uptake by mycelium after peptide treatment was determined by fluorometric measurement using a microplate assay. Conidia (90 μl, 2.5 × 104 conidia/ml) dispensed into wells of microtiter plates were grown in 5% PDB at 24°C for 24 h. Subsequently, 10 μl of peptides from 10× stock solutions were added as described above and allowed to grow for 24 h in the presence of peptides. After this additional incubation, SG was added to a final concentration of 0.2 μM, and samples were incubated four additional hours in the dark at 24°C. Three replicates were prepared for each treatment. After incubation, fluorescence emission was measured with a microplate reader (Fluoroskan Ascent FL; Labsystems, Finland) at an excitation wavelength of 485 nm and an emission wavelength of 538 nm. Growth inhibition was determined in the same samples as an increase in the OD over the values before the addition of peptides. IC50s and MICs for P. digitatum mycelium determined in this set of experiments were found to be different from those found for conidia (see Table 1 and Results).
Time course analysis of SG uptake was conducted following essentially the same procedure except that peptides and SG were added simultaneously to 24-h-grown mycelium, and immediately after peptide-SG addition, fluorescence emission was recorded at 2-min intervals with a microplate reader (Fluoroskan Ascent FL).
Hemolytic activity assay.
The hemolytic activities of the peptides were determined on human red blood cells. Freshly obtained blood cells were washed three times in phosphate-buffered saline (PBS) and resuspended in four times their original volume of PBS. A 90-μl volume of the suspension was plated into wells of 96-well plates, and 10 μl of a 10× stock solution of each peptide was added to a final concentration of 1, 10, or 100 μM. Samples were incubated at 37°C for 1 h and centrifuged at 1,000 × g for 5 min, and the supernatant was transferred to fresh 96-well plates. Release of hemoglobin was monitored by measurement of absorbance at 415 and 575 nm with a Multiskan Spectrum microplate spectrophotometer. No hemolysis and 100% hemolysis were determined for controls with PBS and 0.1% Triton X-100, respectively. The hemolytic activity of each peptide was calculated as the percentage of total hemoglobin released compared with that released by incubation with 0.1% Triton X-100 by using the following formula: % hemolysis = (A415 in the peptide solution − A415 in PBS)/(A415 in 0.1% Triton X-100 − A415 in PBS) × 100.
EMSAs.
Assays of peptide RNA binding activity were conducted by an electrophoretic mobility shift assay (EMSA) essentially as described previously (17). Two hundred fifty nanograms of S. cerevisiae tRNA (Roche Diagnostics) was incubated for 30 min on ice with increasing molar concentrations of PAF26 in a 20-μl volume of TE (10 mM Tris-HCl [pH 8], 1 mM EDTA). Samples were electrophoresed and stained with ethidium bromide.
RESULTS
Permeation and morphological alterations of Penicillium digitatum mycelium exposed to PAF26.
To characterize the mode of action and interaction of PAF26 with the susceptible fungus P. digitatum, we used fluorescence microscopy and an assay based on the uptake of the fluorescent dye Sytox Green. SG binding to nucleic acids results in a >500-fold enhancement in its fluorescence emission. Since the dye does not penetrate live cells, it has been used to assess the integrity of biological membranes (28) and to characterize natural or synthetic antimicrobial peptides active against fungal pathogens (5, 31). We carried out our experiments on mycelia. PAF peptides added to actively growing mycelia affected growth and had inhibitory properties somewhat different than those observed when conidia were used as starting material in the antimicrobial assays (compare the IC50s and MICs in Table 1). Under these assay conditions, the IC50 of PAF26 toward P. digitatum was slightly reduced, from 2.2 μM to 1.3 μM (1.3 μg/ml), while the MIC increased from 4 μM to 6 μM (Table 1).
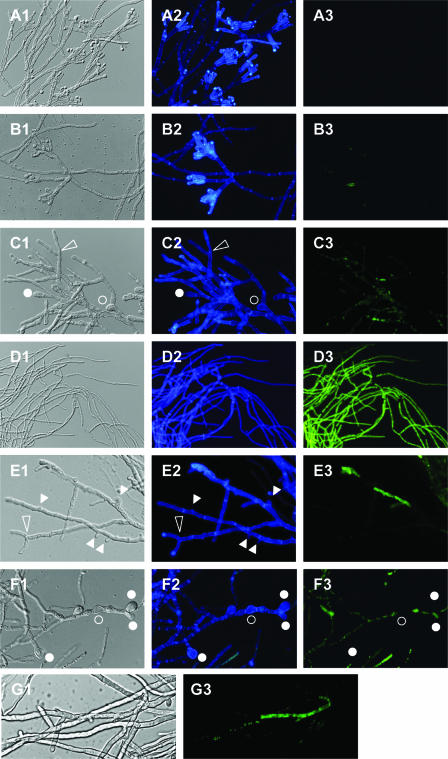
In controls in which P. digitatum mycelium was incubated with the SG probe without pretreatment with peptides, no appreciable fluorescent signal was discerned by using either fluorescence (Fig. 1A3) or confocal laser microscopy (not shown). The dye CFW, which is specific for the cell wall component chitin (24), was used on the same samples and showed a normal deposition of chitin in the absence of PAF26 (Fig. 1A2), with prevalent staining of conidiophores and conidia (located terminally in the hypha), and visualization of septa separating cells at regular distances within hyphae. Incubation with the minimal fungicidal concentration of PAF26 (15 μM) revealed collapsed hyphae with visualizations very similar to those for SG or CFW staining, indicating extensive cell death, wall disorganization, membrane permeation, and absence of sporulation (Fig. 1D). At this high PAF26 concentration, CFW staining of chitin was not localized in regions of high chitin content (as occurred in control samples) but rather occurred all along the hypha (Fig. 1D2). Also, SG staining was very intense and revealed visualization of green fluorescence all along the hypha (Fig. 1D3). Fungal cells seemed to have lost defined separations and exhibited confluent green staining in regions of accumulation of intercellular material. Similar observations with many other short CAMP have led to the conclusion that they act primarily by permeabilizing target microbes.
FIG. 1.
Fluorescence microscopy of P. digitatum mycelium treated with PAF26. Mycelium was incubated in 5% PDB at 24°C without peptide (panels A) or at PAF26 final concentrations of 1 μM (B and E), 3 μM (C, F, and G), or 15 μM (D) for 48 h. After incubation, samples were stained with CFW and SG. Panels represent bright-field images (left; suffix 1 in panel lettering), blue fluorescence indicative of CFW staining (center; suffix 2), and green fluorescence indicative of SG uptake (right; suffix 3) for the same fields. Micrographs A to F were captured with a light fluorescence microscope, while micrograph G was captured with a confocal laser microscope. Circles indicate swollen cells either at the terminus of hyphae (filled circles) or within hyphae (open circles). Open triangles indicate dichotomous tip branching; filled triangles indicate lateral aborted branching.
However, treatment with low sub-MIC concentrations of PAF26 allowed some fungal growth (see also Fig. 2A below), which microscopic observations revealed to be abnormal. At 1 μM (close to the IC50) PAF26, chitin staining was still predominant at terminal conidiophores, but these showed abnormal morphology (Fig. 1B1 and B2). In addition, shorter interseptum distances were observed. These morphological alterations could be visualized in fields in which no or very low permeation was observed by SG staining (Fig. 1B3). Higher magnification illustrated that some hyphal tips showed dichotomous branching and a significant amount of aborted lateral branching at 1 μM PAF26 (Fig. 1E1 and E2). Again, the micrographs show that the latter alterations occurred at regions in which no permeation was detected (Fig. 1E3). Increasing the PAF26 concentration to 3 μM resulted in extensive abnormal branching (Fig. 1C), and CFW staining no longer concentrated at conidiophores and conidia, which were absent as recognizable structures. Instead, balloon-shaped enlargement of individual cells located apically or in the middle of hyphae showed preferential CFW staining (Fig. 1C and F). These balloon structures did not take up SG.
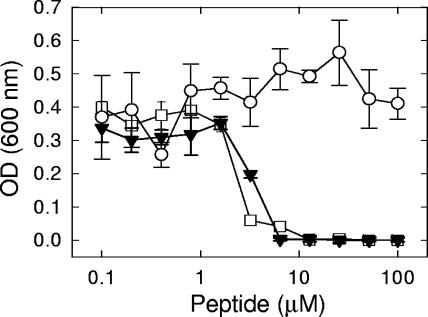
FIG. 2.
Effects on growth and SG uptake by P. digitatum mycelium of peptides PAF26 (top), melittin (center), and mel.subK7I (bottom). Mycelium was incubated with peptides in 5% PDB at 24°C for 24 h. After incubation, SG was added and incubated for an additional 4 h. Dose-response curves show the increase in the OD at 492 nm (left axis; open circles) and the fluorescence intensity (FI) at 538 nm (right axis; filled triangles) after peptide addition for each peptide concentration (μM). Plotted values are means ± standard deviations of three replicate samples. a.u., arbitrary units.
Confocal laser microscopy illustrated in a clearer way a “discontinuous” pattern of SG fluorescence after treatment with sub-MIC concentrations of PAF26, with labeled cells adjacent to nonlabeled ones within the same hypha (Fig. 1G). The confocal micrographs show representative data indicating the presence of cells at different stages of permeabilization, which include dot-like stained spots and staining over all the intracellular space. These micrographs also showed that short aborted hyphal branches emerge from cells not yet permeabilized or at the initial stages of permeabilization after PAF26 exposure.
Dose-response curves were used to quantify permeation and evaluate the correlation between the growth-inhibitory and permeation properties of PAF26 on P. digitatum mycelium (Fig. 2A). Permeation was dependent on peptide concentration, and the response curve correlated roughly with that found for growth inhibition. However, in different experiments it was consistently observed that concentrations of PAF26 below the IC50 had an effect on growth while only minimally inducing permeation (see, for instance, Fig. 2 top, 0.5 μM). This result is in agreement with the poor SG uptake observed at sub-IC50 concentrations by fluorescence microscopy in areas of abnormal fungal growth (Fig. 1B and E). Incubation with the previously characterized peptide PAF20 (13, 15), an inactive analog of PAF26 with 2 residues substituted, resulted in no SG increase of fluorescence (data not shown).
Comparison of the antifungal activities, permeation properties, and hemolytic activities of PAF26 and the cytotoxic peptide melittin.
The observations described above raised the question whether membrane permeation was the primary action of PAF26 on fungi. In order to have a broader view of this question, we compared the antifungal properties of PAF26 with those of melittin, a well-known toxic peptide that induces cell killing by membrane disruption and cytolysis. We have previously reported that PAF26 shows an antifungal activity on conidia similar to that of melittin (see also Table 1) but not the potent antibacterial activity of melittin (15). Dose inhibition and permeation curves of melittin added to mycelia, run in parallel to PAF26 experiments, led to several conclusions (Fig. 2). First, the dose inhibition curve lagged behind that of PAF26, resulting in a slightly higher IC50 for melittin (P = 0.041) (Table 1). Also, we found melittin concentrations at which permeation was observed but fungal biomass was not substantially reduced (1 μM in the experiment shown), a remarkable difference from the observations with PAF26. Finally, we observed that fluorescence intensity values for melittin decreased at high peptide concentrations (Fig. 2B) and also with time of incubation (see below, Fig. 3).
FIG. 3.
Time course of SG uptake by P. digitatum mycelium treated with peptide PAF26 (top), melittin (center), or mel.subK7I (bottom). Mycelium was simultaneously exposed to peptides (at different concentrations) and to 0.2 μM SG in 5% PDB. Immediately after peptide and SG addition, fluorescence at 538 nm was recorded at 2-min intervals for 180 min. Curves show mean fluorescence intensity (FI) values in triplicate samples at 10-min intervals for 0.5 (filled circles), 1 (open circles), 3 (filled triangles), 6 (open triangles), 8 (filled squares), 10 (open squares), and 15 (filled diamonds) μM of the different peptides.
Sequence analogs of melittin with altered properties in terms of peptide structure, propensity to interact with lipid membranes, and cell toxicity have been reported. Mel.subK7I is an analog with a single Lys-to-Ile substitution at position 7 (Table 1) that results in reduced interaction with membranes and reduced toxicity to red blood cells, as measured by its hemolytic properties (22). Surprisingly, mel.subK7I initiated P. digitatum permeabilization at concentrations very similar to those of PAF26 and melittin (0.5 to 1 μM) (Fig. 2C) but had reduced antifungal activity toward P. digitatum (Table 1).
Time course analysis of SG uptake was conducted immediately after the addition of peptides and SG; it also revealed differences between the three peptides (Fig. 3). Similarly rapid increases in fluorescence were recorded at concentrations of melittin from 3 to 15 μM. In contrast, in the case of PAF26 or mel.subK7I, this increase in fluorescence was clearly slower, and differences were observed among different peptide concentrations. These data indicate a quicker and more efficient disruption of the fungal membrane by melittin, which allows more effective uptake of SG. A plateau or decline in fluorescence was observed at higher melittin concentrations (similar to the data shown in Fig. 2B [see above]), which we attributed to degradation of intracellular components after the initial very efficient permeation (i.e., pore formation) of melittin. The highest permeabilization was observed for melittin, even though PAF26 had similar or even higher antifungal potency. In fact, it must be stressed that the IC50 of PAF26 (approximately 1 μM) was much less efficient at inducing permeation than the IC50 of melittin (approximately 3 μM) or mel.sub.K7I (approximately 10 μM). Another view of this result is that 1 μM melittin or PAF26 induced very similar (low) permeation, while PAF26 was more effective at inhibiting growth at this concentration (Fig. 2). However, it must be made clear that within the 3-h recording of the experiment shown in Fig. 3, no significant changes in fungal biomass (i.e., no significant increases in the OD) were observed, and thus a direct comparison between permeation and growth inhibition was not possible in this experiment.
Fungicidal activity on nongerminated conidia was compared for PAF26 and melittin. Representative data at 4 to 5 times the MICs of the peptides (30 μM) (Fig. 4) showed that the two peptides differ markedly in their abilities to kill fungal spores. After just 15 min of PAF26 treatment, conidia lost viability by approximately 1 log unit, and more than 99% loss was found after 30 min (Fig. 4). In contrast, conidia were resistant to the action of melittin, and only minimal loss of viability was observed even after 16 h of treatment. These results also point toward clear differences between the two peptides in their interaction/mechanism of action on conidia/mycelium.
FIG. 4.
Killing kinetics of P. digitatum conidia by synthetic peptides. Conidia (104/ml) were incubated in water in the absence of peptide (filled circles) or in the presence of 30 μM PAF26 (open triangles) or 30 μM melittin (open squares). Aliquots were removed at different time points from 15 min to 16 h, diluted, and plated on PDA in the absence of peptide to assay for CFU recovery. Values are means ± standard deviations of three replicate samples.
The hemolytic activities of the peptides were compared and demonstrated a markedly reduced cytolytic activity of PAF26 toward human red blood cells (Table 2). According to the different concentrations of peptides assayed and the extent of lysis detected, the PAF26 peptide was found to be 103 to 104 times less toxic than melittin. Taken together, these results strongly suggest that the primary mode of antifungal action of PAF26 on P. digitatum does not rely solely on its ability to interact with and disrupt biological membranes, despite the propensity of this peptide to interact with membrane mimetics (14).
TABLE 2.
Hemolytic activities of synthetic peptidesa
| Peptide | Hemolytic activity at the following peptide concn (μM):
|
||
|---|---|---|---|
| 1 | 10 | 100 | |
| PAF26 | <0.3 | <0.3 | <0.3 |
| Melittin | <0.3 | 32.7 ± 0.7 | 115.7 ± 2.5 |
| Mel.subK7I | <0.3 | 9.1 ± 0.7 | 30.2 ± 2.8 |
Numbers indicate percentages of hemolysis relative to the 100% control and are means ± standard deviations of three replicate samples. The detection limit of our assay was 0.3% of the hemolysis in the 0.1% Triton X-100 control.
Localization of PAF26 within P. digitatum fungal structures.
Fluorescence labeling and confocal laser microscopy have been used to study the interaction of AMP with microbes (10, 21, 23, 34, 35). PAF26 was synthesized with its N terminus modified by covalent addition of the fluorophore FITC in order to study its interaction with P. digitatum conidia and mycelial cells. Importantly, FITC-PAF26 did not differ in antifungal potency from PAF26, and the FITC fluorophore did not show antimicrobial activity (Fig. 5), indicating that the addition of FITC had minimal effect on bioactivity. Nongerminated conidia were incubated with 0.3, 3, and 30 μM FITC-PAF26 for different times. Only the highest peptide concentration (30 μM) gave a clear positive signal, which was mostly localized at the periphery over the conidial wall, after 30 min of exposure (Fig. 6A). Minor amounts of peptide were detected inside the spore (Fig. 6A2), which could explain the fungicidal properties of PAF26 (Fig. 4). Much longer incubation times (i.e., 16 h) were necessary to show that FITC-PAF26 was completely internalized and located inside the spore and was notably excluded from the outer wall (Fig. 6B). Under the latter conditions, 42% of the visualized conidia had internalized FITC-PAF26.
FIG. 5.
Dose-response curves of the effects on the growth of P. digitatum of peptides PAF26 (filled triangles) and FITC-PAF26 (open squares) and the fluorophore FITC (open circles). Conidia (2.5 × 104/ml) in 5% PDB were exposed to different concentrations of compounds and incubated at 24°C. Dose-response curves show mean ODs at 600 nm ± standard deviations after 48 h of incubation. The IC50s of PAF26 and FITC-PAF26 did not differ significantly (P > 0.05).
FIG. 6.
Interaction of FITC-PAF26 with P. digitatum. (A and B) Confocal laser microscope localization of FITC-PAF26 in P. digitatum conidia. Quiescent conidia were exposed in distilled water to 30 μM FITC-PAF26 for 30 min (A) or 16 h (B). Bright-field (panels 1), fluorescence (panels 2), and overlay (panels 3) micrographs are shown. Arrowhead points to a minimal fluorescence signal observed inside the spore. (C to F) Confocal laser microscope localization of FITC-PAF26 in germ tubes and hyphae of P. digitatum. Mycelium was exposed in distilled water to 0.3 μM FITC-PAF26 for less than 2 min (C and D), approximately 5 min (E), or approximately 15 min (F). (G) Confocal laser microscope image of P. digitatum mycelium simultaneously exposed to 0.3 μM FITC and 0.3 μM PAF26 for approximately 15 min. In panels C, bright-field, fluorescence, and overlay micrographs are shown as described above, whereas in panels D to G, only bright-field and fluorescence micrographs are shown.
A remarkably distinct result was obtained upon incubation of P. digitatum mycelial cells with FITC-PAF26. The peptide interacted quickly (in less than 2 to 3 min) with the fungal cell wall/membrane at concentrations as low as 0.3 μM (Fig. 6C and D). In germinating conidia, the interaction occurred with the elongating germ tube and later diffused to the cell membrane components inside the germinated spore (compare Fig. 6D with 6E). In some samples, preferential staining at the germ tip was observed (data not shown). After an additional short time, the labeled peptide was localized inside the cells (Fig. 6E), thus demonstrating uptake of FITC-PAF26 at concentrations at which no effect was observed on fungal growth (Fig. 5 and data not shown) or SG uptake (Fig. 3). Inside staining was initially dispersed in the cytoplasm and later concentrated onto numerous discrete structures that could not be identified (Fig. 6F). Higher concentrations of FITC-PAF26 (i.e., 3 and 30 μM) interacted so efficiently with mycelium that in less than 2 min, all the staining was intensely concentrated inside cells (data not shown).
Mycelium treated simultaneously with 0.3 μM FITC and 0.3 μM PAF26 did not show a fluorescence signal in cell envelopes or inside cells (Fig. 6G), demonstrating that the internalization of FITC required the covalent linkage in cis to the peptide. This result also confirms that at this low concentration, PAF26 does not make cells permeable to molecules such as FITC (Fig. 6G) or SG (Fig. 2 top and 3) (data not shown).
Nucleic acid binding properties of PAF26.
Given the membrane translocation capability of PAF26 peptide, its location inside cells, and its cationic properties, we were interested in exploring its putative nucleic acid binding activity. PAF26 bound nonspecifically to S. cerevisiae tRNA, as demonstrated by EMSA (Fig. 7). Binding was recorded as disappearance of free RNA and was observed at concentrations of 8 μM PAF26 and higher; at 32 to 64 μM, more than 80% of free tRNA had disappeared. Bound RNA appeared as a low-mobility smear, indicative of nonspecific binding of the peptide to the nucleic acid.
FIG. 7.
Analysis of RNA-binding properties of PAF26 by EMSA. Yeast tRNA (250 ng) was incubated with no peptide (rightmost lane) or with increasing amounts of peptide (given above the gel) and analyzed by EMSA. The positions of the origin (O) and of free tRNA are indicated on the left.
DISCUSSION
Many AMP induce membrane permeation of microbes, and this fact has been taken as an indication that their mode of action is through disruption of biological membranes. However, recent data for an increasing number of peptides suggest that they might use more sophisticated mechanisms of antimicrobial action (2, 6, 38). Thus, there is a need for clarification of the mechanism of action of selected antimicrobial peptides, information that will be useful in the optimization and design of new antimicrobial sequences.
We showed that PAF26 treatment results in permeation of the membrane of P. digitatum mycelium (Fig. 1, 2, and 3), and in this sense our study parallels previous findings with different short AMP and antifungal plant proteins (5, 26, 27, 31). However, the differences are noteworthy, since in previous reports fungal spore permeation was readily detected upon incubation with other short AMP (5, 27). In our case, SG uptake by conidia was observed only by microscopic visualization (not by fluorescence quantification) and upon incubation of P. digitatum conidia with high PAF26 concentrations for long times (data not shown), in agreement with the results found with FITC-PAF26 (Fig. 6A and B). In our study, the use of different peptide concentrations and incubation times indicates that PAF26 interacts much more poorly with conidial envelopes than with mycelial cell walls (Fig. 6), confirming the existence of distinct compositions of outer envelopes for these fungal structures that likely mediate distinct affinities for PAF26. Thus, the previously reported inhibition of germination by this peptide (15) reflects inhibition of germ tube elongation at a very early step after conidial wall breakdown.
Microscopic observations (Fig. 1) and dose-response curves (Fig. 2A) revealed very poor SG uptake at low sub-MIC concentrations of PAF26 that inhibit growth and cause morphological alterations, indicating that permeabilization is minimal at concentrations that affect fungal growth. Previous reports have discussed the fact that there is not always a complete correlation between the ability of selected CAMP to permeate membranes and their antimicrobial activity (4, 36). For instance, screening of an octapeptide combinatorial library targeted toward inhibition of yeast membrane proton ATPases has led to the identification of the octa- and decapeptides BM0 and BM2 (18). These peptides have a remarkable sequence similarity with PAF26. Low concentrations of BM2 that are growth inhibitory did not cause permeabilization of target yeast cells and did not induce hemolysis (18).
We have also reported that PAF26 is not synergistic with benzimidazole and imidazole (16), which also indicates that it does not facilitate the internalization of these fungicides through permeabilized membranes at the sub-IC50 concentrations of peptide that are required to evaluate synergism. In this lack of synergy PAF26 differs markedly from other synthetic hexa- and heptapeptides, which, as discussed above, easily promote permeation of target spores and are synergic with thiabendazole (5).
Our observations suggest that PAF26 treatment causes alteration and inhibition of P. digitatum growth at an earlier stage than permeation. This conclusion is further supported by the comparison with melittin, for which the SG uptake dose-response curve precedes that of inhibition (Fig. 2). Melittin is a pore-forming peptide and is expected to act on fungal mycelium by a lytic mechanism, as reported with many other microorganisms. Melittin causes Penicillium permeation (Fig. 3) and hemolysis (Table 2) much more efficiently than PAF26 but has inhibitory properties toward the fungus similar to those of the hexapeptide (Table 1). Also, melittin has no fungicidal properties on conidia, as opposed to PAF26 (Fig. 4). We conclude that the growth-inhibitory mechanisms of melittin and PAF26 on P. digitatum are different, neatly illustrating that not all AMP function the same way on a given target.
Morphological alterations following treatment with sub-MIC PAF26 concentrations include dichotomous tip branching and alterations of branch emergence (Fig. 1E), which are phenocopies of filamentous fungal mutants in which establishment and maintenance of polarity are altered (7). Polar growth requires the recruitment of the morphogenetic machinery to specific sites for localized cell wall deposition. Also, swelling of hyphal cells with abnormal deposition of chitin, as determined by CFW staining (Fig. 1C and F), is indicative of alterations in cell wall structure. These results indicate that the P. digitatum cell wall is not solely the first line of interaction with PAF26 but is also a fungal structure that is responsive to peptide exposure. Interestingly, most of the mycelial areas with such alterations are not permeated and can even be surrounded by cells permeable to SG (Fig. 1F).
The different models that explain the activity of most membrane disturbing peptides propose that at low peptide-to-membrane lipid ratios, the peptide tends to adsorb to the membrane surface (2, 9, 29). As the peptide concentration increases, the peptide-to-lipid ratio reaches a threshold value at which the peptide disturbs the membrane and leads to permeation, following different models of peptide-lipid interaction. In contrast to melittin, PAF26 is far too short to span biological membranes as a monomer, and thus higher-order interactions among monomers (at high PAF26 concentrations) would be needed to explain any membrane permeation properties. The small size of PAF26 also implies that there is a theoretical (low) concentration at which PAF26 would adsorb to the membrane but not yet disrupt it. Fluorescent labeling has been used to show internalization of short cationic peptides across cell membranes or model vesicles (10, 21, 34, 35). Importantly, FITC-labeled PAF26 at subinhibitory concentrations interacts with the mycelial cell and quickly translocates across the plasma membrane without apparently altering its morphological or permeation properties. FITC-PAF26 is internalized by mycelium at 0.3 μM (Fig. 6C to F), a concentration fivefold below its IC50 (Table 1) and 10 times below the IC50 and permeation concentration of melittin (Table 1; Fig. 2 and 3). At this low concentration of peptide, no membrane permeation was observed, as determined by uptake of SG (Fig. 2 and 3) or FITC (Fig. 6G); the peptide was not inhibitory (Fig. 5); and neither the morphology of hyphal cells nor the integrity of membranes seemed to be altered (Fig. 6 and data not shown). Similarly, the 21-amino-acid-residue AMP buforin II was also found to accumulate inside E. coli even below its MIC (20). The specificity of the FITC-PAF26 interaction was supported by experiments in which, at concentrations of 0.3 or 3 μM, FITC-PAF26 did not interact with S. cerevisiae and was not translocated inside the yeast cells (data not shown). These experiments were similar to previously reported experiments that demonstrated surface interaction of a distinct fluorophore-labeled hexapeptide with S. cerevisiae (18).
Several antimicrobial peptides are translocated across cell membranes and locate inside cells, wherein they can induce a diversity of inhibitory activities that disrupt normal cell functions not primarily linked to cell permeation (2). Some of the CAMP with internalization properties have amino acid sequences with similarities to protein domains—so-called “penetratins” or “protein transduction domains” (PTD)—which are internalized by a process(es) that does not involve classical endocytosis (11, 25). These domains are cationic, and specific Trp and Phe residues are critical for the cell entry function. PAF26 shares these amino acid sequence properties. We propose that these CAMP concentrate properties relevant for internalization and antimicrobial activity within the same molecule. The antimicrobial fragment lactoferricin (Lfcin) is one such cell-penetrating peptide (25) and also translocates and accumulates inside bacterial cells and binds RNA (8, 34). Of note, the PAF26 sequence is similar to the 6-residue antibacterial core of bovine Lfcin and has a profile of antifungal properties similar to that of synthetic bovine Lfcin-derived peptides (16, 19). Thus, on the basis of the data reported here and previous results of analogous peptide sequences in the literature, we propose that PAF26 is a CAMP that belongs to this class of cell-permeating peptides. Conversely, penetratin or PTD peptides are expected to be a rich source for the development of novel AMP. Another interesting consequence of our work is that the efficient translocation of PAF26 opens the way to its use as a targeting domain that could direct heterologous toxic peptides in cis to filamentous fungi, following reports of similar experiments conducted on human-pathogenic bacteria (3).
Once inside the target cell, CAMP might exert functions that include binding to nucleic acids and/or inhibition of the synthesis of macromolecules, enzymatic activities, or cell walls (2, 6, 38). For instance, cationic AMP with nucleic acid binding properties are known (20, 39), as are Trp-rich CAMP such as indolicidin and LfcinB, which inhibit the intracellular synthesis of nucleic acids and proteins (30, 33). PAF26 is a hexapeptide that exhibits nonspecific tRNA binding in vitro (Fig. 7) and therefore could interfere with cellular RNA functions once inside the fungal cell. In this view, this step of PAF26 action would be rather nonspecific, and the PAF26 profile of specificity against filamentous fungi would be a consequence of distinct interactions with outer envelopes of microbes, as exemplified by the differences in its interactions with conidial and hyphal P. digitatum cells.
Acknowledgments
This work was supported by grant BIO2003-00927 from the Spanish Ministry of Education and Science. B.L.-G. is the recipient of a postdoctoral research contract from the “Juan de la Cierva” program of the Spanish Ministry of Education and Science.
We acknowledge the “Instituto de Biología Molecular y Celular de Plantas” (IBMCP) (UPV-CSIC, Valencia, Spain) and M. Dolores Gómez of its microscopy core facility for its use and for help with the micrographs. We also acknowledge Andrew MacCabe (IATA-CSIC) for critical reading of the manuscript. The excellent technical assistance of M. José Pascual is also acknowledged.
REFERENCES
- 1.Broekaert, W. F., F. R. G. Terras, B. P. A. Cammue, and J. Vanderleyden. 1990. An automated quantitative assay for fungal growth-inhibition. FEMS Microbiol. Lett. 69:55-59. [Google Scholar]
- 2.Brogden, K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238-250. [DOI] [PubMed] [Google Scholar]
- 3.Eckert, R., F. X. Qi, D. K. Yarbrough, J. He, M. H. Anderson, and W. Y. Shi. 2006. Adding selectivity to antimicrobial peptides: rational design of a multidomain peptide against Pseudomonas spp. Antimicrob. Agents Chemother. 50:1480-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epand, R. M., and H. J. Vogel. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1462:11-28. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez, C. F., E. M. Provin, L. Zhu, and D. J. Ebbole. 2002. Independent and synergistic activity of synthetic peptides against thiabendazole-resistant Fusarium sambucinum. Phytopathology 92:917-924. [DOI] [PubMed] [Google Scholar]
- 6.Hancock, R. E. 2001. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 1:156-164. [DOI] [PubMed] [Google Scholar]
- 7.Harris, S. D., and M. Momany. 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41:391-400. [DOI] [PubMed] [Google Scholar]
- 8.Haukland, H. H., H. Ulvatne, K. Sandvik, and L. H. Vorland. 2001. The antimicrobial peptides lactoferricin B and magainin 2 cross over the bacterial cytoplasmic membrane and reside in the cytoplasm. FEBS Lett. 508:389-393. [DOI] [PubMed] [Google Scholar]
- 9.Huang, H. W. 2000. Action of antimicrobial peptides: two-state model. Biochemistry 39:8347-8352. [DOI] [PubMed] [Google Scholar]
- 10.Kragol, G., R. Hoffmann, M. A. Chattergoon, S. Lovas, M. Cudic, P. Bulet, B. A. Condie, K. J. Rosengren, L. J. Montaner, and L. Otvos. 2002. Identification of crucial residues for the antibacterial activity of the proline-rich peptide, pyrrhocoricin. Eur. J. Biochem. 269:4226-4237. [DOI] [PubMed] [Google Scholar]
- 11.Leifert, J. A., and J. L. Whitton. 2003. ‘Translocatory proteins’ and ‘protein transduction domains’: a critical analysis of their biological effects and the underlying mechanisms. Mol. Ther. 8:13-20. [DOI] [PubMed] [Google Scholar]
- 12.Lohner, K., and S. E. Blondelle. 2005. Molecular mechanisms of membrane perturbation by antimicrobial peptides and the use of biophysical studies in the design of novel peptide antibiotics. Comb. Chem. High Throughput Screen. 8:241-256. [DOI] [PubMed] [Google Scholar]
- 13.López-García, B., L. González-Candelas, E. Pérez-Payá, and J. F. Marcos. 2000. Identification and characterization of a hexapeptide with activity against phytopathogenic fungi that cause postharvest decay in fruits. Mol. Plant-Microbe Interact. 13:837-846. [DOI] [PubMed] [Google Scholar]
- 14.López-García, B., J. F. Marcos, C. Abad, and E. Pérez-Payá. 2004. Stabilisation of mixed peptide/lipid complexes in selective antifungal hexapeptides. Biochim. Biophys. Acta 1660:131-137. [DOI] [PubMed] [Google Scholar]
- 15.López-García, B., E. Pérez-Payá, and J. F. Marcos. 2002. Identification of novel hexapeptides bioactive against phytopathogenic fungi through screening of a synthetic peptide combinatorial library. Appl. Environ. Microbiol. 68:2453-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López-García, B., A. Veyrat, E. Pérez-Payá, L. González-Candelas, and J. F. Marcos. 2003. Comparison of the activity of antifungal hexapeptides and the fungicides thiabendazole and imazalil against postharvest fungal pathogens. Int. J. Food Microbiol. 89:163-170. [DOI] [PubMed] [Google Scholar]
- 17.Marcos, J. F., M. Vilar, E. Pérez-Payá, and V. Pallás. 1999. In vivo detection, RNA-binding properties and characterization of the RNA-binding domain of the p7 putative movement protein from carnation mottle carmovirus (CarMV). Virology 255:354-365. [DOI] [PubMed] [Google Scholar]
- 18.Monk, B. C., K. Niimi, S. Lin, A. Knight, T. B. Kardos, R. D. Cannon, R. Parshot, A. King, D. Lun, and D. R. K. Harding. 2005. Surface-active fungicidal D-peptide inhibitors of the plasma membrane proton pump that block azole resistance. Antimicrob. Agents Chemother. 49:57-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muñoz, A., and J. F. Marcos. Activity and mode of action against fungal phytopathogens of bovine lactoferricin-derived peptides. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 20.Park, C. B., H. S. Kim, and S. C. Kim. 1998. Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 244:253-257. [DOI] [PubMed] [Google Scholar]
- 21.Park, C. B., K. S. Yi, K. Matsuzaki, M. S. Kim, and S. C. Kim. 2000. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl. Acad. Sci. USA 97:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez-Payá, E., R. A. Houghten, and S. E. Blondelle. 1994. Determination of the secondary structure of selected melittin analogues with different haemolytic activities. Biochem. J. 299:587-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers, J. P. S., M. M. Martin, D. L. Goosney, and R. E. W. Hancock. 2006. The antimicrobial peptide polyphemusin localizes to the cytoplasm of Escherichia coli following treatment. Antimicrob. Agents Chemother. 50:1522-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pringle, J. R. 1991. Staining of bud scars and other cell-wall chitin with Calcofluor. Methods Enzymol. 194:732-735. [DOI] [PubMed] [Google Scholar]
- 25.Prochiantz, A. 2000. Messenger proteins: homeoproteins, TAT and others. Curr. Opin. Cell Biol. 12:400-406. [DOI] [PubMed] [Google Scholar]
- 26.Reed, J. D., D. L. Edwards, and C. F. Gonzalez. 1997. Synthetic peptide combinatorial libraries: a method for the identification of bioactive peptides against phytopathogenic fungi. Mol. Plant-Microbe Interact. 10:537-549. [DOI] [PubMed] [Google Scholar]
- 27.Rioux, D., V. Jacobi, M. Simard, and R. C. Hamelin. 2000. Structural changes of spores of tree fungal pathogens after treatment with the designed antimicrobial peptide D2A21. Can. J. Bot. 78:462-471. [Google Scholar]
- 28.Roth, B. L., M. Poot, S. T. Yue, and P. J. Millard. 1997. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 63:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shai, Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1462:55-70. [DOI] [PubMed] [Google Scholar]
- 30.Subbalakshmi, C., and N. Sitaram. 1998. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 160:91-96. [DOI] [PubMed] [Google Scholar]
- 31.Thevissen, K., F. R. Terras, and W. F. Broekaert. 1999. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl. Environ. Microbiol. 65:5451-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 33.Ulvatne, H., O. Samuelsen, H. H. Haukland, M. Kramer, and L. H. Vorland. 2004. Lactoferricin B inhibits bacterial macromolecular synthesis in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Lett. 237:377-384. [DOI] [PubMed] [Google Scholar]
- 34.van der Kraan, M. I. A., J. van Marle, K. Nazmi, J. Groenink, W. Van't Hof, E. C. I. Veerman, J. G. M. Bolscher, and A. V. N. Arnerongen. 2005. Ultrastructural effects of antimicrobial peptides from bovine lactoferrin on the membranes of Candida albicans and Escherichia coli. Peptides 26:1537-1542. [DOI] [PubMed] [Google Scholar]
- 35.Vives, E., P. Brodin, and B. Lebleu. 1997. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272:16010-16017. [DOI] [PubMed] [Google Scholar]
- 36.Wu, M., E. Maier, R. Benz, and R. E. Hancock. 1999. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38:7235-7242. [DOI] [PubMed] [Google Scholar]
- 37.Yang, L., T. A. Harroun, T. M. Weiss, L. Ding, and H. W. Huang. 2001. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 81:1475-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeaman, M. R., and N. Y. Yount. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27-55. [DOI] [PubMed] [Google Scholar]
- 39.Yonezawa, A., J. Kuwahara, N. Fujii, and Y. Sugiura. 1992. Binding of Tachyplesin-I to DNA revealed by footprinting analysis—significant contribution of secondary structure to DNA-binding and implication for biological action. Biochemistry 31:2998-3004. [DOI] [PubMed] [Google Scholar]
- 40.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]