Abstract
Nucleoside reverse transcriptase inhibitors are an important class of drugs for treatment of human immunodeficiency virus type 1 (HIV-1) infection. Resistance to these drugs is often the result of mutations that increase the transfer of chain-terminating nucleotides from blocked DNA termini to a nucleoside triphosphate acceptor, resulting in the generation of an unblocked DNA chain and synthesis of a dinucleoside polyphosphate containing the chain-terminating deoxynucleoside triphosphate analogue. We have synthesized and purified several dinucleoside tetraphosphates (ddAp4ddA, ddCp4ddC, ddGp4ddG, ddTp4ddT, Ap4ddG, 2′(3′)-O-(N-methylanthraniloyl)-Ap4ddG, and AppNHppddG) and show that these compounds can serve as substrates for DNA chain elongation and termination resulting in inhibition of DNA synthesis. Thymidine analogue-resistant mutants of reverse transcriptase are up to 120-fold more sensitive to inhibition by these compounds than is wild-type enzyme. Drugs based on the dinucleoside tetraphosphate structure could delay or prevent the emergence of mutants with enhanced primer unblocking activity. In addition, such drugs could suppress the resistance phenotype of mutant HIV-1 that is present in individuals infected with resistant virus.
Chronic treatment of human immunodeficiency virus type 1 (HIV-1)-infected individuals with nucleoside reverse transcriptase inhibitors (NRTIs) such as 3′-azido-2′,3′-dideoxythymidine (AZT, or zidovudine) usually leads to selection of drug resistance mutations in the viral reverse transcriptase (RT) that may reduce or abolish the effectiveness of NRTI therapy (9, 11, 13, 14, 34). A major class of NRTI resistance mutations (designated thymidine analogue mutations, or TAMs, which include M41L, D67N, T69S-XX, K70R, L210W, T215F or Y, and K219Q or E) confers increased removal of chain-terminating nucleotides, primarily thymidine analogues and also, to a lesser extent, several other NRTIs, from blocked DNA chains (1, 3, 15, 16, 18, 21-23, 27). This unblocking reaction occurs through transfer of the chain-terminating nucleotide to a nucleoside triphosphate (NTP) acceptor in a reaction that is related to pyrophosphorolysis (23). The products of the transfer reaction are a dinucleoside tetraphosphate of the form Np4ddN and an extendable primer terminus. In vitro, any NTP, deoxynucleoside triphosphate (dNTP), or dideoxyribonucleoside triphosphate (ddNTP) can serve as acceptor, whereas in vivo the acceptor is most likely ATP, resulting in the synthesis of Ap4ddN (31). The mechanism by which TAMs give rise to primer unblocking has not been determined, but it has been proposed on the basis of crystal structure and substrate specificity data that the residues affected by TAMs play a role in interaction with the ATP substrate or with the adenosine portion of an intermediate formed during the excision reaction (3, 5, 22, 24).
The widespread use of AZT and other NRTIs for antiretroviral therapy has resulted in a high prevalence of mutant HIV-1 containing TAMs, which were present in more than 25% of patients failing therapy in a recent survey (6). Therapies that are effective against these mutant viruses will continue to be needed. The increased ability of RT containing TAMs to utilize NTPs as substrates for primer unblocking suggests that it may be possible to target inhibitors to the mutant enzymes using the Np4ddN interaction surface.
We have used an enzymatic strategy to synthesize several different dinucleoside tetraphosphates containing ddNTP components, and we show that HIV-1 RT can use these compounds as substrates for DNA elongation resulting in chain termination. Mutant RTs containing TAMs are as much as 120-fold more sensitive than wild-type (WT) RT to chain termination by these compounds. Dinucleoside tetraphosphates provide novel tools for further studies to explore the effects of TAMs on HIV-1 RT, and they may serve as a foundation for the development of new drugs that target DNA synthesis and nucleotide excision by this enzyme.
MATERIALS AND METHODS
Expression and purification of HIV-1 RT. His-tagged HIV-1 RT containing wild-type (RTWT), K65R (RTK65R), W88G (RTW88G), M41L/T215Y (RTM41L/T215Y), D67N/K70R/T215Y/K219Q (RTAZT) or M41L/T69S-insertionAG/L210W/R211K/L214F/T215Y/K219Q mutations (RTMDR) were expressed and purified as previously described (26). The specific RNA-dependent DNA polymerase activities of HIV-1 RTWT, RTK65R, RTW88G, RTM41L/T215Y, RTAZT, and RTMDR were 35,000, 16,000, 19,000, 27,000 7,600, and 27,000 U/mg, respectively, where 1 U is equal to the amount of enzyme required for incorporation of 1.0 nmol of [3H]dTMP in 10 min when using poly(rA)/oligo(dT) (Amersham Biosciences) as the substrate (32). These specific activities are consistent with values reported elsewhere in the literature (4, 7, 12, 17, 32). Oligonucleotides were obtained from Sigma Genosys; M13 DNA was from New England Biolabs. [α-32P]dNTPs and [γ-32P]ATP were obtained from PerkinElmer, and [α-33P]ddNTPs and [α-32P]ddATP were from Amersham Biosciences (GE Healthcare).
Synthesis and purification of dinucleoside tetraphosphates.
A 10 μM concentration of L32 primer (5′-CTACTAGTTTTCTCCATCTAGACGATACCAGA-3′) was annealed to an equal amount of one of the templates listed below and incubated with 1 μM HIV-1 RTMDR and the ddNTP corresponding to the first unpaired base on the primer/template (P/T). Each reaction was carried out in 1,000 μl of reaction buffer (40 mM Na-HEPES, pH 7.5, 20 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol, 2.5% glycerol) containing 160 μg of bovine serum albumin (BSA) and 0.5 U of inorganic pyrophosphatase (Roche Applied Science). Templates and ddNTPs used were as follows: WL50 template (5′-GAGTGCTGAGGTCTTCATTCTGGTATCGTCTAGATGGAGAAAACTAGTA G-3′) with 3.2 mM ddATP and 10 μCi of [α-33P]ddATP; WL50-33G template (5′-GAGTGCTGAGGTCTTCAGTCTGGTATCGTCTAGATGGAGAAAACTAGTAG-3′) with 3.2 mM ddCTP and 10 μCi of [α-33P]ddCTP; WL50-33C template (5′-GAGTGCTGAGGTCTTCACTCTGGTATCGTCTAGATGGAGAAAACTAGTAG-3′) with 3.2 mM ddGTP and 10 μCi of [α-33P]ddGTP; and WL50-33A-34T template (5′-GAGTGCTGAGGTCTTCTATCTGGTATCGTCTAGATGGAGAAAACTAGTAG-3′) with 3.2 mM ddTTP and 10 μCi of [α-33P]ddTTP. All nucleotides were pretreated with thermostable pyrophosphatase as previously described (26). The mixture was overlaid with 200 μl of mineral oil and incubated at 37°C for 7 days. After heat inactivation of the RT at 90°C for 5 min, 100 U of calf intestinal alkaline phosphatase (CIP) (New England Biolabs) was added, followed by incubation at 37°C for 2 h. One hundred units of CIP was added a second time, and incubation continued at 37°C for an additional 2 h, followed by heat inactivation at 90°C for 10 min. The CIP-treated products were loaded onto a 5-ml anion exchange column, HiTrap Q HP (Amersham Biosciences) preequilibrated with 10 mM triethylammonium bicarbonate buffer pH 8.5 (Sigma), and eluted with a gradient of triethylammonium bicarbonate buffer up to 1 M. Fractions containing 33P-labeled dinucleoside polyphosphates were vacuum dried and resuspended in reaction buffer. The concentration of each compound was determined by incubation first with snake venom phosphodiesterase to cleave dinucleoside polyphosphates to free nucleotides, followed by spectrophotometric determination of the total concentration of nucleotide. The synthesis and purification of the dinucleoside tetraphosphate Ap4ddG, doubly labeled with 32P and 33P (A-P-P-32P-33P-ddG), were identical to the synthesis and purification of ddGp4ddG except for the following modifications: 10 μM L32 primer annealed to WL50-33C template was incubated with 1 μM HIV-1 RTMDR with 10 mM ATP, 20 μCi of [γ-32P]ATP, 1 mM ddGTP, and 10 μCi of [α-33P]ddGTP. Following anion exchange chromatography, Ap4ddG was separated from ddGp4ddG by reverse-phase high-pressure liquid chromatography using a C18 column (Amersham). The synthesis of 2′(3′)-O-(N-methylanthraniloyl)-(MANT)-Ap4ddG and AppNHppddG was the same as the synthesis of Ap4ddG except that MANT-ATP (Jena Bioscience) or AppNHp (Jena Bioscience), respectively, was included in the reaction instead of ATP.
Inhibition of DNA polymerization by ddNTPs or ddNp4ddNs.
M13 single-stranded DNA (New England Biolabs) annealed to a DNA primer (5′-AAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGAC-3′) (2.5 nM) was incubated with excess HIV-1 RT (100 nM) in 20 μl of reaction buffer containing 80 μg/ml of BSA; a 2.5 μM concentration of each dNTP; and 20 μCi of [α-32P]dATP, [α-32P]dCTP, [α-32P]dGTP, or [α-32P]dTTP in the absence or presence of various concentrations of ddATP or ddAp4ddA, ddCTP or ddCp4ddC, ddGTP or ddGp4ddG, or ddTTP or ddTp4ddT, respectively, at 37°C for 30 min. The reaction was terminated by heating at 90°C for 5 min. Five microliters of reaction mixture was mixed with 5 μl of gel loading dye and separated by electrophoresis through a 10% denaturing polyacrylamide gel. The radioactivity of the products in the dried gel was visualized by exposure on KODAK BioMax MR Film. For quantification of incorporated radioactive nucleotide, the remaining 15 μl of the reaction mixture was spotted on 3MM Whatman filter circles and soaked three times (45 min, 30 min, and 30 min) at 4°C in large excess of 5% trichloroacetic acid containing 20 mM sodium pyrophosphate, followed by two 5-min washes in ethanol at 4°C. The precipitable radioactivity in each sample was quantified by scintillation counting. The amount of inhibition of DNA synthesis was plotted versus inhibitor concentration using SigmaPlot 8.0 to obtain the 50% inhibitory concentration (IC50). IC50s for mutant enzymes were adjusted for differences in activity between mutant and WT RT to correct for effects due to the formation of products of different average lengths (28) using the following formula: IC50 = (uncorrected IC50 × mutant RT activity)/WT RT activity.
Electrophoretic mobility shift assay.
L31 primer (5′-CTACTAGTTTTCTCCATCTAGACGATACCAG-3′) annealed to a fourfold excess of WL50-33C DNA template was terminated with [32P]ddAMP as previously described (23). A 5 nM concentration of L31-[32P]ddAMP/WL50-33C was incubated with 100 nM HIV-1 RT and various concentrations of either ddGTP or ddGp4ddG, as indicated in the legend of Fig. 4, in 10 μl of reaction buffer containing 80 μg/ml BSA for 15 min at 37°C. Samples were incubated at 4°C for 5 min, followed by the addition of 3 μl of heparin and loading dye (0.03 U heparin/μl, 30% sucrose, 0.25% bromophenol blue, and 0.25% xylene cyanol) in reaction buffer to dissociate binary complexes. The samples were separated by electrophoresis through an 8% native polyacrylamide gel at 200 V for 1 h at 4°C using Tris-taurine buffer (90 mM Tris-30 mM taurine). The radioactivity was quantitated by phosphorimaging. The percentage of DNA in complex, after deduction of complex formed in the absence of dNTP, was plotted against the ligand concentration. The apparent dissociation constant [KD(app)] for dead-end complex was obtained by fitting the data to a one-ligand binding curve using SigmaPlot 8.0.
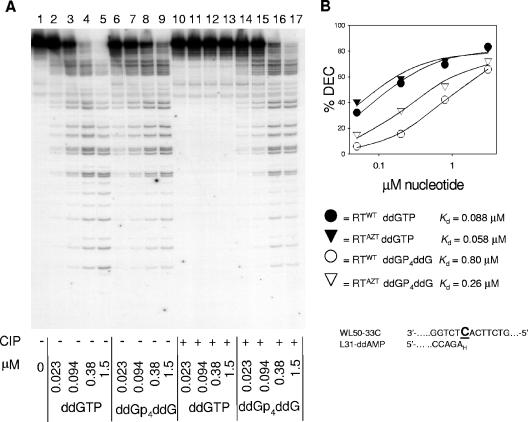
FIG. 4.
Inhibition of DNA synthesis and formation of stable complexes with RTWT and RTAZT by ddGTP or ddGp4ddG. (A) M13 single-stranded DNA annealed to a cDNA primer (2.5 nM) was incubated with HIV-1 RTAZT (100 nM), a 2.5 μM concentration of each dNTP, 50 μCi of [α-32P]dGTP, and the indicated concentration of ddGTP or ddGp4ddG that was either untreated (−) or had been pretreated with CIP (+) for 30 min at 37°C. The samples were separated by 20% urea-polyacrylamide gel electrophoresis. (B) Electrophoretic mobility shift assay for the ability of RTWT or RTAZT to form a stable dead-end complex (DEC) with ddAMP-terminated primer/template. Enzyme was incubated with WL50-33C/L31-[32P]ddAMP template/primer in the presence of the indicated concentration of either ddGTP or ddGp4ddG, and stable complexes were separated from free DNA by nondenaturing electrophoresis as described in Materials and Methods. KD values were determined from plots of the percentage of total radioactivity in complex versus substrate concentration. DNA P/T sequence near the primer terminus is given at the bottom of the panel.
RESULTS
Synthesis and purification of dinucleoside tetraphosphates containing identical 2′,3′-dideoxynucleoside moieties (ddNp4ddN).
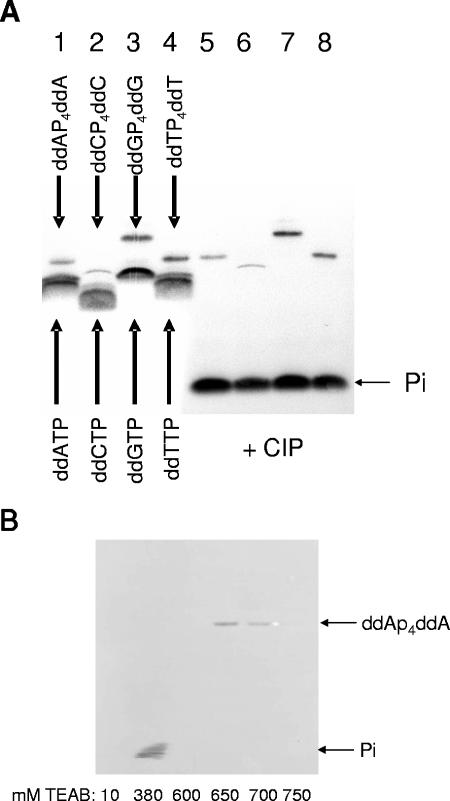
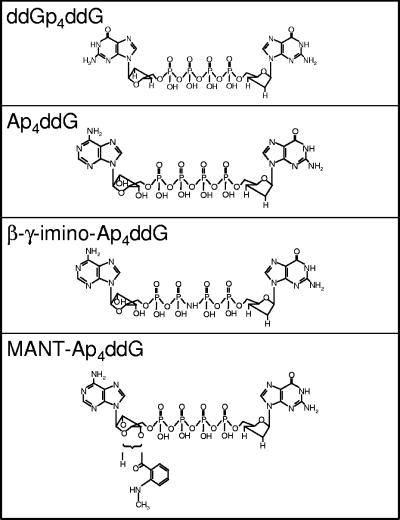
A recycling reaction using multinucleoside-resistant HIV-1 RT (HIV-1 RTMDR) containing M41L/T69S-insertionAG/L210W/R211K/L214F/T215Y/K219Q mutations (which has a high level of ATP-dependent removal activity [16, 21]) was used for synthesis of dinucleoside tetraphosphates as outlined in Fig. 1 and in the Materials and Methods section. Over time, this reaction converted a portion of the ddNTP into ddNp4ddN (Fig. 2A). The reaction mixture was treated with CIP to degrade the remaining ddNTP. The ddNp4ddN, which is resistant to CIP cleavage, was separated from free phosphate and 2′,3′-dideoxynucleosides by anion exchange chromatography (Fig. 2B). The structures of four dinucleoside polyphosphates are shown in Fig. 3.
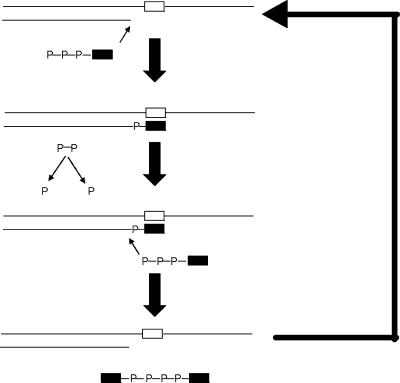
FIG. 1.
Schematics for synthesis of ddNp4ddNs. A 10 μM concentration of DNA P/T was incubated with a 3.2 mM concentration of the ddNTP (a small portion of which was α-33P labeled) that is complementary to the first single-stranded position on the template, 1 μM multinucleoside-resistant HIV-1 RT (HIV-1 RTMDR), and 0.5 units of inorganic pyrophosphatase. Incorporation of the 2′,3′-dideoxynucleoside monophosphate into the primer terminus leads to chain termination. The PPi generated by the incorporation was cleaved by pyrophosphatase, and the chain terminator was transferred from the blocked DNA to ddNTP, leading to formation of the dinucleoside polyphosphate ddNp4ddN and unblocked P/T available for another round of synthesis.
FIG. 2.
Synthesis and purification of ddNp4ddNs. (A) A 10 μM concentration of one of four different DNA P/Ts (see Materials and Methods for sequences) was incubated with HIV-1 RTMDR, inorganic pyrophosphatase, and 3.2 mM ddATP (lanes 1 and 5), ddCTP (lanes 2 and 6), ddGTP (lanes 3 and 7), or ddTTP (lanes 4 and 8) for 7 days at 37°C. Following heat inactivation of the RT, the reaction mixture was incubated with CIP (lanes 5 to 8). (B) The CIP-treated reaction mixture shown in lane 5 in panel A was loaded onto an anion exchange column and eluted with a 10 to 1,000 mM gradient of triethylammonium bicarbonate (TEAB) buffer, pH 8.5. Aliquots of reaction mixtures or selected column fractions were separated by electrophoresis through 20% urea-polyacrylamide gel electrophoresis.
FIG. 3.
Structures of four of the dinucleoside tetraphosphates that were synthesized.
Inhibition of HIV-1 RT-mediated DNA synthesis by ddNTPs and ddNp4ddNs.
In order to test the ability of WT and mutant HIV-1 RT to use ddNp4ddNs as dNTP analogues, we determined the inhibition of DNA-dependent DNA synthesis by these compounds using M13 P/T as a substrate. DNA synthesis was measured by incorporation of [32P]dNMP from [α-32P]dNTP. Incorporation of ddNMP from either ddNTP or ddNp4ddN resulted in chain termination, leading to a decrease in the total amount of incorporated radioactive nucleotide and the appearance of shorter, chain-terminated products. A portion of the reaction mixture was separated by denaturing gel electrophoresis to visualize the extended products, while the majority was spotted onto filter circles and precipitated with trichloroacetic acid to allow quantification of incorporated radioactivity. Figure 4A shows the ability of either ddGTP or ddGp4ddG to inhibit DNA polymerization by HIV-1 RT containing D67N/K70R/T215Y/K219Q mutations (HIV-1 RTAZT). In the absence of inhibitor (Fig. 4, lane 1), a large majority of M13 primers were extended to a similar position, observed as a strong radioactive band at the top of the gel. In the presence of increasing concentrations of ddGTP (lanes 2 to 5), there was a concomitant decrease in the amount of total incorporated radioactive label and a decrease in the average lengths of the extended products that were chain terminated at G-incorporation sites. A similar pattern was observed at increasing concentrations of ddGp4ddG (lanes 6 to 9), although the enzyme was slightly less sensitive to inhibition by ddGp4ddG than to inhibition by ddGTP. Pretreatment of ddGTP with CIP abolished its inhibitory activity (lanes 10 to 13), while similar treatment of ddGp4ddG had minimal affect on its ability to inhibit DNA synthesis (lanes 14 to 17). Therefore, ddGp4ddG was an efficient substrate for DNA synthesis by HIV-1 RTAZT. Figure 4B shows that heparin-resistant complex formed with ddAMP-terminated primer/template by RTAZT and RTWT had similar stability with ddGTP, but the complexes formed with ddGp4ddG were about threefold more stable with RTAZT than with RTWT (KD of 0.26 μM versus 0.80 μM). While the thymidine analogue mutations selectively enhanced interaction with the tetraphosphate substrate, the affinity with the mutant enzyme was still about fourfold less than for ddGTP, in agreement with the sensitivity of this enzyme to inhibition by ddGTP (Fig. 4A).
Mechanism for transfer of ddNMP from a dinucleoside tetraphosphate onto the primer terminus by HIV-1 RT.
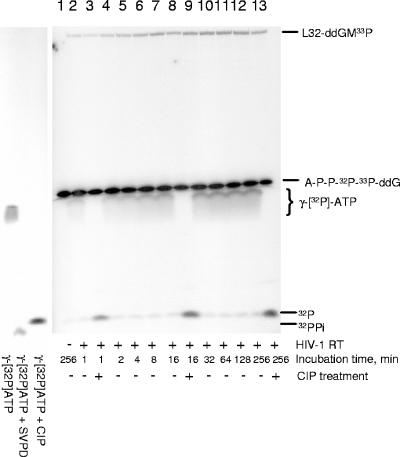
There are two possible reaction mechanisms, a direct and an indirect one, for the ability of HIV-1 RT to use dinucleoside tetraphosphates as substrates for ddNMP incorporation. In the direct mechanism, the dinucleoside tetraphosphate itself serves as the dNTP analogue. After incorporation of a ddNMP moiety from Np4ddN into the primer terminus, this pathway would yield one molecule of NTP. In the indirect pathway, the RT first cleaves the Np4ddN to yield a molecule of NMP and a molecule of ddNTP, which would subsequently be incorporated at the primer terminus. This pathway would yield one molecule of NMP and one molecule of PPi for each incorporation event. To distinguish between these two possibilities, the ability of HIV-1 RTAZT to use doubly labeled Ap4ddG (A-P-P-32P-33P-ddG) as a dGTP analogue was measured. As can be seen in Fig. 5, increased incubation times led to an increased amount of the labeled product L32-ddGM-33P, indicating that Ap4ddG can serve as a dGTP analogue. Concomitant with the increase in L32-ddGM-33P was an increase in [γ-32P]ATP and, importantly, no increase in PPi, indicating that ddGMP was incorporated into the primer terminus by HIV-1 RT directly from Ap4ddG. In addition, a similar incorporation efficiency of ddGMP from Ap4ddG was observed when the reaction was performed in the continuous presence of CIP (data not shown), indicating that no CIP-sensitive intermediate was formed during the reaction, which further supports the direct incorporation mechanism.
FIG. 5.
Products formed during incorporation of ddGMP from doubly labeled Ap4ddG. A 10 μM concentration of unlabeled WL50-33C/L32 template/primer was incubated with 2 μM HIV-1 RTMDR and 200 μM Ap4ddG (A-P-P-32P-33P-ddG) for the indicated times at 37°C, followed by heat inactivation of the RT (5 min at 95°C). Some samples were then treated with 4 U of CIP (+CIP) for 30 min, followed by heat inactivation (5 min at 95°C) of the CIP. The reaction products were separated by 20% urea-polyacrylamide gel electrophoresis, and the radioactivity was visualized by phosphorimaging. Standards for free phosphate and PPi were generated by incubating [γ-32P]ATP with CIP or snake venom phosphodiesterase (SVPD), respectively.
Sensitivity of DNA polymerization by WT and mutant HIV-1 RTs to inhibition by ddNTPs, ddNp4ddNs, or Np4ddNs.
The IC50s were determined for DNA polymerization by WT and mutant RTs for ddATP, ddAp4ddA, ddCTP, ddCp4ddC, ddGTP, ddGp4ddG, ddTTP, and ddTp4ddT (Table 1). HIV-1 RTWT, RTK65R, and RTW88G were much more sensitive (15- to 83-fold) to inhibition by ddNTPs than by ddNp4ddNs. RTK65R and RTW88G were less sensitive to inhibition by either ddNTPs or ddNp4ddNs than RTWT. RTs containing TAMs (RTM41L/T215Y, RTAZT, and RTMDR) had increased sensitivity to ddNp4ddNs compared with RTWT, but there was little difference in sensitivity to ddNTPs. The ratio for inhibition of DNA synthesis (IC50 for ddNp4ddN/IC50 for ddNTP) was 15- to 72-fold for RTWT, 4.8- to 25-fold for RTM41L/T215Y, 1.8- to 4.7-fold for RTAZT, and 0.95- to 4.8-fold for RTMDR. Of the dinucleoside tetraphosphates tested, all six enzymes were more sensitive to inhibition by ddCp4ddC and ddGp4ddG. The IC50s for ddTp4ddT and ddAp4ddA were 1.2- to 8.5-fold higher than for ddCp4ddC and ddGp4ddG.
TABLE 1.
Inhibition of HIV-1 RT-mediated DNA-dependent DNA synthesis by ddNTPs or ddNp4ddNs
| ddNa | Enzyme | IC50 (μM)b
|
IC50 (ddNp4ddN)/IC50 (dNTP) [WT/mutant]c | |
|---|---|---|---|---|
| ddNTP (WT/mutant)c | ddNp4ddN (WT/mutant)c | |||
| ddA | RTWT | 0.11 ± 0.01 (1.0) | 6.5 ± 0.4 (1.0) | 61 (1.0) |
| RTK65R | 0.59 ± 0.09 (0.19) | >36 (<0.18) | >61 (<1.0) | |
| RTW88G | 0.31 ± 0.13 (0.35) | >15 (<0.43) | >48 (<1.3) | |
| RTM41L/T215Y | 0.22 ± 0.03 (0.50) | 2.9 ± 1.0 (2.2) | 13 (4.7) | |
| RTAZT | 0.16 ± 0.01 (0.69) | 0.65 ± 0.12 (10) | 4.2 (15) | |
| RTMDR | 0.25 ± 0.03 (0.44) | 1.2 ± 0.2 (5.4) | 4.8 (13) | |
| ddC | RTWT | 0.079 ± 0.009 (1.0) | 2.1 ± 0.4 (1.0) | 27 (1.0) |
| RTK65R | 0.44 ± 0.05 (0.18) | 11 ± 2 (0.19) | 25 (1.1) | |
| RTW88G | 0.28 ± 0.06 (0.28) | 9.2 ± 1.2 (0.23) | 33 (0.82) | |
| RTM41L/T215Y | 0.095 ± 0.012 (0.83) | 1.1 ± 0.1 (1.9) | 12 (2.2) | |
| RTAZT | 0.10 ± 0.01 (0.79) | 0.18 ± 0.02 (12) | 1.8 (15) | |
| RTMDR | 0.12 ± 0.01 (0.66) | 0.28 ± 0.03 (7.5) | 2.3 (12) | |
| ddG | RTWT | 0.12 ± 0.01 (1.0) | 1.8 ± 0.2 (1.0) | 15 (1.0) |
| RTK65R | 0.49 ± 0.10 (0.24) | 9.5 ± 1.2 (0.19) | 19 (0.79) | |
| RTW88G | 0.24 ± 0.04 (0.50) | 6.0 ± 0.9 (0.30) | 25 (0.60) | |
| RTM41L/T215Y | 0.082 ± 0.010 (1.5) | 0.39 ± 0.11 (4.6) | 4.8 (3.1) | |
| RTAZT | 0.092 ± 0.017 (1.3) | 0.20 ± 0.03 (9.0) | 2.2 (6.8) | |
| RTMDR | 0.21 ± 0.05 (0.57) | 0.20 ± 0.02 (9.0) | 0.95 (16) | |
| ddT | RTWT | 0.12 ± 0.02 (1.0) | 8.6 ± 2.0 (1.0) | 72 (1.0) |
| RTK65R | 0.66 ± 0.12 (0.18) | 42 ± 14 (0.20) | 64 (1.1) | |
| RTW88G | 0.41 ± 0.05 (0.29) | 34 ± 12 (0.25) | 83 (0.87) | |
| RTM41L/T215Y | 0.13 ± 0.02 (0.92) | 3.3 ± 0.4 (2.6) | 25 (2.9) | |
| RTAZT | 0.20 ± 0.02 (0.60) | 0.92 ± 0.09 (9.3) | 4.7 (15) | |
| RTMDR | 0.34 ± 0.06 (0.35) | 0.34 ± 0.02 (25) | 1.0 (72) | |
ddN indicates the dideoxynucleoside component of ddNTP and both dideoxynucleoside components of ddNp4ddN compounds tested as inhibitors of DNA synthesis.
IC50s were determined on M13 P/T and adjusted for equivalent specific activity for each enzyme as described in Materials and Methods. Results are the means of three experiments ± standard deviations.
Ratio of IC50 for RTWT to the IC50 for each of the enzymes.
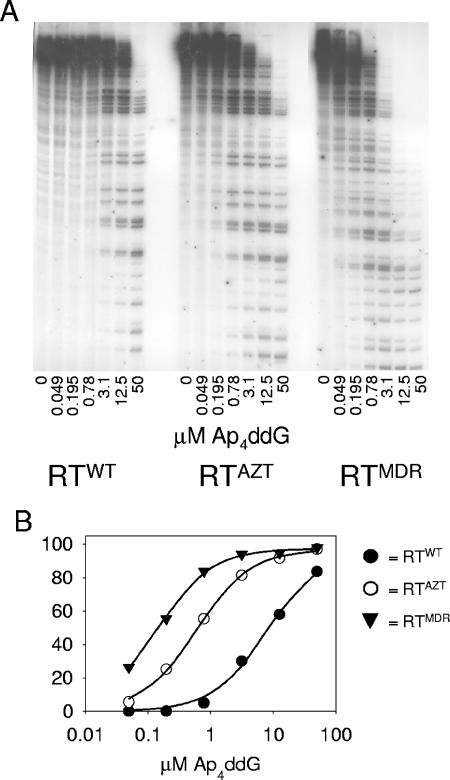
Next, we characterized the ability of Ap4ddG or Ap4ddG-derived compounds containing modifications of the sugar or phosphate chains to inhibit DNA polymerization (Fig. 6 and Table 2). RTMDR used Ap4ddG as a dGTP analogue even more efficiently than ddGp4ddG, while RTWT was about fourfold less sensitive to Ap4ddG than to ddGp4ddG, resulting in a 50-fold higher sensitivity of DNA polymerization by RTMDR than polymerization by RTWT to inhibition by Ap4ddG (Fig. 6 and Tables 1 and 2). With MANT-Ap4ddG the difference in sensitivity between these enzymes was over 100-fold. DNA polymerization by RTMDR was also sensitive to inhibition by AppNHppddG (IC50 of 1.7 μM), a compound to which all other enzymes tested were highly resistant (IC50s of ≥28 μM).
FIG. 6.
Inhibition of DNA-dependent DNA synthesis through incorporation of ddGMP from Ap4ddG by RTWT, RTAZT, or RTMDR. (A) M13 single-stranded DNA annealed to a DNA primer (2.5 nM) was incubated with HIV-1 RTWT or HIV-1 RTAZT (100 nM), a 2.5 μM concentration of each dNTP, 50 μCi of [α-32P]dGTP, and the indicated concentrations of Ap4ddG for 30 min at 37°C. The samples were separated by 20% urea-polyacrylamide gel electrophoresis. (B) Percent inhibition of [32P]dGMP incorporation by Ap4ddG was determined as described in Materials and Methods. A summary of IC50s derived from several experiments is given in Table 2.
TABLE 2.
Inhibition of HIV-1 RT-mediated DNA-dependent DNA synthesis by Ap4ddG, MANT-Ap4ddG, or AppNHppddG
| Enzyme | IC50 (μM)a
|
||
|---|---|---|---|
| Ap4ddG (WT/mutant)b | MANT-Ap4ddG (WT/mutant)b | AppNHppddG (WT/mutant)b | |
| RTWT | 6.8 ± 0.7 (1.0)b | 16 ± 5 (1.0) | >50 (1.0) |
| RTD67N/K70R | 1.4 ± 0.1 (4.9) | 2.5 ± 0.4 (6.4) | >41 (NC)c |
| RTM41L/T215Y | 1.95 ± 0.31 (3.5) | 3.8 ± 0.3 (4.2) | >37 (NC) |
| RTAZT | 0.74 ± 0.05 (9.2) | 0.83 ± 0.09 (19) | 28 ± 9 (>1.8) |
| RTMDR | 0.13 ± 0.03 (52) | 0.13 ± 0.13 (120) | 1.7 ± 0.6 (>29) |
IC50s were determined as in Table 1. Results are the means of three experiments ± standard deviations.
Ratio of IC50 for RTWT to the IC50 for each of the enzymes.
NC, not calculated.
In summary, TAMs and the T69S-XX insertion mutation, which confer an increase in ATP-dependent removal of nucleotide analogues from blocked DNA, also conferred an increased ability to use dinucleoside polyphosphates as dNTP analogues during DNA synthesis. This resulted in greater sensitivity of HIV-1 RT containing these mutations to inhibition by ddNp4ddNs. In contrast, mutations K65R and W88G, which are associated with decreased PPi- and nucleotide-dependent removal activity (25, 30), conferred decreased sensitivity to inhibition by either ddNTPs or ddNp4ddNs.
DISCUSSION
Our results show that AZT-resistant HIV-1 RTs that contain TAMs and have enhanced nucleotide excision activity also have increased ability to use dinucleoside tetraphosphates of the form Np4ddN or ddNp4ddN as substrates for DNA elongation and chain termination. By contrast, when ddNTPs were provided as substrates, the efficiency of chain termination was about the same for TAM-containing RTs and WT RT. The difference between WT and mutant RTs was greatest when MANT-Ap4ddG was used as a substrate. Chain termination with this substrate by RTMDR was 120-fold more efficient than by RTWT. Chain termination with MANT-Ap4ddG by RTAZT was 19-fold more efficient than by RTWT. Other dinucleoside tetraphosphate compounds showed smaller but still substantial differences between mutant and WT RT. Incorporation of ddNMP occurred without prior formation of a ddNTP intermediate, in agreement with a previous report (33). Our results provide tools for further mechanistic studies and development of drugs that target this class of HIV-1 mutants.
Our results suggest that dinucleoside tetraphosphates have the ability to interact with two adjacent nucleoside binding surfaces on RT: one identified as the binding site for the incoming dNTP in the polymerase reaction and the second identified by the residues mutated in TAMs and lying just distal to the end of the phosphate chain of the incoming dNTP. The dNTP binding site is known from the crystal structure of RTWT-P/T-dTTP ternary complex (10), and the locations for the second nucleoside and additional phosphate residue have been inferred by modeling in an ATP molecule with the β and γ phosphates superimposed on the γ and β phosphates of the dTTP (3, 5). In the proposed structure, the adenine base forms π-π interactions with tyrosine or phenylalanine side chains at residue 215 in the mutant RT. Tests of excision substrate specificity also provide support for this model (24). The enhanced rate of Np4ddN-mediated chain termination by TAM-containing RTs can be explained by more favorable binding of Np4ddN to the mutant enzyme, as observed with ddGp4ddG (Fig. 4). While these results support the ability of the tetraphosphates to interact with adjacent sites on RT, our data indicate that functional binding of ddGTP is favored over ddGp4ddG for TAM-containing RTs such as RTAZT. This suggests that binding of Np4ddN compounds to the ddNTP site is severely restricted in RTWT and that TAMs confer increased ability to bind these compounds, possibly by removing steric constraints or by contributing binding interactions that can compensate for limited accessibility of the tetraphosphate compounds to the binding site. The enhanced rate of nucleotide-dependent excision by TAM-containing RTs is largely due to effects on catalytic rate that occur after substrate binding since TAMs have little effect on the KD for the nucleotide substrate (29). It has been proposed that the T215Y or F mutations increase the rate of nucleophilic attack by controlling the alignment of the pyrophosphate component of the acceptor substrate with respect to the bond to be cleaved in the nascent DNA chain (8, 19, 20). Similar considerations may explain the ability of these mutations to enhance the use of dinucleoside tetraphosphates as substrates for incorporation.
Incorporation of ddGMP or ddCMP using ddGp4ddG or ddCp4ddC as a substrate was more efficient than incorporation of ddAMP or ddTMP using the corresponding dinucleoside tetraphosphate substrates. This suggests that a G-C base pair was preferred during the reaction. The difference in ddGMP incorporation between RTMDR and RTWT was greatest when MANT-Ap4ddG was used as a substrate, suggesting that RTMDR is better able to accommodate a bulky substituent on the ribose than is RTWT. AppNHppddG was a poor substrate for chain termination with all of the enzymes tested, indicating that the precise structure of the phosphate chain is important.
These results suggest that it may be possible to identify bifunctional compounds that have high affinity for RT and preferentially bind to the TAM-containing enzymes. Dinucleoside tetraphosphate derivatives that contain hydrolyzable phosphate links have the advantage that they may be able to bypass the need for intracellular phosphorylation to lead to chain termination. This would expand the range of nucleoside structures that can be used for chain termination by HIV-1 since current NRTIs are limited to molecules that are phosphorylated by cellular nucleoside kinases. On the other hand, the presence of phosphate residues may limit the ability of these compounds to enter mammalian cells and increase their sensitivity to degradative enzymes such as phosphodiesterases. However, modification of the phosphate residues might improve bioavailability and stability of dinucleoside polyphosphates (2).
The ability of dinucleoside tetraphosphates to serve as substrates for DNA chain elongation and termination by HIV-1 RT and the strong preference of these compounds for use by TAM-containing mutant enzymes provide a useful tool for analysis of the effects of these mutations on RT activities. In addition, our studies provide evidence for an interaction surface on the mutant enzymes that recognizes both nucleoside components of the Np4ddN structure and may be an attractive target for future drug development.
Acknowledgments
We thank Johan Lennerstrand and Brendan Larder for providing the expression plasmid for HIV-1 RTMDR, John Mellors for the expression plasmid for HIV-1 RTAZT, and Antero So for insightful suggestions on the manuscript.
This work was supported by NIH grant AI-39973 (W.A.S.), and amfAR postdoctoral fellowship 70567-31-RF (P.R.M.), and American Heart Association predoctoral fellowships 0215087B and 0415255B (A.J.S.).
Footnotes
Published ahead of print on 28 August 2006.
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Ballatore, C., C. McGuigan, E. De Clercq, and J. Balzarini. 2001. Synthesis and evaluation of novel amidate prodrugs of PMEA and PMPA. Bioorg. Med. Chem. Lett. 11:1053-1056. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canard, B., K. Chowdhury, R. Sarfati, S. Doublié, and C. C. Richardson. 1999. The motif D loop of human immunodeficiency virus type 1 reverse transcriptase is critical for nucleoside 5′-triphosphate selectivity. J. Biol. Chem. 274:35768-35776. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain, P. P., J. Ren, C. E. Nichols, L. Douglas, J. Lennerstrand, B. A. Larder, D. I. Stuart, and D. K. Stammers. 2002. Crystal structures of zidovudine- or lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215. J. Virol. 76:10015-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, P. K., B. Wynhoven, and P. R. Harrigan. 2004. 2004: Which HIV-1 drug resistance mutations are common in clinical practice? AIDS Rev. 6:107-116. [PubMed] [Google Scholar]
- 7.Clark, P. K., A. L. Ferris, D. A. Miller, A. Hizi, K.-W. Kim, S. M. Deringer-Boyer, M. L. Mellini, A. D. Clark, Jr., G. F. Arnold, W. B. Lebherz III, E. Arnold, G. M. Muschik, and S. H. Hughes. 1990. HIV-1 reverse transcriptase purified from a recombinant strain of Escherichia coli. AIDS Res. Hum. Retrovir. 6:753-764. [DOI] [PubMed] [Google Scholar]
- 8.Goldschmidt, V., and R. Marquet. 2004. Primer unblocking by HIV-1 reverse transcriptase and resistance to nucleoside RT inhibitors (NRTIs). Int. J. Biochem. Cell Biol. 36:1687-1705. [DOI] [PubMed] [Google Scholar]
- 9.Hooker, D. J., G. Tachedjian, A. E. Solomon, A. D. Gurusinghe, S. Land, C. Birch, J. L. Anderson, B. M. Roy, E. Arnold, and N. J. Deacon. 1996. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J. Virol. 70:8010-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 11.Kellam, P., C. A. B. Boucher, and B. A. Larder. 1992. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc. Natl. Acad. Sci. USA 89:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krebs, R., U. Immendörfer, S. H. Thrall, B. M. Wöhrl, and R. S. Goody. 1997. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3-TC. Biochemistry 36:10292-10300. [DOI] [PubMed] [Google Scholar]
- 13.Larder, B. A., S. Bloor, S. D. Kemp, K. Hertogs, R. L. Desmet, V. Miller, M. Sturmer, S. Staszewski, J. Ren, D. K. Stammers, D. I. Stuart, and R. Pauwels. 1999. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob. Agents Chemother. 43:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larder, B. A., P. Kellam, and S. D. Kemp. 1991. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS 5:137-144. [DOI] [PubMed] [Google Scholar]
- 15.Lennerstrand, J., K. Hertogs, D. K. Stammers, and B. A. Larder. 2001. Correlation between viral resistance to zidovudine and resistance at the reverse transcriptase level for a panel of human immunodeficiency virus type 1 mutants. J. Virol. 75:7202-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lennerstrand, J., D. K. Stammers, and B. A. Larder. 2001. Biochemical mechanism of human immunodeficiency virus type 1 reverse transcriptase resistance to stavudine. Antimicrob. Agents Chemother. 45:2144-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majumdar, C., J. Abbotts, S. Broder, and S. H. Wilson. 1988. Studies on the mechanism of human immunodeficiency virus reverse transcriptase. Steady-state kinetics, processivity, and polynucleotide inhibition. J. Biol. Chem. 263:15657-15665. [PubMed] [Google Scholar]
- 18.Mas, A., B. M. Vázquez-Álvarez, E. Domingo, and L. Menéndez-Arias. 2002. Multidrug-resistant HIV-1 reverse transcriptase: involvement of ribonucleotide-dependent phosphorolysis in cross-resistance to nucleoside analogue inhibitors. J. Mol. Biol. 323:181-197. [DOI] [PubMed] [Google Scholar]
- 19.Matamoros, T., J. Deval, C. Guerreiro, L. Mulard, B. Canard, and L. Menéndez-Arias. 2005. Suppression of multidrug-resistant HIV-1 reverse transcriptase primer unblocking activity by α-phosphate-modified thymidine analogues. J. Mol. Biol. 349:451-463. [DOI] [PubMed] [Google Scholar]
- 20.Matamoros, T., S. Franco, B. M. Vázquez-Álvarez, A. Mas, M. A. Martínez, and L. Menéndez-Arias. 2004. Molecular determinants of multi-nucleoside analogue resistance in HIV-1 reverse transcriptases containing a dipeptide insertion in the fingers subdomain: effect of mutations D67N and T215Y on removal of thymidine nucleotide analogues from blocked DNA primers. J. Biol. Chem. 279:24569-24577. [DOI] [PubMed] [Google Scholar]
- 21.Meyer, P. R., J. Lennerstrand, S. E. Matsuura, B. A. Larder, and W. A. Scott. 2003. Effects of dipeptide insertions between codons 69 and 70 of human immunodeficiency virus type 1 reverse transcriptase on primer unblocking, deoxynucleoside triphosphate inhibition, and DNA chain elongation. J. Virol. 77:3871-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, P. R., S. E. Matsuura, A. A. Tolun, I. Pfeifer, A. G. So, J. W. Mellors, and W. A. Scott. 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, P. R., S. E. Matsuura, D. Zonarich, R. R. Chopra, E. Pendarvis, H. Z. Bazmi, J. W. Mellors, and W. A. Scott. 2003. Relationship between 3′-azido-3′-deoxythymidine resistance and primer unblocking activity in foscarnet-resistant mutants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 77:6127-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer, P. R., A. J. Smith, S. E. Matsuura, and W. A. Scott. 2004. Effects of primer-template sequence on ATP-dependent removal of chain-terminating nucleotide analogues by HIV-1 reverse transcriptase. J. Biol. Chem. 279:45389-45398. [DOI] [PubMed] [Google Scholar]
- 27.Naeger, L. K., N. A. Margot, and M. D. Miller. 2002. ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quan, Y., C. Liang, P. Inouye, and M. A. Wainberg. 1998. Enhanced impairment of chain elongation by inhibitors of HIV reverse transcriptase in cell-free reactions yielding longer DNA products. Nucleic Acids Res. 26:5692-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ray, A. S., E. Murakami, A. Basavapathruni, J. A. Vaccaro, D. Ulrich, C. K. Chu, R. F. Schinazi, and K. S. Anderson. 2003. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry 42:8831-8841. [DOI] [PubMed] [Google Scholar]
- 30.Sluis-Cremer, N., D. Arion, N. Kaushik, H. Lim, and M. A. Parniak. 2000. Mutational analysis of Lys65 of HIV-1 reverse transcriptase. Biochem. J. 348:77-82. [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, A. J., P. R. Meyer, D. Asthana, M. R. Ashman, and W. A. Scott. 2005. Intracellular substrates for the primer-unblocking reaction by human immunodeficiency virus type 1 reverse transcriptase: detection and quantitation in extracts from quiescent- and activated-lymphocyte subpopulations. Antimicrob. Agents Chemother. 49:1761-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan, C.-K., J. Zhang, Z.-Y. Li, W. G. Tarpley, K. M. Downey, and A. G. So. 1991. Functional characterization of RNA-dependent DNA polymerase and RNase H activities of a recombinant HIV reverse transcriptase. Biochemistry 30:2651-2655. [DOI] [PubMed] [Google Scholar]
- 33.Victorova, L., V. Sosunov, A. Skoblov, A. Shipytsin, and A. Krayevsky. 1999. New substrates of DNA polymerases. FEBS Lett. 453:6-10. [DOI] [PubMed] [Google Scholar]
- 34.Winters, M. A., R. W. Shafer, R. A. Jellinger, G. Mamtora, T. Gingeras, and T. C. Merigan. 1997. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob. Agents Chemother. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]