Abstract
Fungal pathogens form biofilms that are highly recalcitrant to antimicrobial therapy. The expression of multidrug resistance pumps in young biofilms has been linked to increased resistance to azoles, but this mechanism does not seem to underlie the resistance of mature biofilms that is a model of in vivo infection. The mechanism of drug resistance of mature biofilms remains largely unknown. We report that biofilms formed by the major human pathogen Candida albicans exhibited a strikingly biphasic killing pattern in response to two microbicidal agents, amphotericin B, a polyene antifungal, and chlorhexidine, an antiseptic, indicating that a subpopulation of highly tolerant cells, termed persisters, existed. The extent of killing with a combination of amphotericin B and chlorhexidine was similar to that observed with individually added antimicrobials. Thus, surviving persisters form a multidrug-tolerant subpopulation. Interestingly, surviving C. albicans persisters were detected only in biofilms and not in exponentially growing or stationary-phase planktonic populations. Reinoculation of cells that survived killing of the biofilm by amphotericin B produced a new biofilm with a new subpopulation of persisters. This suggests that C. albicans persisters are not mutants but phenotypic variants of the wild type. Using a stain for dead cells, rare dark cells were visible in a biofilm after amphotericin B treatment, and a bright and a dim population were physically sorted from this biofilm. Only the dim cells produced colonies, showing that this method allows the isolation of yeast persisters. Given that persisters formed only in biofilms, mutants defective in biofilm formation were examined for tolerance of amphotericin B. All of the known mutants affected in biofilm formation were able to produce normal levels of persisters. This finding indicates that attachment rather than formation of a complex biofilm architecture initiates persister formation. Bacteria produce multidrug-tolerant persister cells in both planktonic and biofilm populations, and it appears that yeasts and bacteria have evolved analogous strategies that assign the function of survival to a small part of the population. In bacteria, persisters are dormant cells. It remains to be seen whether attachment initiates dormancy that leads to the formation of fungal persisters. This study suggests that persisters may be largely responsible for the multidrug tolerance of fungal biofilms.
Candida albicans is an opportunistic pathogen that is a common member of the human oral and gastrointestinal microflora. Biofilms of C. albicans often form on indwelling devices, such as blood and urinary catheters and heart valves. These infections are essentially untreatable by antifungals and can develop into life-threatening disease with a mortality rate approaching 40%. The recalcitrance of these infections to antifungals is puzzling, since planktonic populations of the same strain can be susceptible to a range of antifungals, including azole compounds, echinocandins, and amphotericin B. A number of excellent reviews have recently been published on this subject (2, 10, 14, 16, 20, 24, 25, 27, 29, 33, 35, 40, 44).
The biofilm forms when single cells attach to a surface and grow into microcolonies, which then merge and produce a complex three-dimensional (3D) structure that is held together by hyphae and an exopolymer matrix (7). The biofilm contains a mixture of yeast, hyphae, and pseudohyphae. Several factors may account for the high resistance of biofilm cells to antimicrobial drugs. For example, the exopolymer matrix restricts penetration by immune system components (15, 51). Since in vivo, antimicrobials act in concert with the immune system to eradicate infections, the biofilm exopolymer is evidently an important component of recalcitrance. However, the exopolymer does not appreciably hinder the penetration of antifungal drugs (3, 46). Quiescence can be a factor in drug resistance, but the majority of cells in C. albicans biofilms are metabolically active, based on studies with XTT and FUN-1 metabolic indicators (21-23). Genes encoding multidrug resistance (MDR) transporters, MDR1, CDR1 and CDR2, are upregulated upon attachment of C. albicans cells to a surface, and this accounts for the resistance of young biofilms to azole antibiotics (34). However, the high level of drug resistance of mature biofilms (≥48 h) was not affected by deletion of all three of these genes, either singly or in combination, including an mdr1Δ cdr1Δ cdr2Δ triple mutant (26, 34, 39). Decreased ergosterol content (26, 34) and a diminished level of ergosterol biosynthetic gene expression (11) have been reported in mature Candida biofilms and may contribute to drug resistance. Indeed, azoles act by inhibiting ergosterol biosynthesis, and amphotericin B binds to ergosterol. However, ergosterol is unlikely to be involved in the actions of echinocandins that inhibit the synthesis of cell wall β-glucan (8) or of chlorhexidine, a membrane-active antiseptic that is very effective against bacteria that lack sterols. In essence, the mechanism of C. albicans biofilm antifungal resistance remains largely unknown.
It is noteworthy that many features of fungal biofilms are shared by the better-studied bacterial biofilms. In both cases, biofilms are often formed by merging of initial founder microcolonies, are protected from the immune system by exopolymers, are composed mainly of slowly growing cells, and exhibit multidrug tolerance (27, 28). Bacterial biofilms produce dormant persister cells that are largely responsible for multidrug tolerance (6, 19, 48). Persisters are able to survive despite the presence of antibiotics at concentrations well above the MIC and are phenotypic variants of the wild type, rather than mutants (19). Persisters are formed by all bacterial species studied and are present at 0.1 to 1% in the biofilms of Pseudomonas aeruginosa, Escherichia coli, and Staphylococcus aureus, for example (19).
Given their similarity to bacterial biofilms, we reasoned that fungal biofilms may similarly produce persister cells. In this study, we report that C. albicans biofilms contain highly drug-tolerant persisters that may largely account for biofilm multidrug tolerance.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The yeast strains used in this study are described in Table 1. C. albicans was streaked onto yeast extract, peptone, dextrose (YPD) agar from frozen stocks (−80°C) and incubated at 37°C for 24 to 48 h. When necessary (CAI4 derivatives), the growth medium was supplemented with 80 μg/ml uridine. Biofilms were grown essentially as described by Ramage et al. (41). Briefly, cells were grown overnight at 30°C in YPD liquid medium. Samples were harvested by centrifugation, washed twice in sterile phosphate-buffered saline (PBS), pH 7.4, and resuspended in RPMI 1640 with l-glutamine and 0.165 M MOPS (morpholinepropanesulfonic acid) (BioWhittaker). The optical density of the culture (at 600 nm) was adjusted to 0.1, which is equivalent to approximately 1 × 106 cells/ml, by dilution with RPMI 1640. One hundred microliters of this suspension was aliquoted into 300-μl wells of a flat-bottom microtiter plate (Corning Costar 3370). The plates were incubated for 48 h at 37°C on a microtiter plate shaker (Lab-Line Instruments; model 4625) at approximately 100 rpm. Exponential- and stationary-phase cultures inoculated from single colonies were grown in RPMI 1640 with l-glutamine and 0.165 M MOPS at 37°C for 12 and 72 h, respectively.
TABLE 1.
Strains used in this study and tested for persisters
| Strain | Genotype | Biofilm architecture | Persistersa | Source/reference |
|---|---|---|---|---|
| 3153A | Wild-type laboratory strain | Robust 3D wild type | +++ | 41 |
| CKY357 | CAI-4 mkc1Δ::hisG/mkc1Δ::hisG mkc1::pCK70 (URA3) | Reduced filamentation | ++ | 26 |
| CAI4 | SC5314 Δura3::λimm434/Δura3::λimm434 | Robust 3D wild type | + | 26 |
| CKY136 | CAI-4 efg1::hisG/efg1::hisG ade2::pDBI52 (URA3) | Filamentation defect; sparse monolayer of cells | +++ | 12 |
| CKY138 | CAI-4 efg1::hisG/efg1::hisG cph1Δ::hisG/cph1Δ::hisG ade2::pDBI52 (URA3) | Filamentation defect; sparse monolayer of cells | ++ | 12 |
| MC191 | ura3Δ::λimm434/ura3Δ::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG flo8::ARG4/flo8::HIS1 ade2::URA3/ADE2 | Functionally defective hyphae | +++ | Microbia |
| MC195 | ura3Δ::λimm434/ura3Δ::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG flo8::ARG4/flo8::HIS1 ade2::URA3::FLO8-2/ADE2 | Robust 3D wild type | + | Microbia |
| MC245 | ura3Δ::λimm434/ura3Δ::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG flo8::ARG4/FLO8 ade2::URA3/ADE2 HIS::his/his | Robust 3D wild type | ++ | Microbia |
| DAY185 | Δura3::λimm434/Δura3::λimm434 arg4::hisG/arg4::hisG/pARG4-URA3 his1::hisG/his1::hisG/pHIS1 | Robust 3D wild type | ++ | 45 |
| DAY286 | Δura3::λimm434/Δura3::λimm434 arg4::hisG/arg4::hisG/pARG4-URA3 his1::hisG/his1::hisG | Robust 3D wild type | ++ | 45 |
| GKO443 | Δura3::λimm434/Δura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG suv3::Tn7-UAU1/suv3::Tn7-URA3 | Biofilm defect; decreased biomass | ++ | 45 |
| GKO798 | Δura3::λimm434/Δura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG kem1::Tn7-UAU1/kem1::Tn7-URA3 | Biofilm defect; decreased biomass | ++ | 37 |
| GKO814 | Δura3::λimm434/Δura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG nup85::Tn7-UAU1/nup85::Tn7-URA3 | Biofilm defect; decreased biomass | ++ | 45 |
| GKO9 | Δura3::λimm434/Δura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG mds3::Tn7-UAU1/mds3::Tn7-URA3 | Biofilm defect; decreased biomass | ++ | 45 |
| CJN702 | Δura3::λimm434/Δura3::λimm434arg4::hisG/arg4::hisG his1::hisG::pHIS1/his1::hisG bcr1::ARG4/bcr1::URA3 | Functionally defective hyphae | ++ | 37 |
| CJN698 | Δura3::λimm434/Δura3::λimm434arg4::hisG/arg4::hisG his1::hisG::pHIS1-BCR1/his1::hisG bcr1::ARG4/bcr1::URA3 | Robust 3D wild type | ++ | 37 |
+++, 1 to 2% survival; ++, 0.1 to 1% survival; +, 0.05 to 0.1% survival.
Antibiotic susceptibility testing.
Amphotericin B and chlorhexidine were obtained from Fisher Scientific, and stock solutions were prepared in dimethyl sulfoxide. Caspofungin was obtained from Merck. Antifungals were dissolved in growth medium. Biofilms were washed twice with sterile PBS in order to remove nonadherent cells prior to antifungal challenge. Antifungals were then added at 100 μl per well. Biofilms were disrupted by scraping them and were vortexed vigorously for 30 seconds before serial dilution and plating. Microscopic observation showed that the resulting suspension consisted of clumps and individual cells in approximately equal numbers, with the size of clumps being ≤10 cells. The plates were incubated for 24 to 48 h prior to counting of the colonies.
One hundred-microliter samples of exponentially growing (12-h) or stationary-phase (72-h) planktonic cultures were harvested by centrifugation, washed twice with PBS, and challenged in microtiter plates with antifungals in 100 μl of RPMI 1640 with l-glutamine and 0.165 M MOPS growth medium. The cultures containing antifungals were incubated at 37°C for 24 h, washed twice with PBS by centrifugation and aspiration of the supernatant, and resuspended in 100 μl PBS. The cultures were then 10-fold serially diluted in PBS and plated for CFU determination.
For sequential challenge, biofilms were treated with 100 μg/ml amphotericin B or chlorhexidine as described above. After 24 h, the medium was aspirated, 100 μl RPMI 1640 with l-glutamine and 0.165 M MOPS containing 100 μg/ml of amphotericin B and chlorhexidine was added to each well, and the plates were incubated for an additional 24 h.
Progeny biofilms were formed by inoculating samples from antibiotic-challenged, resuspended biofilms into fresh YPD medium and repeating the biofilm growth procedure described above.
Live-dead-cell determination using fluorescein diacetate staining and epifluorescence microscopy.
Biofilms of strain 3153A were grown and resuspended in RPMI 1640 with l-glutamine and 0.165 M MOPS with 100 μg/ml fluorescein diacetate alone (control) or with amphotericin B (100 μg/ml) and fluorescein diacetate (100 μg/ml) for 24 h. One-microliter samples of biofilms, scraped from the microtiter well at various time points, were placed onto a glass slide under a glass coverslip and viewed with a 100× oil immersion lens using a Zeiss Axioskop 2 plus microscope with a standard fluorescein isothiocyanate filter. Photographs were taken using an AxioCam (Carl Zeiss) black-and-white charge-coupled device camera with an accompanying 0.63× lens using Axiovision version 4.5. Live and dead control populations of budding yeast cells were obtained from an overnight 3153A culture grown in YPD medium at 30°C. Samples were pelleted and resuspended in YPD medium with 100 μg/ml fluorescein diacetate alone or 100 μg/ml amphotericin B and fluorescein diacetate for 24 h. Planktonic samples were viewed and photographed in a manner similar to the biofilm cells described above.
Isolation of persisters.
Biofilms from strain MC 191 were grown and resuspended in RPMI 1640 with l-glutamine and 0.165 M MOPS with fluorescein diacetate (Fisher Scientific; 100 μg/ml) with or without amphotericin B (100 μg/ml). The biofilms were disrupted by scraping and vortexing them and were washed five times with PBS. Samples were analyzed using a MoFlo (Dako) cell sorter. A homogeneous population of cells was isolated using a gate based on forward and side scatter. Cells that passed the forward and side scatter gate were counted as single events and subsequently sorted into two distinct populations based on high and low green fluorescence. Single events from these populations were then physically sorted directly onto separate YPD agar plates in the form of a 96-well grid and were incubated for 48 h. Photographs of the plates were taken using an Olympus Camedia C-4040 digital camera.
RESULTS
Detection of persisters in C. albicans biofilms.
Dose-dependent killing provides a straightforward approach to detect persisters. A biphasic killing curve revealing a subpopulation of cells that survive low doses of antimicrobials indicates the presence of persisters. Several compounds, including amphotericin B, chlorhexidine, and caspofungin, have been reported to kill Candida biofilms (23, 42, 49).
Mature, 48-h biofilms formed in wells of microtiter plates were rinsed with fresh growth medium and challenged with antifungals for 24 h. In parallel, antifungals were added to exponentially growing and stationary planktonic cultures, as well. Caspofungin, a β-glucan synthesis inhibitor of the cell wall, had a rather limited effect on biofilms, producing ≤10-fold killing (data not shown), and was therefore unsuitable for this study.
Amphotericin B, a compound that binds to ergosterol and forms an oligomeric pore in the membrane (1), effectively killed exponentially growing and stationary cells, with little indication of surviving cells (Fig. 1A). By contrast, strikingly biphasic killing was observed in Candida biofilms, with the majority of the population killed at low concentrations (but above the MIC) while the remaining cells were unaffected by higher concentrations of the drug (Fig. 1A). About 1% of the cells appeared completely invulnerable to amphotericin B, indicating the presence of persisters in the yeast biofilm. This is very different from our previous observations with bacteria, where stationary planktonic populations produce more persisters than the biofilm (48). It was also rather surprising to see complete resistance to killing by amphotericin B, which makes “holes” in the membrane.
FIG. 1.
Survival of C. albicans 3153A biofilm and exponential- and stationary-phase cells. Biofilms were cultured in RPMI medium for 48 h, scraped, vortexed, resuspended in 100 μl PBS, and plated for CFU determination. Exponential- and stationary-phase cultures were obtained by growth in the same medium. The experiment was performed in triplicate, and the error bars represent standard deviations. (A) Amphotericin B. (B) Chlorhexidine.
Similarly to amphotericin B, chlorhexidine, a membrane-acting antiseptic used in oral antifungal applications (5), produced distinctly biphasic killing of the biofilm, while cells in both exponential- and stationary-phase cultures were completely eliminated (Fig. 1B). At higher concentrations (above 100 μg/ml), killing of persisters became evident, and the biofilm was completely sterilized at 1,000 μg/ml, a concentration twofold lower than what is commonly used in mouthwash and as a therapy for treatment of oral thrush caused by C. albicans (0.2%).
Persister cells are multidrug tolerant.
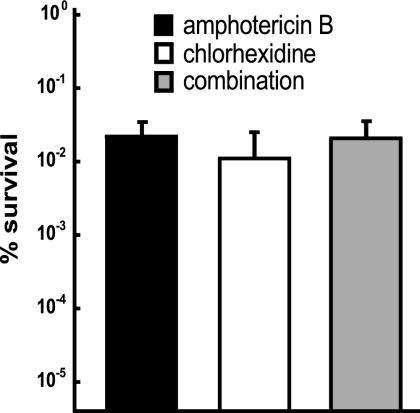
In bacteria, persisters are dormant cells that are tolerant of multiple antibiotics. We wanted to learn whether yeast persisters were similarly multidrug tolerant. Mature, 48-h biofilms were challenged with 100 μg/ml amphotericin B, 100 μg/ml chlorhexidine, or a combination of the two drugs. No additional killing was detected when biofilms were treated with both amphotericin B and chlorhexidine compared to cells treated with individual antimicrobials (Fig. 2). Similarly, the numbers of persisters were essentially the same (1 to 3%) when biofilms were treated sequentially for 24 h with amphotericin B and then chlorhexidine or vice versa (data not shown). These experiments indicated the presence of a single uniform persister population.
FIG. 2.
Survival of C. albicans biofilms challenged with amphotericin B and chlorhexidine. Biofilms were treated with 100 μg/ml amphotericin B, 100 μg/ml chlorhexidine, or a combination of the two antifungals for 24 h. The biofilms were washed and sampled for CFU determination before and after antibiotic treatment. The experiment was performed in triplicate, and the error bars indicate standard deviations.
To examine the possible roles of drug efflux transporters in resistance to these two antimicrobials, the susceptibilities of the wild-type strain and a mutant lacking Cdr1p, Cdr2p, Mdr1p, and Flu1p were tested. The MICs for the wild type and the quadruple mutant DSY1024 (cdr1Δ::hisG/cdr1Δ::hisG cdr2Δ::hisG/cdr2Δ::hisG camdr1Δ::hisG/camdr1Δ::hisG flu1Δ::hisG/flu1Δ::hisG-URA3-hisG) (30) were the same, 1 μg/ml for amphotericin B and 8 μg/ml for chlorhexidine. This suggests that resistance to amphotericin B and chlorhexidine is not influence by C. albicans efflux transporters, and therefore, that efflux does not contribute to persister survival.
Persisters are phenotypic variants of the wild type.
We next wanted to learn whether cells that resisted killing by drugs were phenotypic variants of the wild type or mutants. Biofilms were treated with amphotericin B or chlorhexidine (100 μg/ml), after which they were disrupted by being vortexed, washed, and reinoculated. The new biofilm, derived from persisters that survived drug treatment, was again treated with antifungals, and the procedure was repeated a total of three times (Fig. 3). It is apparent that the population produced by surviving persisters is not more resistant to drugs, but rather gives rise to a new persister subpopulation. If the surviving cells were mutants, we would have seen either complete resistance upon reapplication of the antifungal or a progressive increase in the numbers of surviving cells with each treatment cycle. Thus, we conclude that C. albicans persisters, similar to their bacterial counterparts, are phenotypic variants of the wild type that arise in a clonal population of genetically identical cells.
FIG. 3.
Heritability of persister formation. Biofilms were treated with 100 μg/ml amphotericin B or 100 μg/ml chlorhexidine for 24 h, after which they were disrupted by being vortexed, washed, and reinoculated in order to form new biofilms. The biofilms were sampled for CFU determination before and after antibiotic treatment. The procedure was repeated a total of three times. The experiment was performed in triplicate, and the error bars indicate standard deviations.
Isolation of persisters.
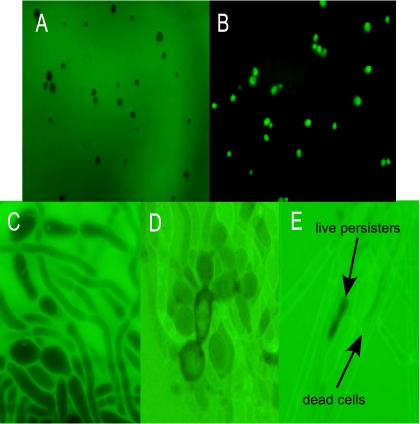
Several dyes have been reported to discriminate between live and dead fungal cells, including fluorescein, which was recently reported to specifically stain dead S. cerevisiae cells (52). Exponentially growing C. albicans cells, untreated or killed with amphotericin B, stained with fluorescein diacetate in the expected manner (Fig. 4 A and B). We then stained a biofilm with fluorescein diacetate (Fig. 4C to E). After addition of amphotericin B, there was a visible decrease in the number of live (dark) cells, and their morphology became aberrant (Fig. 4D). After 48 h of amphotericin B treatment, there were only a small number of unstained cells, and importantly, they appeared to have normal morphology. These cells looked like regular pseudohyphae or yeast and were indistinguishable from morphologically normal untreated cells. This finding is similar to that for bacterial persisters, which look like regular cells (47).
FIG. 4.
Live-dead staining of C. albicans with fluorescein diacetate. Planktonic or biofilm cells were stained with 100 μg/ml fluorescein diacetate and examined by fluorescence microscopy. (A) Live planktonic cells. (B) Dead planktonic cells after treatment with 100 μg/ml amphotericin B (×400 magnification). (C, D, and E) Biofilms (×1,000 magnification) of untreated control and after 18 and 48 h of amphotericin B treatment (100 μg/ml), respectively.
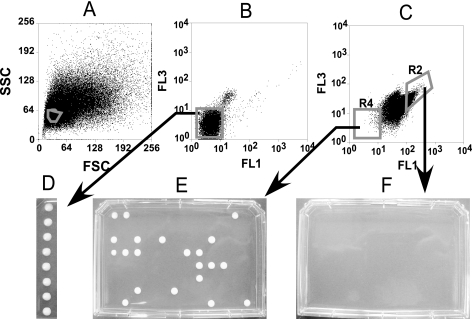
Are the dim C. albicans cells in the amphotericin B-treated biofilm live persisters or rather dead cells that for some reason happened to be unstained? The excellent difference observed between live and dead fluorescein diacetate-stained cells suggested that putative persisters could be sorted physically. In order to minimize size heterogeneity in the sorting process, we took advantage of the flo8 (riv1) null mutant strain that is defective in both hyphal and higher-order biofilm formation. This strain produces a sparse layer of attached yeast cells and formed a normal level of persisters (Table 1). The absence of hyphae in the strain made it a good choice for sorting. Mature, 48-h biofilms were stained with fluorescein diacetate, disrupted, and sorted with a MoFlo instrument. The sample contained debris from disrupted biofilms, and light scattering was used to determine the predominant particle group, which represented yeast cells. This group was then isolated with a scatter gate and analyzed for fluorescent properties (Fig. 5A). Cells representing the mean in the fluorescence distribution (Fig. 5B) from a biofilm stained with fluorescein diacetate but not treated with an antibiotic were sorted directly onto a plate with YPD medium, and after a 48-h incubation, produced colonies. The sorting worked well under these conditions, with eight cells producing eight colonies (Fig. 5D). Next, we sorted cells from a biofilm treated with amphotericin B. There was a strong shift of the bulk of the population to higher fluorescence (Fig. 5C), reflecting the staining of dead cells with fluorescein diacetate. Two subpopulations, a bright (R2) and a dim (R4) one, were then chosen for physical sorting onto YPD plates. Ninety-six of the sorted dim particles produced 21 colonies. This shows that a considerable part of the dim cells observed after treatment with amphotericin B were indeed live persisters. The imperfect correlation between sorted events and the colonies they produced in this case may have been caused by a considerable background of debris. Indeed, the concentration of live cells was low, and debris is not likely to exhibit bright-green fluorescence. Therefore, debris would be sorted and appear disproportionately represented in the dim subpopulation. Alternatively, it is possible that some of the dim cells were actually dead. There was no growth from over 6,000 sorted bright cells (Fig. 5F).
FIG. 5.
Isolation of persister cells from a biofilm. C. albicans MC191 was grown as a biofilm for 48 h in RPMI 1640 medium in microtiter plate wells. A homogenous population of cells from disrupted biofilms was obtained by applying a forward scatter and a side scatter gate, as shown in panel A, for all subsequent analyses. (B) A biofilm was stained with 100 μg/ml fluorescein diacetate for 24 h, disrupted, washed three times with PBS, and analyzed with a MoFlo cell sorter. Single events representing individual cells were physically sorted directly on YPD medium and incubated for 48 h (D). (C) A biofilm was treated with 100 μg/ml amphotericin B, stained with fluorescein diacetate, and similarly analyzed with the cell sorter. Two distinct populations were separated based on green fluorescence intensity, as shown. (E) Particles representing 96 events from the dim population, R4, were sorted onto YPD agar and incubated for 48 h. (F) Particles representing over 6,000 events from R2 were sorted onto YPD agar and incubated for 48 h.
Persisters in biofilm-deficient strains.
Given that persisters were produced only in the biofilm, it was interesting to test biofilm-defective mutants for persister production. Several mutants, e.g., strains lacking the transcription factors Efg1p and Cph1p or strains lacking the MAP kinase Mkc1p, have been reported to produce defective biofilms. A collection of such mutants was tested for persister formation. All strains tested produced biofilms under the conditions of these experiments, though their biomass in many cases appeared to be distinctly less than that of the wild type. After challenge with 100 μg/ml amphotericin B, the biofilms were disrupted and plated for colony counting. Somewhat unexpectedly, all tested strains produced normal levels of persisters (Table 1). Of particular interest was strain CKY138, lacking Efg1p and Cph1p, positive transcriptional regulators of filamentation. This strain is severely defective for biofilm formation, showing reduced adherence and a failure to produce the elaborate three-dimensional structure that characterizes wild-type biofilms. Nevertheless, an efg1Δ/cph1Δ double-mutant strain gave rise to persisters. Therefore, persister formation is not dependent on the formation of a complex biofilm structure. Apparently, the ability to attach to a surface, which all of these strains exhibit, is sufficient to promote persister production. This is consistent with previously published work, which suggested that attachment alone is sufficient to confer resistance to fluconazole on a young biofilm (32) and that resistances to fluconazole and amphotericin B were similar in sessile wild-type and efg1Δ/cph1Δ cells, as measured by XTT reduction (43).
Given that attachment rather than biofilm architecture appeared to be important for persister formation, it was especially interesting to test a strain deficient in Mkc1p, a recently described contact-dependent kinase that affects biofilm formation and invasiveness (26). However, a strain lacking Mkc1p was able to produce normal levels of persisters (Table 1). Therefore, our analysis of biofilm mutants suggests that known genes are not involved in persister formation. The isolation of persisters, as described in this study, opens the possibility of obtaining their transcription profile and thus identifying genes involved in the persister phenotype.
DISCUSSION
The discovery of persisters in bacterial biofilms helped explain the puzzling resistance of biofilms to killing (19). A similar paradox exists in the field of fungal biofilms—planktonic cells may be highly susceptible to a range of antimicrobials, while mature biofilms appear to be resistant to killing by “everything” compared to planktonic populations. A biphasic pattern of killing is a defining feature demonstrating the presence of persisters in bacterial populations. In this study, we found that C. albicans biofilms exhibit this striking pattern of killing, with the majority of cells being fairly susceptible and a small subpopulation, 10−3 to 10−2 cells, highly tolerant of antimicrobials. Our analysis was limited by necessity to highly microbicidal antifungals, amphotericin B and chlorhexidine, which can effectively kill slowly growing or nongrowing cells; both of these compounds are membrane acting. Persisters appeared to be completely invulnerable to amphotericin B, which was rather unexpected. It has been reported that biofilm cells have a decreased level of ergosterol, to which amphotericin B binds prior to forming a pore in the membrane (34). It is possible that persisters have an even lower level of ergosterol than the bulk of biofilm cells and that other factors, such as dormancy, contribute to their survival against amphotericin B. Importantly, an experiment with the nonspecific antiseptic chlorhexidine suggests that C. albicans persisters are not merely cells with diminished ergosterol levels. A prominent subpopulation of cells tolerant of chlorhexidine was observed, as the majority of cells were killed at a low concentration of 10 μg/ml and additional killing did not occur until the concentration reached a very high dose of 500 μg/ml. At even higher concentrations, chlorhexidine was able to completely eradicate the biofilm. Interestingly, this biofilm eradication concentration of chlorhexidine, 1 mg/ml, is comparable to the 2-mg/ml concentration found in mouthwash used to treat C. albicans oral thrush infections. It is important to note that neither amphotericin B nor chlorhexidine is a substrate of Cdr1p, Cdr2p, Mdr1p, or any known drug efflux pump. Our experiments showed that the MIC of the wild type to these antimicrobials was the same as that of a strain with multiple deletions in the MDRs.
The biphasic killing produces an impression of the presence of resistant mutants in a population, and we examined the possible heritability of resistance in the surviving cells. Repeated reinoculation of persisters after amphotericin B treatment restored the original biofilm with a new persister subpopulation. This is similar to the nonheritable nature of bacterial persisters (18).
In order to observe persisters, we used fluorescein diacetate, which specifically stains dead yeast cells. After treatment with amphotericin B, most of the cells became morphologically aberrant (bloated) and then stained with fluorescein diacetate, while a small fraction of cells retained normal morphology and were dark. We then used cell sorting to isolate the dark cells. The dark cells produced colonies, while the bright ones did not. This experiment demonstrates the existence of unique survivor cells within the antifungal-treated biofilm.
The production of fungal persisters by biofilm populations alone was unexpected. We did not find persisters in either exponential- or stationary-phase planktonic cultures of C. albicans. In principle, stimulation by fresh medium or regrowth could have accounted for the presence of persisters in biofilms, but not in planktonic stationary-phase cultures. In our experiments, both biofilms and stationary-phase cultures were washed, and then antibiotics were added in fresh growth medium. It is important to note that stationary-phase yeast cells do not grow when resuspended without dilution in the same volume of fresh medium. Nevertheless, stationary-phase cultures did not contain persisters; persisters were observed only in the biofilm. It seems that the inability of a planktonic stationary-phase culture to form persisters cannot be explained by their disappearance due to stimulation with fresh medium.
Stationary-phase cultures grown in RPMI 1640 medium produce cell aggregates but do not form persisters. Indeed, crowding does not seem to be important—addition of the quorum-sensing molecule farnesol or tyrosol did not affect the numbers of persisters in biofilms (M. LaFleur, unpublished data). These observations suggest that attachment to a substratum is the important factor that determines persister formation in Candida.
In bacteria, the frequency of persisters increases sharply with the cell density of the population (18), and there are actually more persisters in a nongrowing stationary-phase culture than in a slowly growing biofilm (48). It was also surprising to find that persisters were present at normal levels in all of the presently described mutants that produce defective biofilms. These results point to attachment, rather than formation of a complex biofilm structure, as the stimulus for persister formation. At the same time, a mutant in the Mkc1p MAP kinase that is involved in contact sensing and control of the biofilm phenotype produced normal levels of persisters, as well. This suggests that additional, unidentified sensors of attachment are probably responsible for triggering a program that produces persisters.
A transcription profile of isolated persisters has recently been obtained in E. coli. Persisters overexpress several toxin-antitoxin modules. Induction of toxin expression from a controllable promoter caused the production of persisters, and deletion of a particular toxin-antitoxin module, hipBA, caused a decrease in persisters in a biofilm (19). Upon overexpression, these toxins block important targets in the cell, leading to dormancy. Bacterial persisters have characteristics of dormant cells in that they are slowly growing or nongrowing (4) and lack noticeable expression of degradable green fluorescent protein, indicating low levels of transcription (47). It is premature to speculate whether the drug tolerance of surviving C. albicans cells reported in this study is also caused by dormancy, but it is useful to note that dormancy has been documented in fungi. The ascospore of Saccharomyces cerevisiae is the best-studied dormant form of yeast. As a typical spore, it is highly tolerant of a wide range of noxious factors (36). C. albicans produces chlamydospores, a thick-walled asexual form derived from hyphae (31). However, chlamydospores are not known to confer long-term viability and do not appear to be present at sites of infection or in biofilms (17, 31). Another dormant form of yeast, known as quiescence, occurs after 5 to 7 days of incubation of S. cerevisiae at 30°C in complete media (13). Quiescent cells exhibit increased thermotolerance, osmotolerance, and tolerance of a variety of cell wall-acting agents compared to dividing yeasts (9, 13, 38). Several genes are known to be involved in the entry into, the exit from, or the maintenance of quiescence (13). However, it is unknown whether these cells are tolerant of antifungals.
Future studies will show whether yeast persisters are dormant cells that have any connection to the previously described dormant forms. For now, we may say that yeasts and bacteria have evolved analogous survival strategies through convergent evolution that assign the function of survival to a small part of the population. The fact that persisters in yeast are specific to biofilms suggests they are not the result of “mistakes,” such as random overexpression of nonspecific proteins that may lead to stasis, as has recently been suggested for bacteria (50). The finding of a persister subpopulation in C. albicans will help to solve the puzzle of biofilm resistance to antifungals, and the method of isolating persisters reported in this study provides a powerful tool to search for persister genes.
Acknowledgments
We are grateful to Aaron Mitchell and Todd Milne (Microbia) for providing yeast strains and to Merck for a gift of caspofungin. We thank Allen Parmelee of the Tufts School of Medicine laser cytometry core facility for helpful discussion and assistance with cell sorting.
This work was supported by NIH Institute of General Medicine grant 2 R01 GM061162-05A1 to K.L. C.A.K. acknowledges support from NIH grant AI052805.
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Baginski, M., K. Sternal, J. Czub, and E. Borowski. 2005. Molecular modelling of membrane activity of amphotericin B, a polyene macrolide antifungal antibiotic. Acta Biochim. Pol. 52:655-658. [PubMed] [Google Scholar]
- 2.Baillie, G. S., and L. J. Douglas. 1999. Candida biofilms and their susceptibility to antifungal agents. Methods Enzymol. 310:644-656. [DOI] [PubMed] [Google Scholar]
- 3.Baillie, G. S., and L. J. Douglas. 2000. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J. Antimicrob. Chemother. 46:397-403. [DOI] [PubMed] [Google Scholar]
- 4.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 5.Basrani, B., and C. Lemonie. 2005. Chlorhexidine gluconate. Aust. Endod. J. 31:48-52. [DOI] [PubMed] [Google Scholar]
- 6.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick, and M. A. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datry, A., and E. Bart-Delabesse. 2006. Caspofungin: mode of action and therapeutic applications. Rev. Med. Interne 27:32-39. [DOI] [PubMed] [Google Scholar]
- 9.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146:2121-2132. [DOI] [PubMed] [Google Scholar]
- 10.Douglas, L. J. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30-36. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Sanchez, S., S. Aubert, I. Iraqui, G. Janbon, J. M. Ghigo, and C. d'Enfert. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 3:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giusani, A. D., M. Vinces, and C. A. Kumamoto. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray, J. V., G. A. Petsko, G. C. Johnston, D. Ringe, R. A. Singer, and M. Werner-Washburne. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol Rev. 68:187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawser, S., and K. Islam. 2006. Candida, p. 171-203. In J. L. Pace, M. E. Rupp, and R. G. Finch (ed.), Biofilms, infection, and antimicrobial therapy. Taylor & Francis, Boca Raton, Fla.
- 15.Hoyle, B. D., J. Jass, and J. W. Costerton. 1990. The biofilm glycocalyx as a resistance factor. J. Antimicrob. Chemother. 26:1-5. [DOI] [PubMed] [Google Scholar]
- 16.Jabra-Rizk, M. A., W. A. Falkler, and T. F. Meiller. 2004. Fungal biofilms and drug resistance. Emerg. Infect. Dis. 10:14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansons, V. K., and W. J. Nickerson. 1970. Induction, morphogenesis, and germination of the chlamydospore of Candida albicans. J. Bacteriol. 104:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 19.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojic, E. M., and R. O. Darouiche. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17:255-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn, D. M., M. Balkis, J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2003. Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J. Clin. Microbiol. 41:506-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn, D. M., J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect. Immun. 70:878-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn, D. M., T. George, J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob. Agents Chemother. 46:1773-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn, D. M., and M. A. Ghannoum. 2004. Candida biofilms: antifungal resistance and emerging therapeutic options. Curr. Opin. Investig. Drugs 5:186-197. [PubMed] [Google Scholar]
- 25.Kumamoto, C. A. 2002. Candida biofilms. Curr. Opin. Microbiol. 5:608-611. [DOI] [PubMed] [Google Scholar]
- 26.Kumamoto, C. A. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 102:5576-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumamoto, C. A., and M. D. Vinces. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113-133. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, K., A. Spoering, N. Kaldalu, I. Keren, and D. Shah. 2006. Persisters: specialized cells responsible for biofilm tolerance to antimicrobial agents, p. 241-256. In J. L. Pace, M. E. Rupp, and R. G. Finch (ed.), Biofilms, infection, and antimicrobial therapy. Taylor & Francis, Boca Raton, Fla.
- 29.Lopez-Ribot, J. L. 2005. Candida albicans biofilms: more than filamentation. Curr. Biol. 15:R453-R455. [DOI] [PubMed] [Google Scholar]
- 30.Marchetti, O., P. Moreillon, J. M. Entenza, J. Vouillamoz, M. P. Glauser, J. Bille, and D. Sanglard. 2003. Fungicidal synergism of fluconazole and cyclosporine in Candida albicans is not dependent on multidrug efflux transporters encoded by the CDR1, CDR2, CaMDR1, and FLU1 genes. Antimicrob. Agents Chemother. 47:1565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, S. W., L. M. Douglas, and J. B. Konopka. 2005. Cell cycle dynamics and quorum sensing in Candida albicans chlamydospores are distinct from budding and hyphal growth. Eukaryot. Cell. 4:1191-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mateus, C., S. A. Crow, Jr., and D. G. Ahearn. 2004. Adherence of Candida albicans to silicone induces immediate enhanced tolerance to fluconazole. Antimicrob Agents Chemother. 48:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee, P. K., and J. Chandra. 2004. Candida biofilm resistance. Drug Resist. Updat. 7:301-309. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee, P. K., J. Chandra, D. M. Kuhn, and M. A. Ghannoum. 2003. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 71:4333-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee, P. K., G. Zhou, R. Munyon, and M. A. Ghannoum. 2005. Candida biofilm: a well-designed protected environment. Med. Mycol. 43:191-208. [DOI] [PubMed] [Google Scholar]
- 36.Neiman, A. M. 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol Rev. 69:565-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nobile, C. J., and A. P. Mitchell. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150-1155. [DOI] [PubMed] [Google Scholar]
- 38.Plesset, J., J. R. Ludwig, B. S. Cox, and C. S. McLaughlin. 1987. Effect of cell cycle position on thermotolerance in Saccharomyces cerevisiae. J. Bacteriol. 169:779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramage, G., S. Bachmann, T. F. Patterson, B. L. Wickes, and J. L. Lopez-Ribot. 2002. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J. Antimicrob. Chemother. 49:973-980. [DOI] [PubMed] [Google Scholar]
- 40.Ramage, G., S. P. Saville, D. P. Thomas, and J. L. Lopez-Ribot. 2005. Candida biofilms: an update. Eukaryot. Cell. 4:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramage, G., K. Vande Walle, B. L. Wickes, and J. L. Lopez-Ribot. 2001. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 45:2475-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramage, G., K. VandeWalle, S. P. Bachmann, B. L. Wickes, and J. L. Lopez-Ribot. 2002. In vitro pharmacodynamic properties of three antifungal agents against preformed Candida albicans biofilms determined by time-kill studies. Antimicrob. Agents Chemother. 46:3634-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramage, G., K. VandeWalle, J. L. Lopez-Ribot, and B. L. Wickes. 2002. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 214:95-100. [DOI] [PubMed] [Google Scholar]
- 44.Ramage, G., B. L. Wickes, and J. L. Lopez-Ribot. 2001. Biofilms of Candida albicans and their associated resistance to antifungal agents. Am. Clin. Lab. 20:42-44. [PubMed] [Google Scholar]
- 45.Richard, M. L., C. J. Nobile, V. M. Bruno, and A. P. Mitchell. 2005. Candida albicans biofilm-defective mutants. Eukaryot. Cell. 4:1493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samaranayake, Y. H., J. Ye, J. Y. Yau, B. P. Cheung, and L. P. Samaranayake. 2005. In vitro method to study antifungal perfusion in Candida biofilms. J. Clin. Microbiol. 43:818-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah, D. V., Z. Zhang, K. Kurg, N. Kaldalu, A. Khodursky, and K. Lewis. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Theraud, M., Y. Bedouin, C. Guiguen, and J. P. Gangneux. 2004. Efficacy of antiseptics and disinfectants on clinical and environmental yeast isolates in planktonic and biofilm conditions. J. Med. Microbiol. 53:1013-1018. [DOI] [PubMed] [Google Scholar]
- 50.Vazquez-Laslop, N., H. Lee, and A. A. Neyfakh. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 188:3494-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Eiff, C., C. Heilmann, and G. Peters. 1999. New aspects in the molecular basis of polymer-associated infections due to staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 18:843-846. [DOI] [PubMed] [Google Scholar]
- 52.Wysocki, R., and S. J. Kron. 2004. Yeast cell death during DNA damage arrest is independent of caspase or reactive oxygen species. J. Cell Biol. 166:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]